eCry1Gb.1Ig, A Novel Chimeric Cry Protein with High Efficacy against Multiple Fall Armyworm (Spodoptera frugiperda) Strains Resistant to Different GM Traits
Abstract
:1. Introduction
2. Results
2.1. Generation of Cry1Gb-Based Chimeric Proteins
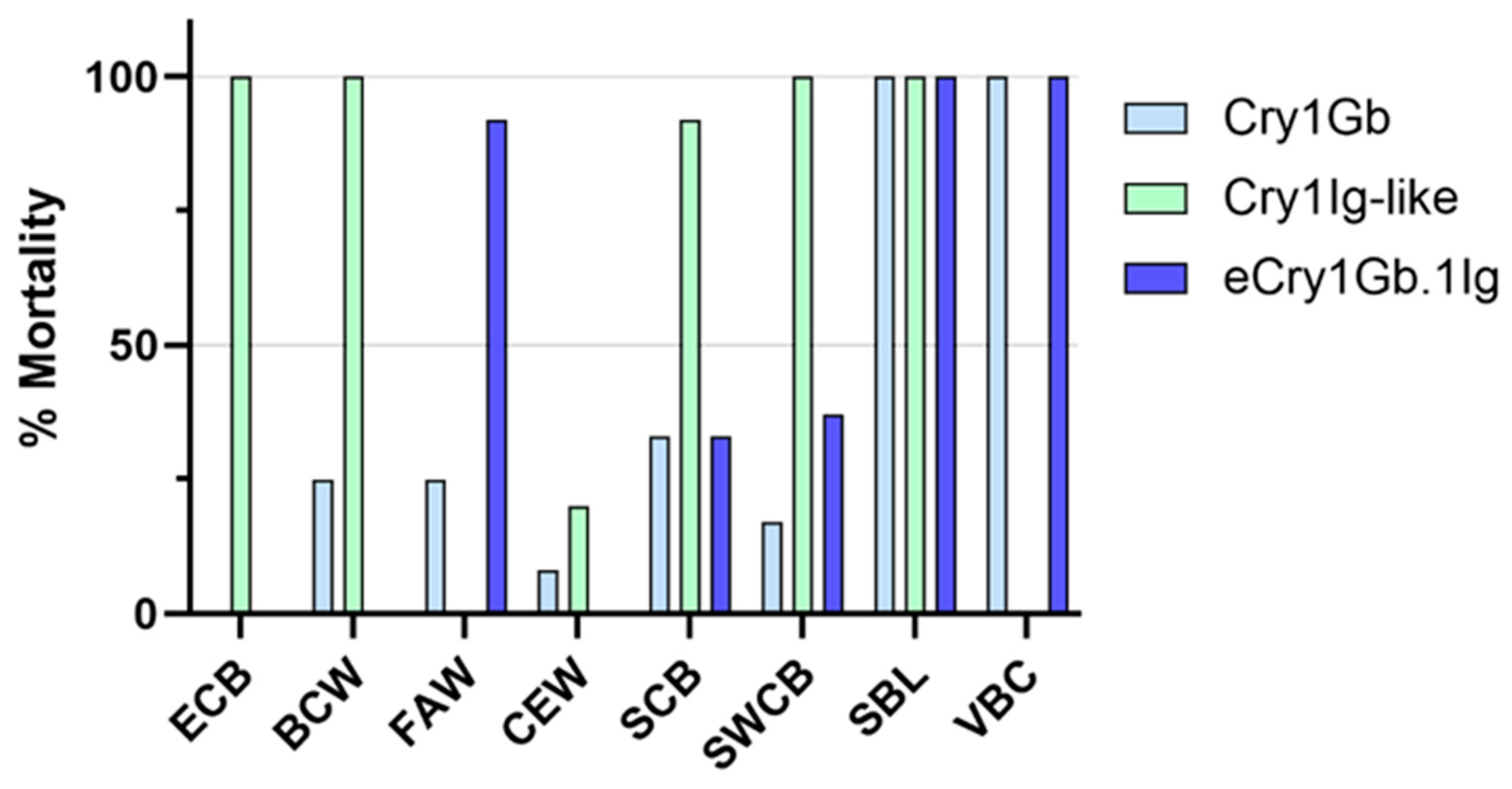
2.2. Screening of Cry1Gb Chimeric Proteins for Insecticidal Activities against FAW and Other Insects
2.3. eCry1Gb.1Ig Is Highly Efficacious against FAW Strains Resistant to Cry1Fa, Vip3Aa and Cry1A.105/Cry2Ab2
2.4. Plants Carrying the eCry1Gb.1Ig Trait Effectively Control Not Just Susceptible, but Also Multiple Resistant FAW Strains
3. Discussion
4. Materials and Methods
4.1. Recombinant DNA and Bt Protein Production
4.2. Insects
4.3. Diet Bioassays
4.4. Leaf Disc and on-Plant Feeding Bioassays
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Skendzic, S.; Zovko, M.; Zivkovic, I.P.; Lesic, V.; Lemic, D. The Impact of Climate Change on Agricultural Insect Pests. Insects 2021, 12, 440. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Carrière, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 2017, 35, 926–935. [Google Scholar] [CrossRef]
- Mallet, J. The evolution of insecticide resistance: Have the insects won? Trends Ecol. Evol. 1989, 4, 336–340. [Google Scholar] [CrossRef]
- Binning, R.R.; Coats, J.; Kong, X.; Hellmich, R.L. Susceptibility and aversion of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Cry1F Bt maize and considerations for insect resistance management. J. Econ. Entomol. 2014, 107, 368–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, F. Resistance of the fall armyworm, Spodoptera frugiperda, to transgenic Bacillus thuringiensis Cry1F corn in the Americas: Lessons and implications for Bt corn IRM in China. Insect Sci. 2021, 28, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, R.N.; Dhanani, I.; Asokan, R.; Mahadevaswamy, H.; Kalleshwaraswamy, C.M.; Meagher, R.L. Genetic characterization of fall armyworm infesting South Africa and India indicate recent introduction from a common source population. PLoS ONE 2019, 14, e0217755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagoshi, R.N.; Koffi, D.; Agboka, K.; Tounou, K.A.; Banerjee, R.; Jurat-Fuentes, J.L.; Meagher, R.L. Comparative molecular analyses of invasive fall armyworm in Togo reveal strong similarities to populations from the eastern United States and the Greater Antilles. PLoS ONE 2017, 12, e0181982. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.F.; Qi, G.J.; Chen, H.; Ma, J.; Liu, J.; Jiang, Y.Y.; Lee, G.S.; Otuka, A.; Hu, G. Overseas immigration of fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae), invading Korea and Japan in 2019. Insect Sci. 2022, 29, 505–520. [Google Scholar] [CrossRef]
- Westbrook, J.; Fleischer, S.; Jairam, S.; Meagher, R.; Nagoshi, R. Multigenerational migration of fall armyworm, a pest insect. Ecosphere 2019, 10, e02919. [Google Scholar] [CrossRef]
- Johnson, S. Migration and the life history strategy of the fall armyworm, Spodoptera frugiperda in the western hemisphere. Int. J. Trop. Insect Sci. 1987, 8, 543–549. [Google Scholar] [CrossRef]
- Adang, M.J.; Crickmore, N.; Jurat-Fuentes, J.L. Diversity of Bacillus thuringiensis crystal toxins and mechanism of action. In Advances in Insect Physiology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 47, pp. 39–87. [Google Scholar]
- Omoto, C.; Bernardi, O.; Salmeron, E.; Sorgatto, R.J.; Dourado, P.M.; Crivellari, A.; Carvalho, R.A.; Willse, A.; Martinelli, S.; Head, G.P. Field-evolved resistance to Cry1Ab maize by Spodoptera frugiperda in Brazil. Pest Manag. Sci. 2016, 72, 1727–1736. [Google Scholar] [CrossRef]
- Banerjee, R.; De Bortoli, C.P.; Huang, F.; Lamour, K.; Meagher, R.; Buntin, D.; Ni, X.; Reay-Jones, F.P.F.; Stewart, S.; Jurat-Fuentes, J.L. Large genomic deletion linked to field-evolved resistance to Cry1F corn in fall armyworm (Spodoptera frugiperda) from Florida. Sci. Rep. 2022, 12, 13580. [Google Scholar] [CrossRef]
- Storer, N.P.; Babcock, J.M.; Schlenz, M.; Meade, T.; Thompson, G.D.; Bing, J.W.; Huckaba, R.M. Discovery and characterization of field resistance to Bt maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico. J. Econ. Entomol. 2010, 103, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Hasler, J.; Meagher, R.; Nagoshi, R.; Hietala, L.; Huang, F.; Narva, K.; Jurat-Fuentes, J.L. Mechanism and DNA-based detection of field-evolved resistance to transgenic Bt corn in fall armyworm (Spodoptera frugiperda). Sci. Rep. 2017, 7, 10877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flagel, L.; Lee, Y.W.; Wanjugi, H.; Swarup, S.; Brown, A.; Wang, J.; Kraft, E.; Greenplate, J.; Simmons, J.; Adams, N.; et al. Mutational disruption of the ABCC2 gene in fall armyworm, Spodoptera frugiperda, confers resistance to the Cry1Fa and Cry1A.105 insecticidal proteins. Sci. Rep. 2018, 8, 7255. [Google Scholar] [CrossRef] [Green Version]
- Boaventura, D.; Ulrich, J.; Lueke, B.; Bolzan, A.; Okuma, D.; Gutbrod, O.; Geibel, S.; Zeng, Q.; Dourado, P.M.; Martinelli, S. Molecular characterization of Cry1F resistance in fall armyworm, Spodoptera frugiperda from Brazil. Insect Biochem. Mol. Biol. 2020, 116, 103280. [Google Scholar] [CrossRef]
- Niu, Y.; Head, G.P.; Price, P.A.; Huang, F. Performance of Cry1A. 105-selected fall armyworm (Lepidoptera: Noctuidae) on transgenic maize plants containing single or pyramided Bt genes. Crop Prot. 2016, 88, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Fatoretto, J.C.; Michel, A.P.; Silva Filho, M.C.; Silva, N. Adaptive potential of fall armyworm (Lepidoptera: Noctuidae) limits Bt trait durability in Brazil. J. Integr. Pest Manag. 2017, 8, 17. [Google Scholar] [CrossRef]
- Bernardi, O.; Bernardi, D.; Ribeiro, R.S.; Okuma, D.M.; Salmeron, E.; Fatoretto, J.; Medeiros, F.C.; Burd, T.; Omoto, C. Frequency of resistance to Vip3Aa20 toxin from Bacillus thuringiensis in Spodoptera frugiperda (Lepidoptera: Noctuidae) populations in Brazil. Crop Prot. 2015, 76, 7–14. [Google Scholar] [CrossRef]
- Yang, F.; Williams, J.; Porter, P.; Huang, F.; Kerns, D.L. F2 screen for resistance to Bacillus thuringiensis Vip3Aa51 protein in field populations of Spodoptera frugiperda (Lepidoptera: Noctuidae) from Texas, USA. Crop Prot. 2019, 126, 104915. [Google Scholar] [CrossRef]
- Heckel, D.G. How do toxins from Bacillus thuringiensis kill insects? An evolutionary perspective. Arch. Insect Biochem. Physiol. 2020, 104, e21673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evdokimov, A.G.; Moshiri, F.; Sturman, E.J.; Rydel, T.J.; Zheng, M.; Seale, J.W.; Franklin, S. Structure of the full-length insecticidal protein C ry1 A c reveals intriguing details of toxin packaging into in vivo formed crystals. Protein Sci. 2014, 23, 1491–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Maagd, R.A.; Kwa, M.S.; van der Klei, H.; Yamamoto, T.; Schipper, B.; Vlak, J.M.; Stiekema, W.J.; Bosch, D. Domain III substitution in Bacillus thuringiensis delta-endotoxin CryIA(b) results in superior toxicity for Spodoptera exigua and altered membrane protein recognition. Appl. Env. Microbiol. 1996, 62, 1537–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, F.S.; deFontes, C.M.; Hart, H.; Warren, G.W.; Chen, J.S. Lepidopteran-active variable-region sequence imparts coleopteran activity in eCry3.1Ab, an engineered Bacillus thuringiensis hybrid insecticidal protein. Appl. Env. Microbiol. 2010, 76, 3082–3088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrero, S.; Bel, Y.; Hernández-Martínez, P.; Ferré, J. Susceptibility, mechanisms of response and resistance to Bacillus thuringiensis toxins in Spodoptera spp. Curr. Opin. Insect Sci. 2016, 15, 89–96. [Google Scholar] [CrossRef]
- Martínez, C.; Caballero, P. Contents of cry genes and insecticidal toxicity of Bacillus thuringiensis strains from terrestrial and aquatic habitats. J. Appl. Microbiol. 2002, 92, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Thammasittirong, A.; Attathom, T. PCR-based method for the detection of cry genes in local isolates of Bacillus thuringiensis from Thailand. J. Invertebr. Pathol. 2008, 98, 121–126. [Google Scholar] [CrossRef]
- Zhu, C.; Niu, Y.; Zhou, Y.; Guo, J.; Head, G.P.; Price, P.A.; Wen, X.; Huang, F. Survival and effective dominance level of a Cry1A. 105/Cry2Ab2-dual gene resistant population of Spodoptera frugiperda (JE Smith) on common pyramided Bt corn traits. Crop Prot. 2019, 115, 84–91. [Google Scholar] [CrossRef]
- Wen, Z. More than 10 years after commercialization, MIR162 remains a powerful tool for controlling major Lepidopteran corn pests. Unpublished (In preparation)
- Hofte, H.; de Greve, H.; Seurinck, J.; Jansens, S.; Mahillon, J.; Ampe, C.; Vandekerckhove, J.; Vanderbruggen, H.; van Montagu, M.; Zabeau, M.; et al. Structural and functional analysis of a cloned delta endotoxin of Bacillus thuringiensis berliner 1715. Eur. J. Biochem. 1986, 161, 273–280. [Google Scholar] [CrossRef]
- Smith, P.e.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Morsello, S.; Head, G.P.; Sansone, C.; Huang, F.; Gilreath, R.T.; Kerns, D.L. F2 screen, inheritance and cross-resistance of field-derived Vip3A resistance in Spodoptera frugiperda (Lepidoptera: Noctuidae) collected from Louisiana, USA. Pest Manag. Sci. 2018, 74, 1769–1778. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Guo, J.; Head, G.P.; Price, P.A.; Huang, F. Phenotypic performance of nine genotypes of Cry1A. 105/Cry2Ab2 dual-gene resistant fall armyworm on non-Bt and MON 89034 maize. Pest Manag. Sci. 2019, 75, 2124–2132. [Google Scholar] [CrossRef] [PubMed]
- Little, N.S.; Mullen, R.M.; Allen, K.C.; Tyler, H.L. Leaf tissue assay for lepidopteran pests of Bt cotton. Southwest. Entomol. 2017, 42, 953–958. [Google Scholar] [CrossRef]
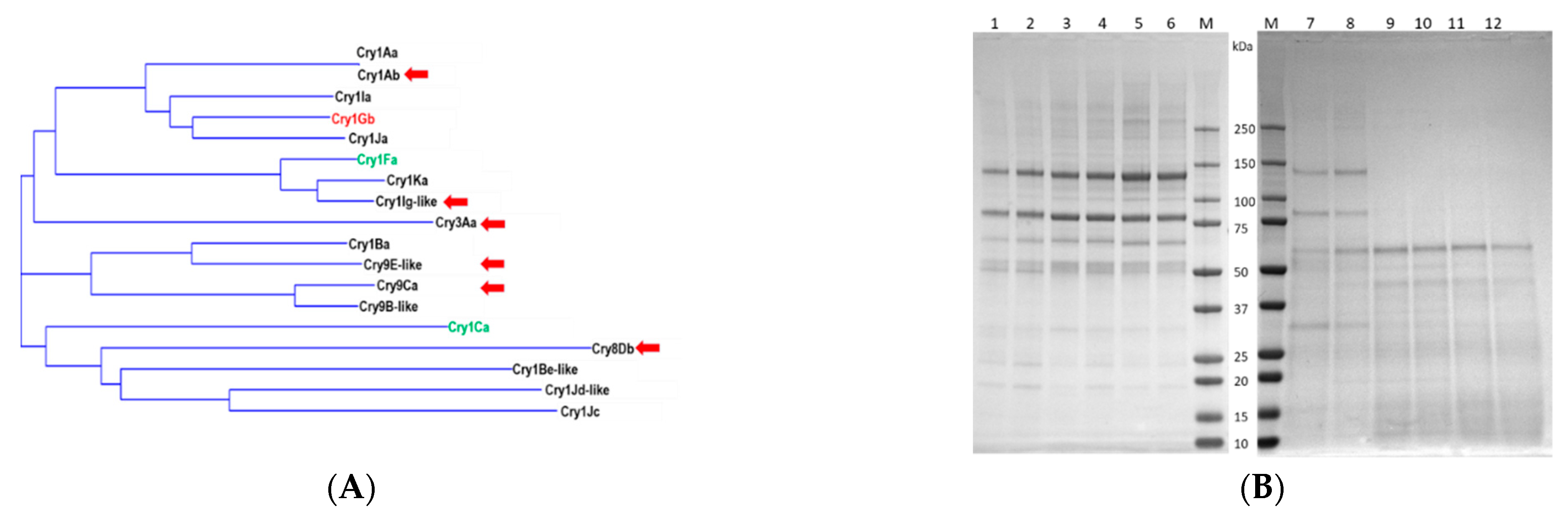


| Chimeric Proteins | Domain 1 | Domain 2 | Domain 3 | Protoxin-Tail |
|---|---|---|---|---|
| Cry1Gb-3A | Cry1Gb | Cry1Gb | Cry3A | Cry1Gb |
| eCry1Gb.1Ig | Cry1Gb | Cry1Gb | Cry1Ig | Cry1Gb |
| Cry1Gb-9E | Cry1Gb | Cry1Gb | Cry9E | Cry1Gb |
| Cry1Gb-1Ab | Cry1Gb | Cry1Gb | Cry1Ab | Cry1Gb |
| Cry1Gb-8D | Cry1Gb | Cry1Gb | Cry8D | Cry1Gb |
| Cry1Gb-9Ca | Cry1Gb | Cry1Gb | Cry9Ca | Cry1Gb |
| Proteins | Concentrations (µg/cm2) | Mortality (%), a * | Mortality (%), b * |
|---|---|---|---|
| Cry1Gb-3A | 3.2 | 25 | 33 |
| eCry1Gb.1Ig | 3.2 | 100 | 83 |
| Cry1Gb-9E | 3.2 | 8 | 17 |
| Cry1Gb-1Ab | 3.2 | 0 | 0 |
| Cry1Fa | 1.0 | 100 | - |
| Buffer 1 | 0 | 8 | - |
| Proteins | Mortality (% Effective Mortality) | ||
|---|---|---|---|
| S. eridania, BR | cosmioides, BR | S. litura, CN | |
| eCry1Gb.1Ig | 8 | 46 | 100 |
| Vip3Aa19 | 100 | 100 | - |
| Buffer 1 | 13 | 4 | 17 |
| PBS | 4 | 4 | - |
| Proteins | FAW Mortality (% Effective Mortality) | |||
|---|---|---|---|---|
| BenFAW | FAWVip3AR | Cry1Fa-R-PR | BrFAW | |
| eCry1Gb.1Ig | 100 | 100 | 100 | 100 |
| Cry1Fa | 100 | 100 | 0 | 8 |
| Vip3Aa19 | 100 | 0 | 100 | 100 |
| Buffer 1 | 0 | 0 | 0 | 0 |
| PBS | 0 | 0 | 0 | 0 |
| FAW Strains | Cry1Fa | Vip3Aa19 | eCry1Gb.1Ig | |||
|---|---|---|---|---|---|---|
| No. Insects * | EC50 (95%CI) | No. Insects * | EC50 (95%CI) | No. Insects * | EC50 (95%CI) | |
| BenFAW | 816 | 72.8 (46.4–107) | 912 | 39.0 (31.5–47.5) | 792 | 3.57 (2.66–4.7) |
| BrFAW | 216 | >>3000 | 1080 | 2.92 (2.3–3.64) | ||
| FAWVip3AR | 360 | >>3000 | 1152 | 54.6 (41.5–70.2) | ||
| FAWLSUS | 1146 | 12.6 (8.0–17.7) | ||||
| FAWDUALR | 1139 | 16.9 (13.6–20.4) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chae, H.; Wen, Z.; Hootman, T.; Himes, J.; Duan, Q.; McMath, J.; Ditillo, J.; Sessler, R.; Conville, J.; Niu, Y.; et al. eCry1Gb.1Ig, A Novel Chimeric Cry Protein with High Efficacy against Multiple Fall Armyworm (Spodoptera frugiperda) Strains Resistant to Different GM Traits. Toxins 2022, 14, 852. https://doi.org/10.3390/toxins14120852
Chae H, Wen Z, Hootman T, Himes J, Duan Q, McMath J, Ditillo J, Sessler R, Conville J, Niu Y, et al. eCry1Gb.1Ig, A Novel Chimeric Cry Protein with High Efficacy against Multiple Fall Armyworm (Spodoptera frugiperda) Strains Resistant to Different GM Traits. Toxins. 2022; 14(12):852. https://doi.org/10.3390/toxins14120852
Chicago/Turabian StyleChae, Hyunsook, Zhimou Wen, Travis Hootman, Jo Himes, Qianqian Duan, Joel McMath, Jesse Ditillo, Richard Sessler, Jared Conville, Ying Niu, and et al. 2022. "eCry1Gb.1Ig, A Novel Chimeric Cry Protein with High Efficacy against Multiple Fall Armyworm (Spodoptera frugiperda) Strains Resistant to Different GM Traits" Toxins 14, no. 12: 852. https://doi.org/10.3390/toxins14120852





