1. Introduction
As forest ecosystems play an important role in global carbon cycling and the mitigation of carbon concentrations in the atmosphere, accurately estimating forest biomass is necessary and has been widely studied [
1,
2,
3,
4,
5,
6,
7,
8]. Due to the difficulty and high cost of collecting field data, especially belowground biomass, most of the existing studies have focused on forest aboveground biomass (AGB) [
9,
10,
11,
12,
13,
14,
15,
16,
17] and the use of remotely sensed data [
18,
19,
20]. However, the estimation accuracy of forest AGB varies depending on many factors, including the remotely sensed images, the independent variables used, and the algorithms used to model the relationship of AGB with the independent variables [
9].
As remote sensing technologies develop, various image data from Landsat, SPOT, QuickBird, IKONOS, WorldView, ASTER, MODIS, AVHRR, Radarsat, and ALOS PALSAR have become available and are used for AGB estimation [
9]. Many studies have also investigated the data fusion of different sensor images for improving AGB estimation [
21,
22]. Given a study area, what kind of sensor data should be selected has become very challenging during the past several decades [
23]. Overall, Landsat imagery has been most widely used for this purpose because of free downloading, a long-time history, large coverage, and medium spatial and temporal resolutions [
9,
21,
24,
25,
26,
27]. Especially, the new Landsat 8 provides images with more spectral bands and a higher radiometric resolution than previous Landsat satellites and, thus, a greater potential for improving AGB estimation [
12,
28].
Selecting spectral variables that have strong relationships with forest AGB is the key to increasing the accuracy of AGB estimation using remote sensing data [
9,
12,
19,
26]. Various spectral bands and other potential variables derived from the bands, such as vegetation indices, various transformations, texture measures, and fractional images have been used to construct the estimation models of forest AGB [
9,
24,
26]. Generally, the models with the combinations of vegetation indices, texture measures, and environmental variables such as elevation, slope, and aspect can lead to higher estimation accuracy than those with original bands, and this is especially true for the forests with complex canopy structures [
9,
12,
19,
21].
Various parametric and nonparametric algorithms have been developed for mapping forest AGB using remotely sensed images [
9,
19]. Simple linear regression models (LMs) and nonlinear models such as power models [
29,
30] and logistic regression models [
31] are often used. The LMs account for the relationships between forest AGB and predictors and are the most popular statistical models [
9,
32]. However, the relationships are often nonlinear and show power, exponential, or logarithmic forms. The nonlinear regression models or the linearization of the nonlinear relationship are common methods to deal with the nonlinear relationships. However, the determination coefficients obtained are often smaller than 0.5 [
27,
33], and the estimates obtained are, thus, not reliable [
32].
Nonparametric models are an alternative approach to improve the estimation accuracy of forest AGB using remote sensing images. Many nonparametric algorithms, such as artificial neural networks (ANN) [
34,
35], random forest (RF) [
36,
37], k-Nearest Neighbors (kNN) [
38], support vector machine (SVM) [
39,
40], and Maximum Entropy (MaxEnt) [
41,
42], have been explored to model the relationships between forest AGB and predictors. However, the model structures derived from these algorithms are often difficult to interpret [
9,
19,
32]. The RF, introduced by Leo Breiman [
43], can be used for either classifying categorical variables or estimating continuous variables such as forest AGB [
43,
44]. It provides the average prediction from a number of regression models created by a subset of training data selected randomly with a random subset of predictors [
45,
46]. Recently RF has been frequently used in AGB estimation by integrating the samples from field inventories, remote sensing, and other predictors [
47,
48]. Moreover, ANN, proposed in 1943, is a mathematical model inspired by biological neural networks. Its algorithm is the product of artificial intelligence as black-box models due to unknown weights and a difficulty in terminating the learning process [
49]. Therefore, it has a strong ability for fitting the data [
50] and provides a robust solution for complex and nonlinear problems due to its universal approximation properties [
24]. Many different neural network models have been developed [
51] and are widely used in various fields [
52]. Unfortunately, the estimation accuracy of the nonparametric models is very much limited by the used sample sizes.
Moreover, forest AGB estimates derived from remote sensing data are associated with uncertainties [
19], which has been widely recognized and for which substantial research has been conducted [
53,
54]. The uncertainties often result in underestimations for forests with great AGB values and overestimations for forests with small AGB values. The underestimations usually occur when AGB reaches 100–150 Mg/ha [
33,
55,
56], while the overestimations often happen for forests with AGB less than 40 Mg/ha [
33]. Many studies have indicated that the uncertainties are related to the structures of forest ecosystems, the topographic characteristics, the remotely sensed data (insensitivity and saturation) and their spatial resolutions, and the methods used [
33,
56,
57,
58,
59,
60,
61,
62]. Especially, spectral reflectance due to data saturation becomes less sensitive to AGB changes in the dense, multilayer, and complex canopy forest ecosystems and often leads to the underestimations of forest AGB [
19]. Few studies have analyzed the saturation values of forest AGB for different forest ecosystems, and only a few reports have demonstrated methods to reduce the impact of data saturation on AGB estimation accuracy [
20,
22,
33].
The heterogeneity of complex forest canopy structures may be the major impact for the data saturation and underestimation for the forests with high AGB values [
22]. The data saturation varies depending on vegetation types because of the characteristics of their surface reflectance, including tree species, ages, and forest canopy structures [
19,
63]. Zhao et al. [
22] analyzed the saturation values using Landsat imagery for different vegetation types, slopes, and aspects in a subtropical region of China; estimated AGB considering the stratification of forest types using a stepwise linear regression; and improved the estimation accuracy of AGB. The stratification of forest types based on tree species and environmental variables can provide the potential of improving the estimation of forest AGB [
22,
32,
33]. Moreover, incorporating the tree age into the estimation models can also lead to an improvement of the AGB/carbon sequestration estimates and can reduce the overestimation and underestimation [
64,
65,
66,
67,
68].
Iizuka and Tateishi [
64] and Sanga-Ngoie et al. [
68] introduced tree volume-derived age into tree carbon estimation models for improving the estimation accuracy of tree carbon sequestration using optical remote sensing imagery. Moreover, Zheng et al. [
65] used 60 sample trees to obtain a forest stand age map in which the ages for most of the forests were less than 21 years old, then developed a polynomial model with spectral variables from Landsat ETM+ images and stand age involved, and obtained a high estimation accuracy of AGB. Lefsky et al. [
66] mapped the forest stand age using the time series of Landsat images, then estimated the AGB of young stands using Lidar data in western Oregon, USA, and found that the estimation models were appropriate for the forests with the classes of ages 14.5 to 20.5 years but resulted in overestimations for the smaller age classes. Based on the years in which the trees were planted, Liu et al. [
67] also obtained and added the age variable into their linear regression models to estimate the AGB of plantations using Landsat images. The studies indicate that taking tree ages into account would obtain a considerable improvement in estimating AGB. The studies dealt with reducing the overestimation of AGB for young forests, but the reports related to the improvement of the underestimation for the forests with high AGB values are still lacking.
Overall, the overestimation and underestimation of forest AGB commonly exist when optical remote sensing imagery is used. Reducing the overestimation and underestimation is critical to increase the estimation accuracy of forest AGB but is greatly challenging. The objective of this study was to develop a novel method of reducing the overestimation and underestimation to improve the accuracy of estimating forest AGB when optical images were utilized. The improvement was explored first by using Landsat 8 OLI images and four models to estimate AGB, including two parametric models: LM and a LM with combined variables (LMC), and two nonparametric models: RF and ANN. The stand group age as a dummy variable was then introduced into the four models, which led to four new models with the age dummy variable involved. Moreover, the eight models were compared for the estimation performance of forest AGB, and the significant contributions of adding the age dummy variable into the models to mitigating the overestimation and underestimation were statistically examined. The analyses and examinations were conducted in Pinus densata forests distributed in Yunnan of Southwestern China.
2. Materials and Methods
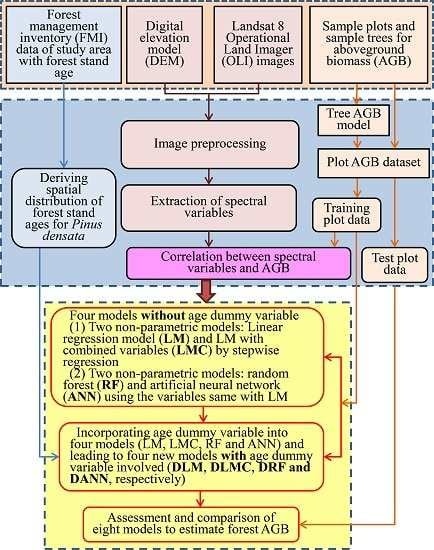
In
Figure 1, the methodologic framework of this study is illustrated, consisting of the following steps: 1) the selection of the study area; 2) the collection of the sample plot and tree biomass data; 3) the calculation of the tree and plot AGB; 4) the acquisition, preprocessing, and analysis of the Landsat 8 OLI images; 5) the development of the models by incorporating the age dummy variable; and 6) the assessment and comparison of the forest AGB predictions from the models with and without the age dummy variable.
2.1. Study Area
The study area is located in Shangri-La City, northwestern Yunnan of Southwestern China, and the AGB of
Pinus densata forests were mapped (
Figure 2). The study area is characterized by a cold temperate zone with the mean altitude of 3459 m above sea level. The annual mean temperature is about 5.4 °C with the monthly highest and lowest temperature of 13.3 °C in July and −3.8 °C in December, respectively. The winters are chilly but sunny because of the high altitude. The annual mean precipitation is about 607 mm. The seasonal distribution of precipitation is uneven with 70% in the rainy season (June–September). The evaporation is about 1671 mm per year, and the relative humidity is about 70%. The soil type is mainly dark brown forest soil [
69]. The study area is dominated by cold, temperate coniferous forests with dominant tree species of
Pinus,
Picea, Larix, and
Abies.
Pinus densata is an endemic and pioneer tree species for reforestation at the subalpine and alpine areas in the Hengduan Mountains [
70]. It is a typical cool-temperature coniferous tree species distributed between cold-temperature coniferous forests dominated by
Picea,
Abies, and
Larix and warm-temperature coniferous forests dominated by
Pinus yunnanensis and
P. armandii. In the study area, most of the forests are pure with a single canopy layer dominated by
Pinus densata or mixed with small amount of
Quercus pannosa,
Pinus armandii,
Picea likiangensis,
Larix potaninii var.
macrocarpa, and
Betula spp., with the altitude ranging from 3000 m to 3700 m.
2.2. Measurement and Calculation of Aboveground Biomass for Sample Trees and Plots
A total of 147 square sample plots were measured in the field in 2017. The plots were selected in the
P. desata pure forests of the study area by considering the stand ages, elevation, slope, and aspect, and the sampling distances between the plots were about 1 km (
Figure 2). Each plot had an area of 30 m × 30 m, and its coordinates of location, elevation, slope, and aspect were measured. Within each plot, the diameter at breast height (1.3 m above ground) (DBH) and height (H) of each tree were recorded. The stand age (SA) of each plot is the mean age of three standard trees having similar DBH values to the average DBH of the plot. The ages of the three trees were measured by counting the annual rings on the wood cores obtained by a growth cone at the tree base.
Moreover, a spatial distribution map of the Pinus densata forest stand ages for the study area was obtained from the Shangri-La City forest inventory conducted in 2016 for forest management and planning. In the forest inventory, a spatial distribution map of forest types, that is, stand compartments, was produced in the field using visual interpretation based on aerial photographs. The compartments had similar dominant tree species and homogeneous canopy structures. Within each of the forest compartments, a certain number of 25.8 m × 25.8 m plots were selected, and within each of the plots, the DBH of each tree was measured and the average DBH was obtained. Then, 3 to 5 trees having a DBH similar to the plot average DBH were selected to measure the tree ages by counting the layers of branches along the tree trunk because the growth of Pinus densata trees is characterized by adding a layer of branches each year.
In addition, a total of 100 sampling trees were selected from the 147 sample plots to measure tree AGB. The selection of the sample trees was based on their DBH classes from 6 cm to 76 cm with a 2 cm interval and on their elevation, slope, and aspect. At least three sample trees were obtained for each of the DBH classes. The values of the tree AGB components including wood, bark, branches, and needles were obtained according to the method of Wang [
71]. The biomass samples of wood and bark for each sample tree were collected by taking a 3 cm thickness disk at a 2 m interval along the tree trunk. The biomass values of each stem and its bark were measured by a method of volume and density. The volumes of the wood segments and barks were calculated using their lengths and diameters of the surfaces and under the barks. The samples of branches and leaves were selected, collected, and weighted by the method of graded branches. All the samples were dried to constant weights at 105 °C using an oven, and the sample density values of wood and bark were measured using a drainage method. Finally, the biomass values of wood and bark for the sample trees were calculated using the volumes and the corresponding sample density values. The branch and needle biomass of the sample trees were obtained using the ratios of fresh weight to the corresponding dry matter.
The AGB data of the individual sample trees (Unit: kg) were fit using a power function, and the AGB of each plot (Unit: Mg/ha) was summed using following Equation (1).
where
AGBs is the AGB of a plot,
AGBi is the estimated AGB of tree
i, and
n is the number of trees within the plot. Although the biomass samples of tree components were used, in fact, the biomass values of the sample trees and plots were associated with uncertainties and should be considered as reference values.
2.3. Remote Sensing Data and Preprocessing
Three Landsat 8 Operational Land Imager (OLI) images obtained from the website of the United States Geological Survey (USGS) were used in this research (
Table 1). The images were geo-referenced to a Universal Transverse Mercator coordinate system with zone 47 north with a root mean square error (RMSE) of less than one pixel. An atmospheric calibration of the images was conducted using the dark object subtraction approach [
72], and their topographic correction was made using the C-correction approach using a digital elevation model (DEM) at a spatial resolution of 30 m × 30 m [
73,
74]. The images were then mosaicked and clipped according to the study area (
Figure 2b).
A total of 231 remote sensing variables were derived (
Table 2), and they included 8 bands, 22 vegetation indices, 9 image transformations, and 192 textural measures (including mean, variance, homogeneity, contrast, dissimilarity, entropy, angular second moment, and correlation on seven bands with three moving window sizes: 3 × 3, 5 × 5, and 7 × 7 pixels). Vegetation indices and image transformations have been widely used in the estimation of forest AGB, and the texture measures were selected to capture the forest canopy structures [
32]. The relationships between the spectral variables and AGB were analyzed using a Pearson correlation analysis, and the spectral variables with significant correlations (
p ≤ 0.05) were selected to build the AGB estimation models.
2.4. Modeling Methods
Four models including LM, LMC, RF, and ANN were compared to estimate the AGB of the
Pinus densata forests. The spectral variables that had statistically significant correlations with plot AGB were selected to develop the LM model by a stepwise regression with the variance inflation factor (VIF) of 10 to test the collinearity of the independent variables. In addition to the spectral variables above, logarithmic, quadratic, and cubic transformations were calculated for each spectral variable and used as the combined variables to construct the LMC model using a stepwise regression with VIF of 10. The introduction of the combined variables provided the potential to model the nonlinear relationship of AGB with the spectral variables. The modeling of RF was carried out by the Random Forest package in R software. Both the number of regression trees (
ntree) and the number of input variables per node (
mtry) were set, the optimal
ntree was determined by bootstrap and RMSE, and the
mtry was tested from one-third of all the independent variables [
43]. Finally, we used the neuralnet package in R software to develop the ANN model. The number of nodes and layers for the models was set according to the default of the package, and the number of hidden layers was adjusted according to the predicted results. In addition, the independent variables used for ANN and RF were the same as those utilized for LM.
2.5. Modeling Methods with Age Dummy Variable
In this study, the sample plots were grouped into young stand, middle age stand, near-mature stand, mature stand, and overmature stand according to the stand ages (SA) of the
Pinus densata forests. The age intervals of young stand, middle age stand, near-mature stand, mature stand, and overmature stand were SA ≤ 20 years, 20 years < SA ≤ 30 years, 30 years < SA ≤ 40 years, 40 years < SA≤ 60 years, and SA > 60 years, respectively. The age intervals of the stand groups were determined according to the growth rate and forest management objectives of
Pinus densata. In this region,
Pinus densata forests with the age range of 40 to 60 years are considered as wood mature with the maximum annual value and can be harvested [
75,
76]. Whether the stand age was used as a dummy variable and included into the models (LM, LMC, ANN, and RF) of AGB prediction and whether the potential combination of neighboring age groups was made were determined by following steps:
(1) The models LM, LMC, ANN, and RF were developed and used to predict the AGB values of both the modeling and test plots;
(2) The predictions of AGB were compared with the reference values from the field plots, and the mean residuals of the predictions were statistically tested for their significant differences from zero at the significance level of 0.05 based on each of the stand age groups;
(3) Given a stand group and a model, the existence of the significant difference implied an overestimation or underestimation, and the age of the stand group as a dummy variable was, thus, added into the model; otherwise, the age dummy variable was not involved;
(4) The models with the age dummy variable were developed, the significant difference tests of the obtained mean residuals from zero were conducted, and the reduction of the error was analyzed; and
(5) When the models without the age dummy variable led to the mean residuals that were not significantly different from zero at the significance level of 0.05 for two neighboring age groups, the sample plots of the neighboring age groups were combined and the above four steps were repeated using the combined dataset.
Based on the results, the forest stands were divided into four groups: young stand, middle-age stand, near-mature stand, and mature and overmature stand in which the stand group ages were used as a dummy variable and added into the models.
2.6. Assessment and Validation of Predictions from Models
The evaluation of the obtained AGB models and the corresponding estimates was conducted using a determination coefficient (R2) and RMSE between the observed values and estimated values of AGB to assess and validate the models based on the plot dataset used for model fitting. Moreover, the applicability of the models was also validated using the test dataset with the mean error (ME) defined as the average value of the residuals (observed values minus estimated values). The mean error of each age group for the different estimation models was statistically tested for its significant difference from zero at the significance level of 0.05 using single sample t-test by SPSS. Among the 147 plots, 100 plots were randomly selected and used for the model fitting and 47 plots were used for the validation of the models.