Exploring the Concept of Lineage Diversity across North American Forests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
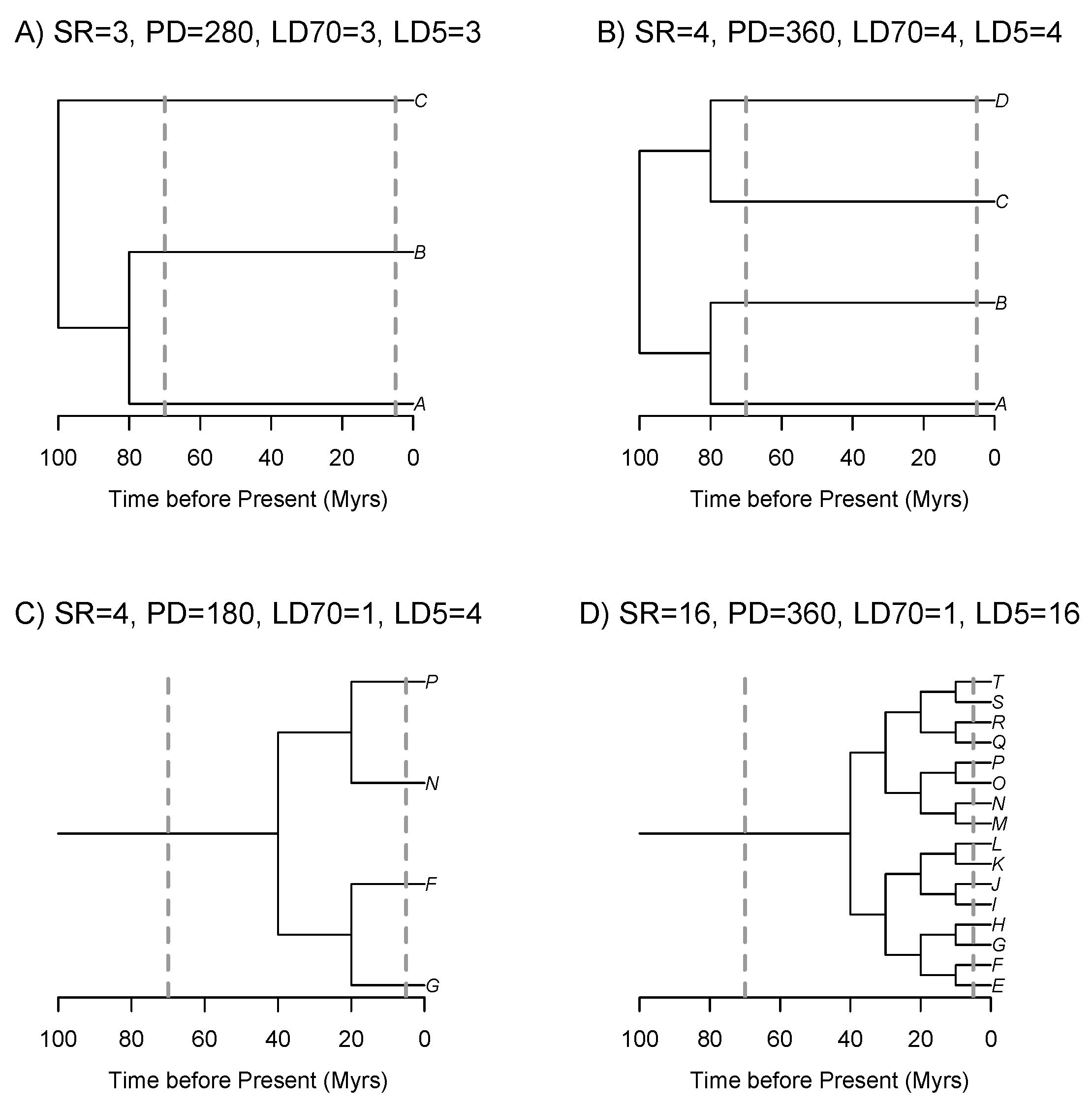
2.2. Assemblage Lineage Diversity (ALD) Metrics
2.2.1. Taxonomic Measures
2.2.2. Phylogenetic Measures
2.3. Statistical Analyses
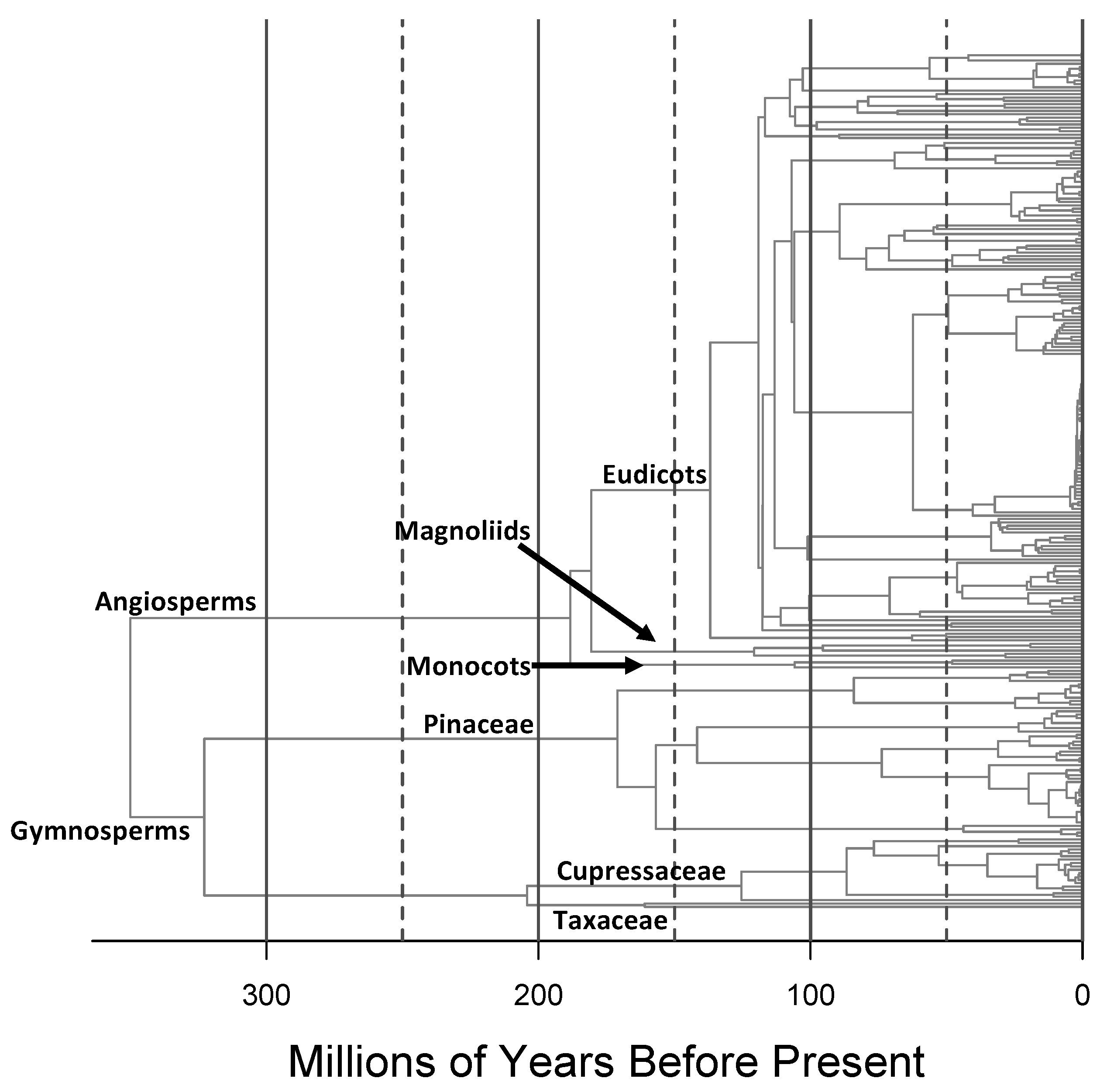
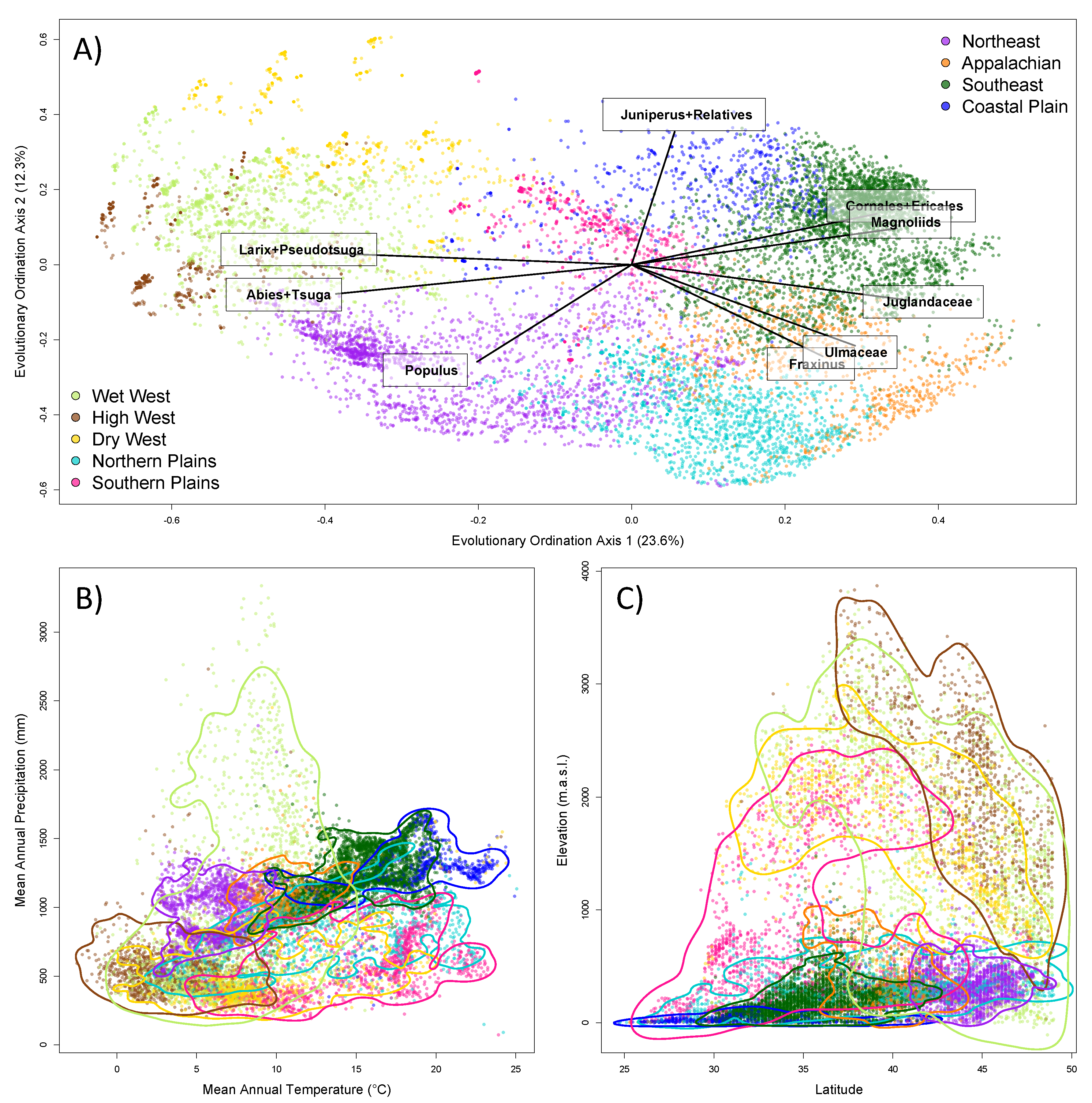
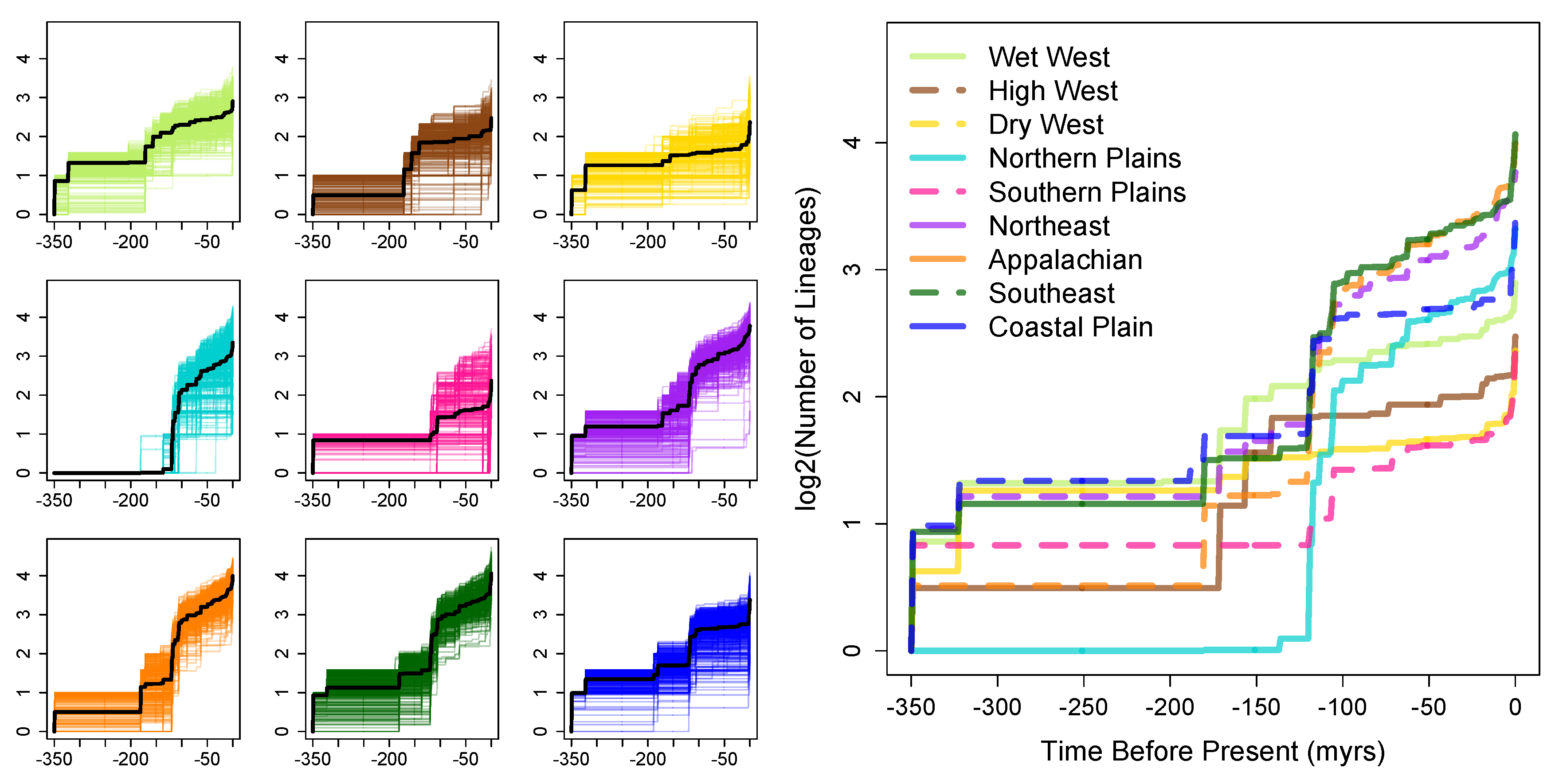
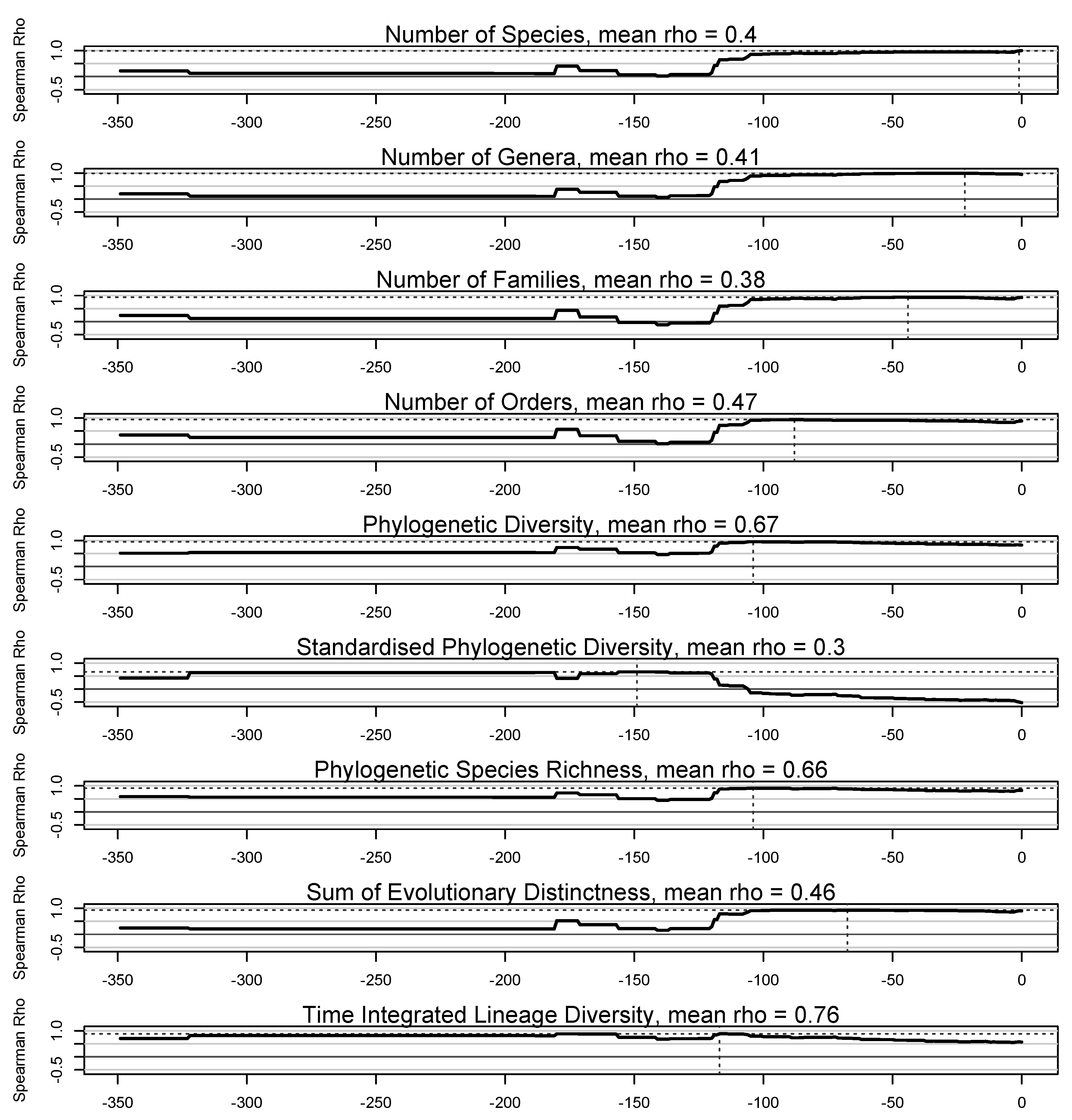
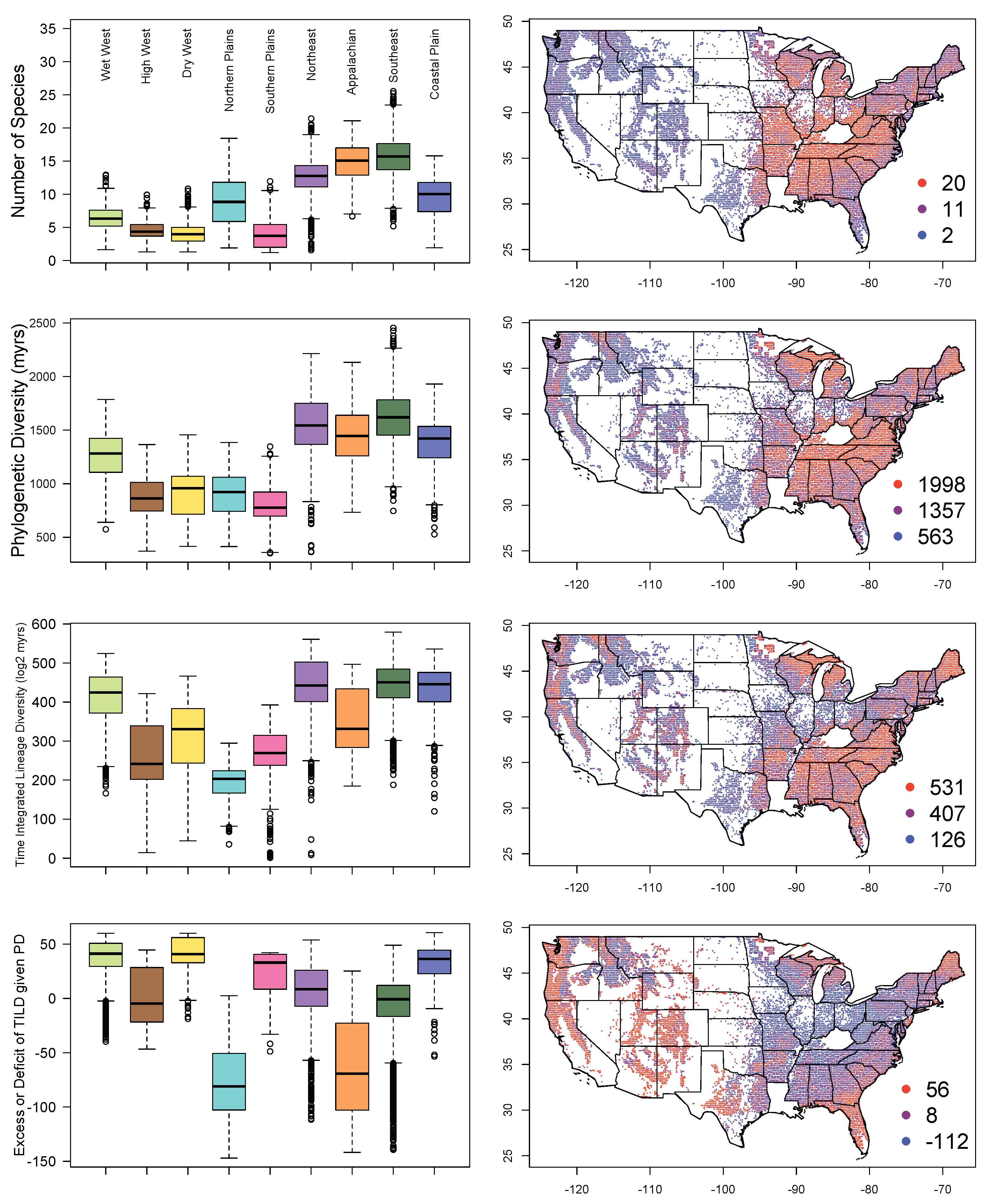
3. Results
4. Discussion
4.1. Taxonomic Measures of Lineage Diversity
4.2. Phylogenetic Measures of Lineage Diversity
4.3. Tree Diversity Patterns across the Contiguous United States
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
Appendix C
References
- De Queiroz, K. The general lineage concept of species, species criteria, and the process of speciation. In Endless Forms: Species and Speciation; Howard, D.J., Berlocher, S., Eds.; Oxford University Press: New York, NY, USA, 1998; pp. 57–75. [Google Scholar]
- Winter, M.; Devictor, V.; Schweiger, O. Phylogenetic diversity and nature conservation: Where are we? Trends Ecol. Evol. 2013, 28, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Geeta, R.; Lohmann, L.G.; Magallón, S.; Faith, D.P.; Hendry, A.; Crandall, K.; De Meester, L.; Webb, C.; Prieur-Richard, A.H.; Mimura, M.; et al. Biodiversity only makes sense in the light of evolution. J. Biosci. 2014, 39, 333–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forest, F.; Crandall, K.A.; Chase, M.W.; Faith, D.P. Phylogeny, extinction and conservation: Embracing uncertainties in a time of urgency. Philos. Trans. R. Soc. B 2015, 370, 20140002. [Google Scholar] [CrossRef] [PubMed]
- Kozak, K.H.; Blaine, R.A.; Larson, A. Gene lineages and eastern North American palaeodrainage basins: Phylogeography and speciation in salamanders of the Eurycea bislineata species complex. Mol. Ecol. 2006, 15, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Oliver, P.; Keogh, J.S.; Moritz, C. New approaches to cataloguing and understanding evolutionary diversity: A perspective from Australian herpetology. Aust. J. Zool. 2015, 62, 417–430. [Google Scholar] [CrossRef]
- Milián-García, Y.; Jensen, E.L.; Madsen, J.; Álvarez Alonso, S.; Serrano Rodríguez, A.; Espinosa López, G.; Russello, M.A. Founded: Genetic Reconstruction of Lineage Diversity and Kinship Informs Ex situ Conservation of Cuban Amazon Parrots (Amazona leucocephala). J. Hered. 2015, 106, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Becking, L.E.; de Leeuw, C.A.; Knegt, B.; Maas, D.L.; De Voogd, N.J.; Suyatna, I.; Peijnenburg, K.T. Highly divergent mussel lineages in isolated Indonesian marine lakes. PeerJ 2016, 4, e2496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moritz, C.C.; Pratt, R.C.; Bank, S.; Bourke, G.; Bragg, J.G.; Doughty, P.; Keogh, J.S.; Laver, R.J.; Potter, S.; Teasdale, L.C.; et al. Cryptic lineage diversity, body size divergence, and sympatry in a species complex of Australian lizards (Gehyra). Evolution 2018, 72, 54–66. [Google Scholar] [CrossRef]
- Linder, H.P.; Eldenas, P.; Briggs, B.G. Contrasting patterns of radiation in African and Australian Restionaceae. Evolution 2003, 57, 2688–2702. [Google Scholar] [CrossRef]
- Jønsson, K.A.; Fabre, P.H.; Ricklefs, R.E.; Fjeldså, J. Major global radiation of corvoid birds originated in the proto-Papuan archipelago. Proc. Natl. Acad. Sci. USA 2011, 108, 2328–2333. [Google Scholar] [CrossRef] [Green Version]
- Honorio-Coronado, E.N.; Dexter, K.G.; Pennington, R.T.; Chave, J.; Lewis, S.L.; Alexiades, M.N.; Alvarez, E.; Alves de Oliveira, A.; Amaral, I.L.; Araujo-Murakami, A.; et al. Phylogenetic diversity of Amazonian tree communities. Divers. Distrib. 2015, 21, 1295–1307. [Google Scholar] [CrossRef]
- Rezende, V.L.; Dexter, K.G.; Pennington, R.T.; Oliveira-Filho, A.T. Geographical variation in the evolutionary diversity of tree communities across southern South America. J. Biogeogr. 2017, 44, 2365–2375. [Google Scholar] [CrossRef]
- Law, C.J.; Slater, G.J.; Mehta, R.S. Lineage diversity and size disparity in Musteloidea: Testing patterns of adaptive radiation using molecular and fossil-based methods. Syst. Biol. 2017, 67, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Leslie, A.B.; Beaulieu, J.; Holman, G.; Campbell, C.S.; Mei, W.; Raubeson, L.R.; Mathews, S. An overview of extant conifer evolution from the perspective of the fossil record. Am. J. Bot. 2018, 105, 1531–1544. [Google Scholar] [CrossRef] [PubMed]
- Magallón, S.; Gómez-Acevedo, S.; Sánchez-Reyes, L.L.; Hernández-Hernández, T. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 2015, 207, 437–453. [Google Scholar] [CrossRef] [PubMed]
- Raup, D.M.; Gould, S.J.; Schopf, T.J.; Simberloff, D.S. Stochastic models of phylogeny and the evolution of diversity. J. Geol. 1973, 81, 525–542. [Google Scholar] [CrossRef]
- Yguel, B.; Jactel, H.; Pearse, I.S.; Moen, D.; Winter, M.; Hortal, J.; Helmus, M.R.; Kühn, I.; Pavoine, S.; Purschke, O.; et al. The evolutionary legacy of diversification predicts ecosystem function. Am. Nat. 2016, 188, 398–410. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Burrill, E.; Wilson, A.; Turner, J.; Pugh, S.; Menlove, J.; Christiansen, G.; Conkling, B.; David, W. The Forest Inventory and Analysis Database: Database Description and User Guide version 8.0 for Phase 2; U.S. Department of Agriculture, Forest Service: Washington, DC, USA, 2018. Available online: www.fia.fs.fed.us/library/database-documentation (accessed on 18 December 2018).
- Maitner, B.S.; Boyle, B.; Casler, N.; Condit, R.; Donoghue, J.; Durán, S.M.; Guaderrama, D.; Hinchliff, C.E.; Jørgensen, P.M.; Kraft, N.J.; et al. The bien r package: A tool to access the Botanical Information and Ecology Network (BIEN) database. Methods Ecol. Evol. 2018, 9, 373–379. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Ma, Z.; Sandel, B.; Svenning, J.C. Phylogenetic assemblage structure of North American trees is more strongly shaped by glacial–interglacial climate variability in gymnosperms than in angiosperms. Ecol. Evol. 2016, 6, 3092–3106. [Google Scholar] [CrossRef]
- Harris, L.W.; Davies, T.J. A Complete Fossil-Calibrated Phylogeny of Seed Plant Families as a Tool for Comparative Analyses: Testing the ‘Time for Speciation’ Hypothesis. PLoS ONE 2016, 11, e0162907. [Google Scholar] [CrossRef] [PubMed]
- Nunn, C.L.; Altizer, S.; Sechrest, W.; Jones, K.E.; Barton, R.A.; Gittleman, J.L. Parasites and the evolutionary diversification of primate clades. Am. Nat. 2004, 164, S90–S103. [Google Scholar] [CrossRef] [PubMed]
- Mazel, F.; Guilhaumon, F.; Mouquet, N.; Devictor, V.; Gravel, D.; Renaud, J.; Cianciaruso, M.V.; Loyola, R.; Diniz-Filho, J.A.F.; Mouillot, D.; et al. Multifaceted diversity–area relationships reveal global hotspots of mammalian species, trait and lineage diversity. Glob. Ecol. Biogeogr. 2014, 23, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Blaimer, B.B.; Brady, S.G.; Schultz, T.R.; Fisher, B.L. Functional and phylogenetic approaches reveal the evolution of diversity in a hyper diverse biota. Ecography 2015, 38, 901–912. [Google Scholar] [CrossRef]
- Tucker, C.M.; Cadotte, M.W.; Carvalho, S.B.; Davies, T.J.; Ferrier, S.; Fritz, S.A.; Grenyer, R.; Helmus, M.R.; Jin, L.S.; Mooers, A.O.; et al. A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. 2017, 92, 698–715. [Google Scholar] [CrossRef] [PubMed]
- Helmus, M.R.; Bland, T.J.; Williams, C.K.; Ives, A.R. Phylogenetic measures of biodiversity. Am. Nat. 2007, 169, E68–E83. [Google Scholar] [CrossRef]
- Tsirogiannis, C.; Sandel, B. PhyloMeasures: A package for computing phylogenetic biodiversity measures and their statistical moments. Ecography 2016, 39, 709–714. [Google Scholar] [CrossRef]
- Safi, K.; Armour-Marshall, K.; Baillie, J.E.; Isaac, N.J. Global patterns of evolutionary distinct and globally endangered amphibians and mammals. PLoS ONE 2013, 8, e63582. [Google Scholar] [CrossRef]
- Isaac, N.J.; Turvey, S.T.; Collen, B.; Waterman, C.; Baillie, J.E. Mammals on the EDGE: Conservation priorities based on threat and phylogeny. PLoS ONE 2007, 2, e296. [Google Scholar] [CrossRef]
- Bryant, J.A.; Lamanna, C.; Morlon, H.; Kerkhoff, A.J.; Enquist, B.J.; Green, J.L. Microbes on mountainsides: Contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. USA 2008, 105, 11505–11511. [Google Scholar] [CrossRef] [Green Version]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Pavoine, S. A guide through a family of phylogenetic dissimilarity measures among sites. Oikos 2016, 125, 1719–1732. [Google Scholar] [CrossRef] [Green Version]
- Legendre, P.; Gallagher, E.D. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, M.; Legendre, P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 1997, 67, 345–366. [Google Scholar] [CrossRef]
- Duong, T. ks: Kernel density estimation and kernel discriminant analysis for multivariate data in R. J. Stat. Softw. 2007, 21, 1–16. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.; et al. Package ‘Vegan’: Community Ecollogy Package. 2018. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 22 June 2019).
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Package ‘cluster’. 2018. Available online: https://cran.r-project.org/web/packages/cluster/index.html (accessed on 22 June 2019).
- Pavoine, S. Package adiv: Analysis of diversity. 2018. Available online: https://cran.r-project.org/web/packages/adiv/index.html (accessed on 22 June 2019).
- Forest, F.; Grenyer, R.; Rouget, M.; Davies, T.; Cowling, R.; Faith, D.; Balmford, A.; Manning, J.; Proches, S.; van der Bank, M. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 2007, 445, 757–760. [Google Scholar] [CrossRef]
- Rapacciuolo, G.; Graham, C.H.; Marin, J.; Behm, J.E.; Costa, G.C.; Hedges, S.B.; Helmus, M.R.; Radeloff, V.C.; Young, B.E.; Brooks, T.M. Species diversity as a surrogate for conservation of phylogenetic and functional diversity in terrestrial vertebrates across the Americas. Nat. Ecol. Evol. 2018, 3, 1. [Google Scholar] [CrossRef]
- Miller, J.T.; Jolley-Rogers, G.; Mishler, B.D.; Thornhill, A.H. Phylogenetic diversity is a better measure of biodiversity than taxon counting. J. Syst. Evol. 2018, 56, 663–667. [Google Scholar] [CrossRef] [Green Version]
- Webb, C. Phylogenies and community ecology. Annu. Rev. Ecol. Evol. S 2002, 33, 475–505. [Google Scholar] [CrossRef]
- Currie, D.J.; Paquin, V. Large-scale biogeographical patterns of species richness of trees. Nature 1987, 329, 326–327. [Google Scholar] [CrossRef]
- Jenkins, C.N.; Van Houtan, K.S.; Pimm, S.L.; Sexton, J.O. US protected lands mismatch biodiversity priorities. Proc. Natl. Acad. Sci. USA 2015, 112, 5081–5086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, E.L. Deciduous Forests of Eastern North America. Soil Sci. 1950, 71, 259–279. [Google Scholar] [CrossRef]
- DeSiervo, M.H.; Jules, E.S.; Kauffmann, M.E.; Bost, D.S.; Butz, R.J. Revisting john sawyer and dale thornburgh’s 1969 vegetation plots in the russian wilderness: A legacy continued. Fremontia 2016, 44, 20. [Google Scholar]
- Hawkins, B.A.; Rueda, M.; Rangel, T.F.; Field, R.; Diniz-Filho, J.A.F. Community phylogenetics at the biogeographical scale: cold tolerance, niche conservatism and the structure of North American forests. J. Biogeogr. 2014, 41, 23–38. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dexter, K.G.; Segovia, R.A.; Griffiths, A.R. Exploring the Concept of Lineage Diversity across North American Forests. Forests 2019, 10, 520. https://doi.org/10.3390/f10060520
Dexter KG, Segovia RA, Griffiths AR. Exploring the Concept of Lineage Diversity across North American Forests. Forests. 2019; 10(6):520. https://doi.org/10.3390/f10060520
Chicago/Turabian StyleDexter, Kyle G., Ricardo A. Segovia, and Andy R. Griffiths. 2019. "Exploring the Concept of Lineage Diversity across North American Forests" Forests 10, no. 6: 520. https://doi.org/10.3390/f10060520




