Aromatic Residues on the Side Surface of Cry4Ba-Domain II of Bacillus thuringiensis subsp. israelensis Function in Binding to Their Counterpart Residues on the Aedes aegypti Alkaline Phosphatase Receptor
Abstract
:1. Introduction
2. Results
2.1. Analysis of Possible Binding Residues between the Cry4Ba Toxin and the Aa-mALP Receptor
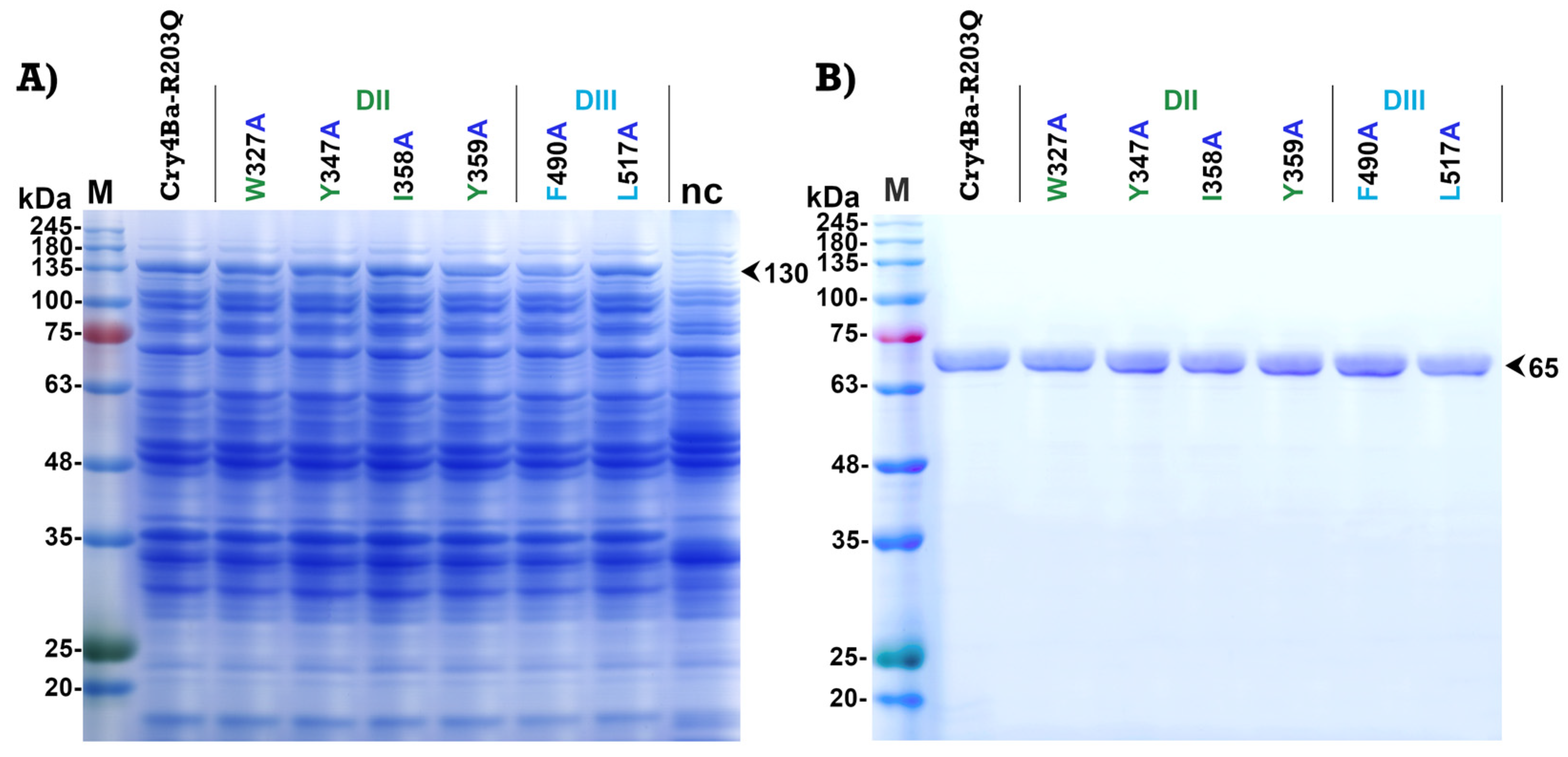
2.2. Expression and Purification of Cry4Ba and Its Mutants
2.3. Larvicidal Activity of Cry4Ba and Its Mutants
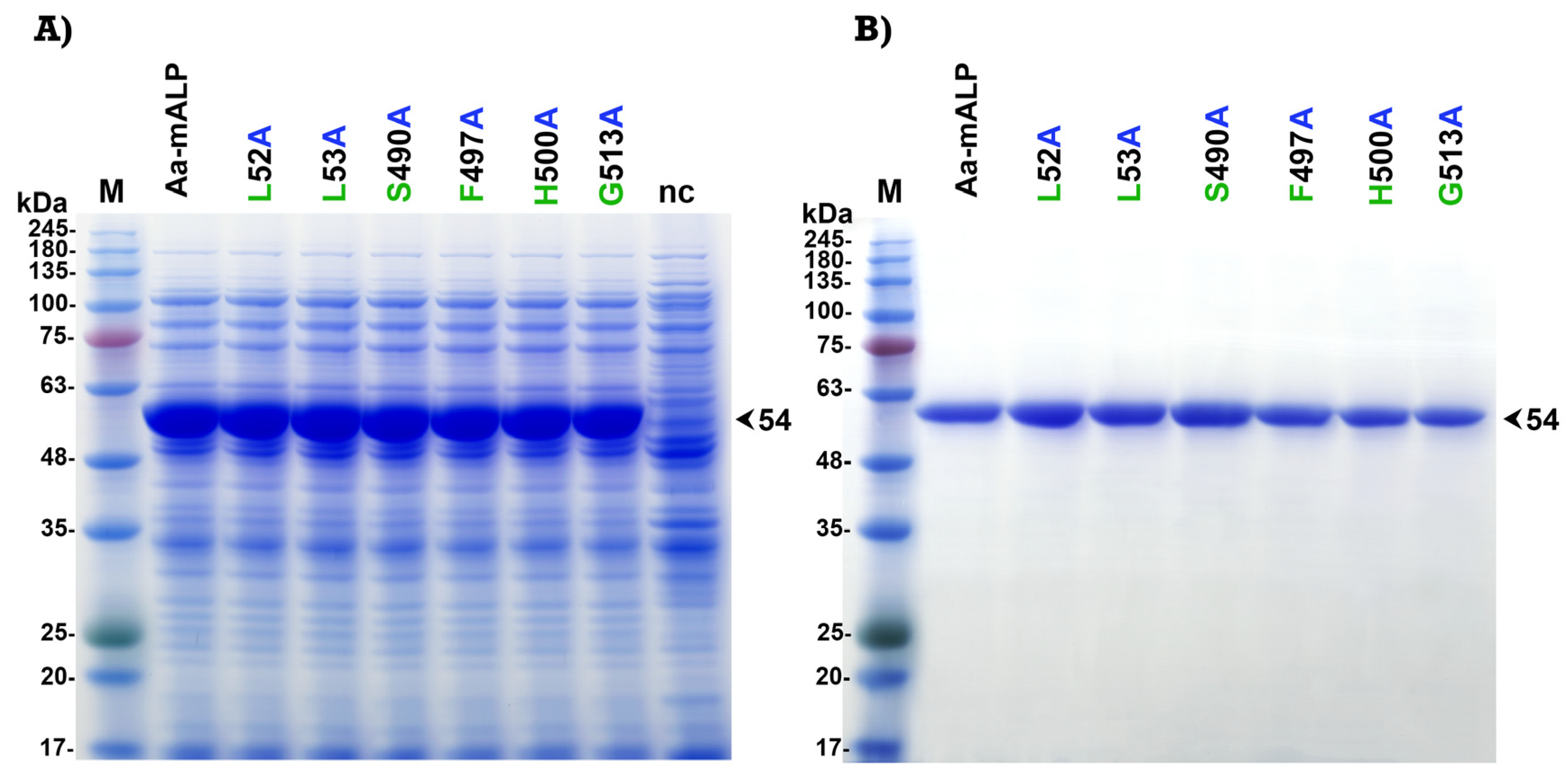
2.4. Expression and Purification of Aa-mALP and Its Mutants
2.5. Quantitative Analysis of Cry4Ba Toxin—Aa-mALP Receptor Interactions
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Materials
5.2. In Silico Binding Analysis of Cry4Ba Toxin—Aa-mALP Receptor Interaction
5.3. Construction of Cry4Ba Mutant Plasmids
5.4. Construction of Aa-mALP Mutant Plasmids
5.5. Expression and Purification of Cry4Ba and Its Mutants
5.6. Expression and Purification of Aa-mALP and Its Mutants
5.7. Mosquito Larvicidal Activity Assays
5.8. Cry4Ba Toxin-Aa-mALP Binding Assays Using ELISA
5.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Dengue Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition; WHO: Geneva, Switzerland, 2009. [Google Scholar]
- Bhatt, P.; Sabeena, S.P.; Varma, M.; Arunkumar, G. Current understanding of the pathogenesis of dengue virus infection. Curr. Microbiol. 2021, 78, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Kularatne, S.A.; Dalugama, C. Dengue infection: Global importance, immunopathology and management. Clin. Med. 2022, 22, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messina, J.P.; Brady, O.J.; Golding, N.; Kraemer, M.U.G.; Wint, G.R.W.; Ray, S.E.; Pigott, D.M.; Shearer, F.M.; Johnson, K.; Earl, L.; et al. The current and future global distribution and population at risk of dengue. Nat. Microbiol. 2019, 4, 1508–1515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Dengue vaccine: WHO position paper, September 2018-Recommendations. Vaccine 2019, 37, 4848–4849. [Google Scholar] [CrossRef]
- Park, J.; Kim, J.; Jang, Y.-S. Current status and perspectives on vaccine development against dengue virus infection. J. Microbiol. 2022, 60, 247–254. [Google Scholar] [CrossRef]
- Lawrence, A.L. Bacillus thuringiensis serovariety israelensis and Bacillus sphaericus for mosquito control. J. Am. Mosq. Control Assoc. 2007, 23, 133–163. [Google Scholar] [CrossRef]
- Silva-Filha, M.; Romão, T.; Rezende, T.; Carvalho, K.; de Menezes, H.G.; Nascimento, N.A.D.; Soberón, M.; Bravo, A. Bacterial toxins active against mosquitoes: Mode of action and resistance. Toxins 2021, 13, 523. [Google Scholar] [CrossRef]
- Tetreau, G.; Stalinski, R.; David, J.-P.; Després, L. Monitoring resistance to Bacillus thuringiensis subsp. israelensis in the field by performing bioassays with each Cry toxin separately. Mem. Inst. Oswaldo Cruz 2013, 108, 894–900. [Google Scholar] [CrossRef]
- Brühl, C.A.; Després, L.; Frör, O.; Patil, C.D.; Poulin, B.; Tetreau, G.; Allgeier, S. Environmental and socioeconomic effects of mosquito control in Europe using the biocide Bacillus thuringiensis subsp. israelensis (Bti). Sci. Total Environ. 2020, 724, 137800. [Google Scholar] [CrossRef]
- Carvalho, K.D.S.; Crespo, M.M.; Araújo, A.P.; Da Silva, R.S.; De Melo-Santos, M.A.V.; De Oliveira, C.M.F.; Silva-Filha, M.H.N.L. Long-term exposure of Aedes aegypti to Bacillus thuringiensis svar. israelensis did not involve altered susceptibility to this microbial larvicide or to other control agents. Parasites Vectors 2018, 11, 673. [Google Scholar] [CrossRef] [PubMed]
- Ben-Dov, E. Bacillus thuringiensis subsp. israelensis and its dipteran-specific toxins. Toxins 2014, 6, 1222–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boonserm, P.; Davis, P.; Ellar, D.J.; Li, J. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J. Mol. Biol. 2005, 348, 363–382. [Google Scholar] [CrossRef] [PubMed]
- Bayyareddy, K.; Andacht, T.M.; Abdullah, M.A.; Adang, M.J. Proteomic identification of Bacillus thuringiensis subsp. israelensis toxin Cry4Ba binding proteins in midgut membranes from Aedes (Stegomyia) aegypti Linnaeus (Diptera, Culicidae) larvae. Insect Biochem. Mol. Biol. 2009, 39, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.I.; Reyes, E.Z.; Cancino-Rodezno, A.; Bedoya-Pérez, L.P.; Caballero-Flores, G.G.; Muriel-Millan, L.F.; Likitvivatanavong, S.; Gill, S.S.; Bravo, A.; Soberón, M. Aedes aegypti alkaline phosphatase ALP1 is a functional receptor of Bacillus thuringiensis Cry4Ba and Cry11Aa toxins. Insect Biochem. Mol. Biol. 2012, 42, 683–689. [Google Scholar] [CrossRef] [Green Version]
- Saengwiman, S.; Aroonkesorn, A.; Dedvisitsakul, P.; Sakdee, S.; Leetachewa, S.; Angsuthanasombat, C.; Pootanakit, K. In vivo identification of Bacillus thuringiensis Cry4Ba toxin receptors by RNA interference knockdown of glycosylphosphatidylinositol-linked aminopeptidase N transcripts in Aedes aegypti larvae. Biochem. Biophys. Res. Commun. 2011, 407, 708–713. [Google Scholar] [CrossRef]
- Dechklar, M.; Tiewsiri, K.; Angsuthanasombat, C.; Pootanakit, K. Functional expression in insect cells of glycosylphosphatidylinositol-linked alkaline phosphatase from Aedes aegypti larval midgut: A Bacillus thuringiensis Cry4Ba toxin receptor. Insect Biochem. Mol. Biol. 2011, 41, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Aroonkesorn, A.; Pootanakit, K.; Katzenmeier, G.; Angsuthanasombat, C. Two specific membrane-bound aminopeptidase N isoforms from Aedes aegypti larvae serve as functional receptors for the Bacillus thuringiensis Cry4Ba toxin implicating counterpart specificity. Biochem. Biophys. Res. Commun. 2015, 461, 300–306. [Google Scholar] [CrossRef]
- Thammasittirong, A.; Dechklar, M.; Leetachewa, S.; Pootanakit, K.; Angsuthanasombat, C. Aedes aegypti membrane-bound alkaline phosphatase expressed in Escherichia coli retains high-affinity binding for Bacillus thuringiensis Cry4Ba toxin. Appl. Environ. Microbiol. 2011, 77, 6836–6840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thammasittirong, A.; Thammasittirong, S.N.-R.; Imtong, C.; Charoenjotivadhanakul, S.; Sakdee, S.; Li, H.-C.; Okonogi, S.; Angsuthanasombat, C. Bacillus thuringiensis Cry4Ba insecticidal toxin exploits Leu615 in its C-terminal domain to interact with a target receptor—Aedes aegypti membrane-bound alkaline phosphatase. Toxins 2021, 13, 553. [Google Scholar] [CrossRef]
- Khaokhiew, T.; Angsuthanasombat, C.; Promptmas, C. Correlative effect on the toxicity of three surface-exposed loops in the receptor-binding domain of the Bacillus thuringiensis Cry4Ba toxin. FEMS Microbiol. Lett. 2009, 300, 139–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Dehouck, Y.; Kwasigroch, J.M.; Rooman, M.; Gilis, D. BeAtMuSiC: Prediction of changes in protein–protein binding affinity on mutations. Nucleic Acids Res. 2013, 41, W333–W339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo, A.L.D.A.; Soccol, V.T.; Soccol, C.R. Bacillus thuringiensis: Mechanism of action, resistance, and new applications: A review. Crit. Rev. Biotechnol. 2016, 36, 317–326. [Google Scholar] [CrossRef]
- Vílchez, S. Making 3D-Cry toxin mutants: Much more than a tool of understanding toxins mechanism of action. Toxins 2020, 12, 600. [Google Scholar] [CrossRef]
- Morrow, J.K.; Zhang, S. Computational prediction of protein hot spot residues. Curr. Pharm. Des. 2012, 18, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Verkhivker, G.M.; Bouzida, D.; Gehlhaar, D.K.; Rejto, P.A.; Freer, S.T.; Rose, P.W. Monte Carlo simulations of the peptide recognition at the consensus binding site of the constant fragment of human immunoglobulin G: The energy landscape analysis of a hot spot at the intermolecular interface. Proteins 2002, 48, 539–557. [Google Scholar] [CrossRef] [PubMed]
- Mattapally, S.; Singh, M.; Murthy, K.S.; Asthana, S.; Banerjee, S.K. Computational modeling suggests impaired interactions between NKX2.5 and GATA4 in individuals carrying a novel pathogenic D16N NKX2.5 mutation. Oncotarget 2018, 9, 13713–13732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almond, B.D.; Dean, D.H. Structural stability of Bacillus thuringiensis delta-endotoxin homolog-scanning mutants determined by susceptibility to proteases. Appl. Environ. Microbiol. 1993, 59, 2442–2448. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Elkayam, T.; Wolfson, H.; Nussinov, R. Protein-protein interactions: Structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl. Acad. Sci. USA 2003, 100, 5772–5777. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, D.; Zhu, J.; Nussinov, R.; Ma, B. Local and global anatomy of antibody-protein antigen recognition. J. Mol. Recognit. 2018, 31, e2693. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.-P.; Lee, K.H.; Jian, J.-W.; Yang, A.-S. Origins of specificity and affinity in antibody–protein interactions. Proc. Natl. Acad. Sci. USA 2014, 111, E2656–E2665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, L.; Bandelac, G.; Volgina, A.; Korostoff, J.; DiRienzo, J.M. Role of aromatic amino acids in receptor binding activity and subunit assembly of the cytolethal distending toxin of Aggregatibacter actinomycetemcomitans. Infect. Immun. 2008, 76, 2812–2821. [Google Scholar] [CrossRef] [Green Version]
- Melton-Witt, J.A.; Bentsen, L.M.; Tweten, R.K. Identification of functional domains of Clostridium septicum alpha toxin. Biochemistry 2006, 45, 14347–14354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singkhamanan, K.; Promdonkoy, B.; Chaisri, U.; Boonserm, P. Identification of amino acids required for receptor binding and toxicity of the Bacillus sphaericus toxin. FEMS Microbiol. Lett. 2010, 303, 84–91. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Verkhivker, G.M.; Agajanian, S.; Oztas, D.Y.; Gupta, G. Comparative perturbation-based modeling of the SARS-CoV-2 spike protein binding with host receptor and neutralizing antibodies: Structurally adaptable allosteric communication hotspots define spike sites targeted by global circulating mutations. Biochemistry 2021, 60, 1459–1484. [Google Scholar] [CrossRef]
- Thamwiriyasati, N.; Sakdee, S.; Chuankhayan, P.; Katzenmeier, G.; Chen, C.-J.; Angsuthanasombat, C. Crystallization and preliminary X-ray crystallographic analysis of a full-length active form of the Cry4Ba toxin from Bacillus thuringiensis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 721–724. [Google Scholar] [CrossRef] [Green Version]
- Sriwimol, W.; Aroonkesorn, A.; Sakdee, S.; Kanchanawarin, C.; Uchihashi, T.; Ando, T.; Angsuthanasombat, C. Potential prepore trimer formation by the Bacillus thuringiensis mosquito-specific toxin: Molecular insights into a critical prerequisite of membrane-bound monomers. J. Biol. Chem. 2015, 290, 20793–20803. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thammasittirong, A.; Thammasittirong, S.N.-R. Aromatic Residues on the Side Surface of Cry4Ba-Domain II of Bacillus thuringiensis subsp. israelensis Function in Binding to Their Counterpart Residues on the Aedes aegypti Alkaline Phosphatase Receptor. Toxins 2023, 15, 114. https://doi.org/10.3390/toxins15020114
Thammasittirong A, Thammasittirong SN-R. Aromatic Residues on the Side Surface of Cry4Ba-Domain II of Bacillus thuringiensis subsp. israelensis Function in Binding to Their Counterpart Residues on the Aedes aegypti Alkaline Phosphatase Receptor. Toxins. 2023; 15(2):114. https://doi.org/10.3390/toxins15020114
Chicago/Turabian StyleThammasittirong, Anon, and Sutticha Na-Ranong Thammasittirong. 2023. "Aromatic Residues on the Side Surface of Cry4Ba-Domain II of Bacillus thuringiensis subsp. israelensis Function in Binding to Their Counterpart Residues on the Aedes aegypti Alkaline Phosphatase Receptor" Toxins 15, no. 2: 114. https://doi.org/10.3390/toxins15020114




