The Role of Yeast-Surface-Display Techniques in Creating Biocatalysts for Consolidated BioProcessing
Abstract
:1. Introduction
1.1. Climate Change and the Bio-Based Economy
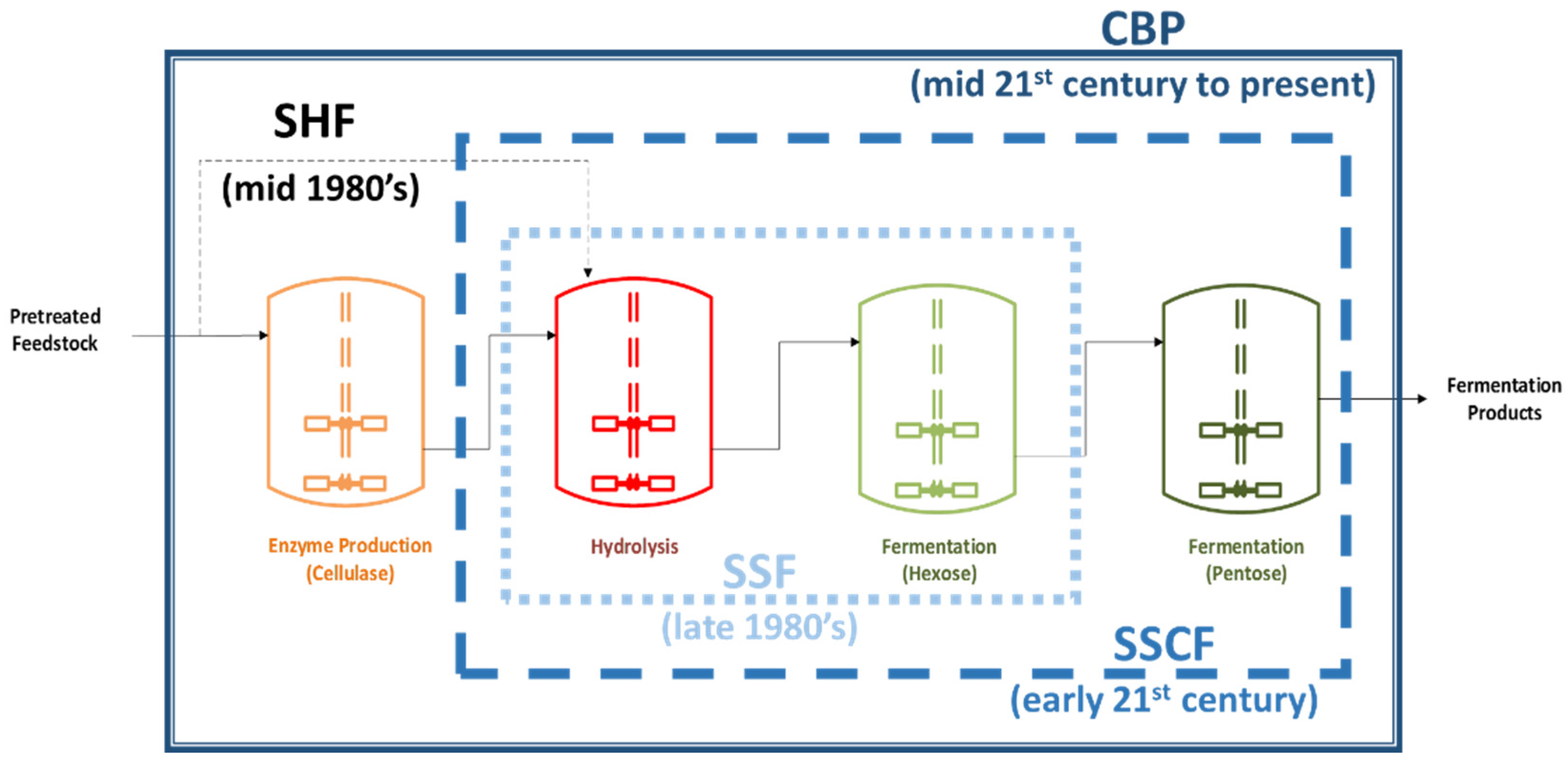
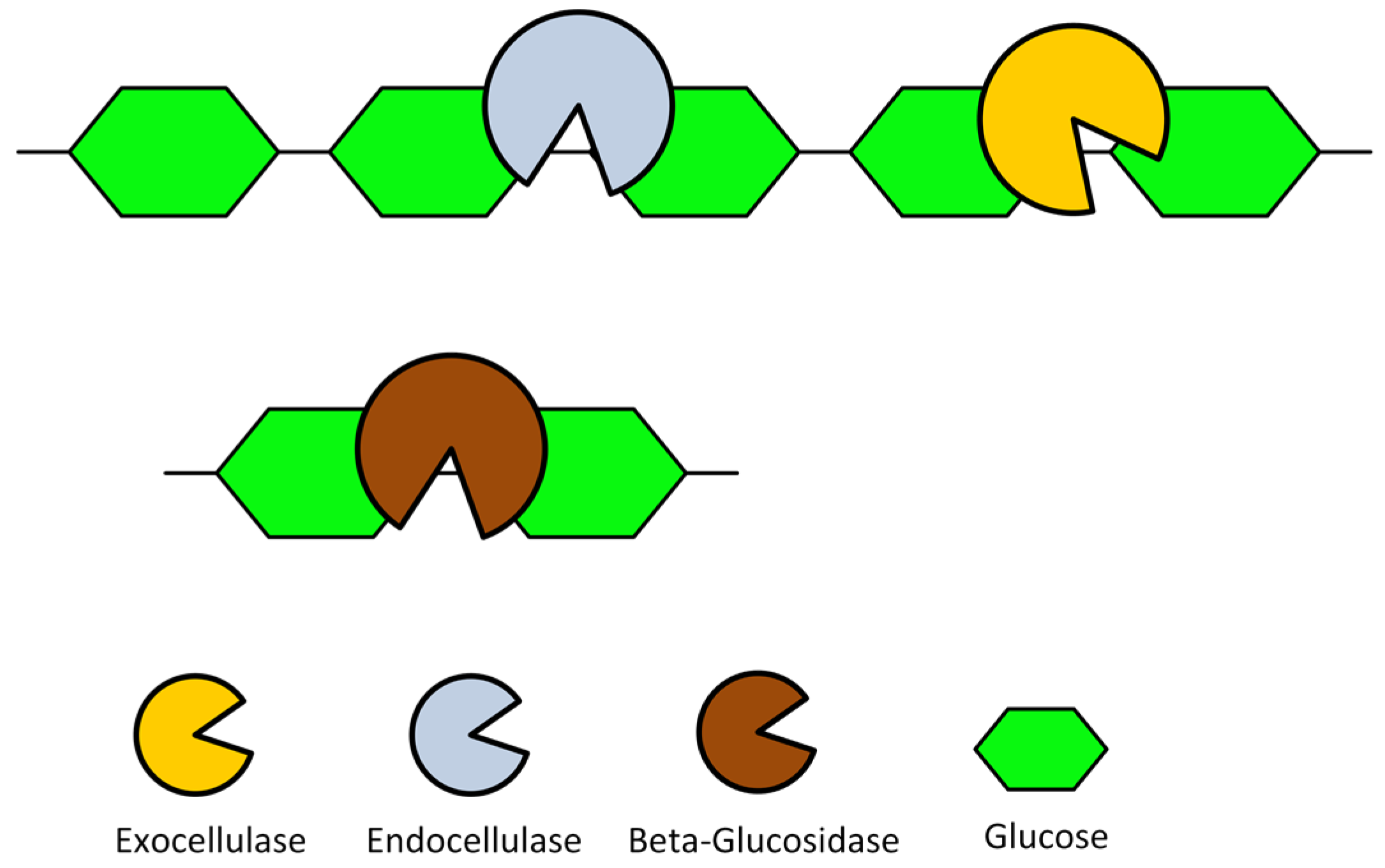
1.2. Lignocellulosic Biomass and Consolidated Bio-Processing
2. The Development of Yeast Cell Surface Display
2.1. Development of the Surface Display Technology
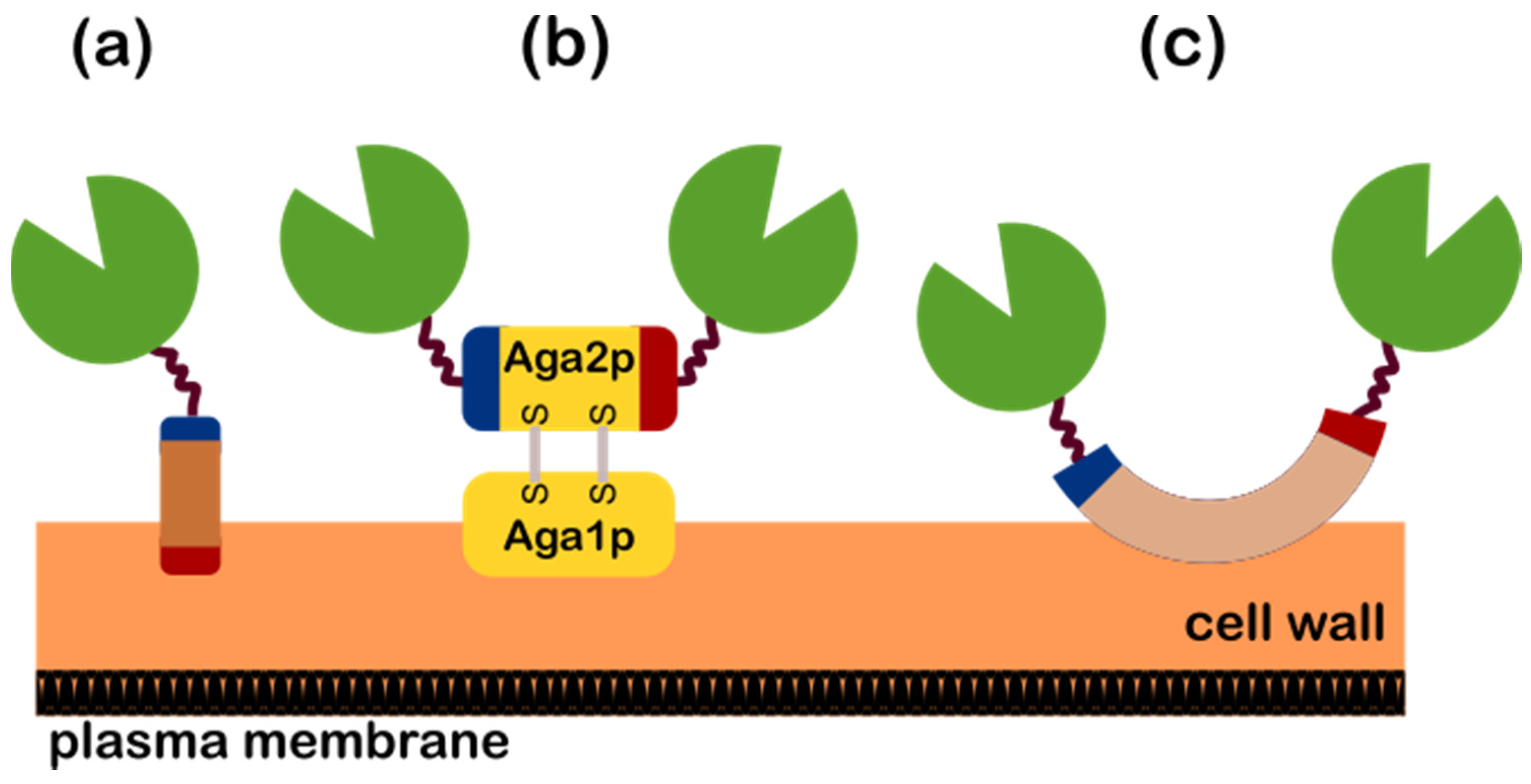
2.2. Yeast Surface Display
2.3. Strategies for the Yeast Surface Display of CBP-Related Enzymes
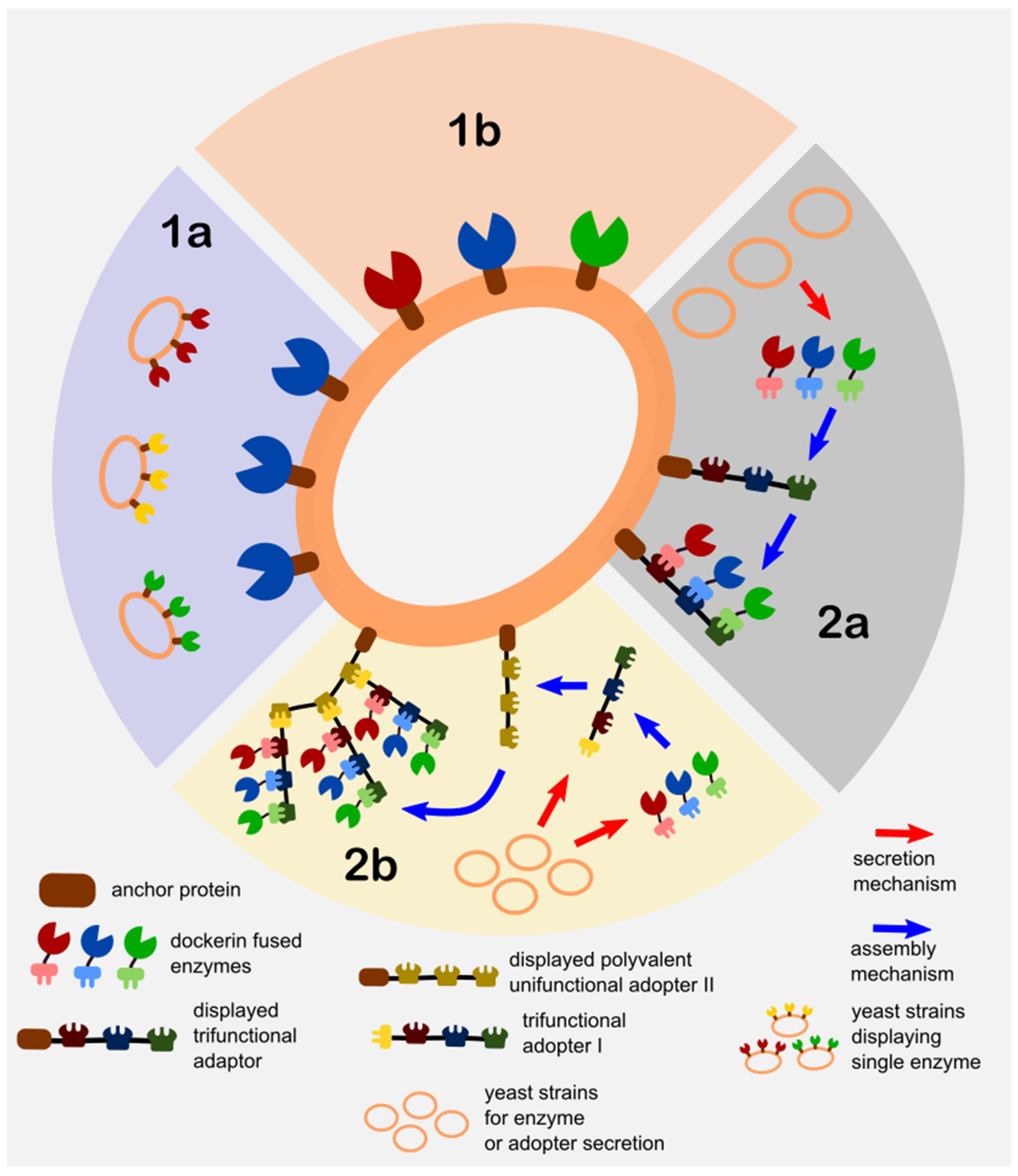
- Direct Yeast Surface Display (DYSD)—this strategy directly involves the fusion of the enzyme of interest/s to the yeast anchor protein for it to be expressed and localize on its surface and is further classified into the following:
- Single enzyme display consortium (SEDC)—a mixed culture (a.k.a synthetic consortium) strategy of different strains with each displaying one type of enzyme. The utilization of the mixed culture technique is adapted as a means to control the multi-enzyme composition or ratios via the culture inoculation ratios.
- Co-display (CD)—the direct display of more than one enzymes on yeast that is achieved by directly cloning the enzyme/s fused with anchor protein into one yeast strain. The control of the displayed enzyme ratios
- Juxtaposed Assembly of Non-Native Adaptors and Secreted Enzymes on Yeast (JANNASEY)—this strategy is developed based on the biomimicry of the “cellulosome” machinery of several anaerobic cellulose-consuming microorganisms (e.g., clostridia, ruminal bacteria) [36,37]. In addition, exploiting the cellulosome’s nature as a reconfigurable platform for immobilization of multiple enzymes including an enzyme cascade configuration, along with the advancement of molecular biology and conjugation techniques, led to the creation of “designer cellulosomes”. These designer cellulosomes typically consists of the cell wall anchoring subunit (or sometimes directly fused to the adaptor subunit) for cell wall attachment, the adaptor subunit (a.k.a. scaffoldin or miniscaffoldin), which consists of the configurable multi-cohesin along with the cellulose binding domains that either bind to the anchoring subunit or onto another adaptor subunit, and finally the secreted enzyme subunit which consists of enzyme/s each fused to a specific dockerin domain that corresponds to the cohesin/s in the adaptor subunit [38]. The term “minicellulosomes” is often used to distinguish the designer cellulosomes that contain engineered adaptor subunits (a.k.a. miniscaffoldin to be consistent with the minicellulosome) with non-native cohesin and cellulose-binding domains. Other researchers’ even use the term “truncated cellulosome” that is synonymous to the minicellulosome to even give it a clearer distinction with the cellulosome [37,39]. In order to prevent the inappropriate use of the terms related to cellulosomes in this article, this yeast display strategy is generally classified as the juxtaposed assembly of non-native adaptors and secretion enzymes on yeast in reference to its structured and precise self-assembly on yeast. Furthermore, the various assembly techniques of the cellulosome components are classified as follows:
- Single multi-functional adaptor assembly (SMFAA)—this strategy typically involves the consortium of two sets of strains wherein one strain displays the chimeric adaptor subunit on its surface, and another set of strains that secrete the dockerin fused enzymes that assemble in situ onto the displayed adaptor subunit of another strain. The multi-functionality this system is derived from the fact that the adaptor subunit it contains varying cohesin domains from different microorganisms for specificity.
- Complex adaptor assembly (CAA)—this is a direct biomimicry of the native bacterial cellulosome assembly where one or more adaptor subunits are assembled on the bacterial surface. When applied to yeast surface display, it involves a consortium of at least three sets of strains where one strain displays the anchoring subunit, another set of strains that secrete one or more adaptor subunits (one of which binds to the anchoring subunit and another type of binds to the cell-surface-anchored adaptor), and finally the strains that secrete the dockerin fused enzymes, which assemble onto the adaptors. The complex assembly is regulated by the valence (monovalent if it contains a single cohesin and polyvalent if it contains more than one) of the adaptor subunit. Adaptor polyvalence can also be achieved by specifically engineering the adaptor to contain multiple cohesin types from different microorganisms for regulating the enzyme loading via cohesin-dockerin specificity.
3. Yeast Surface Display Studies for Bioethanol Production from Lignocellulosic Biomass
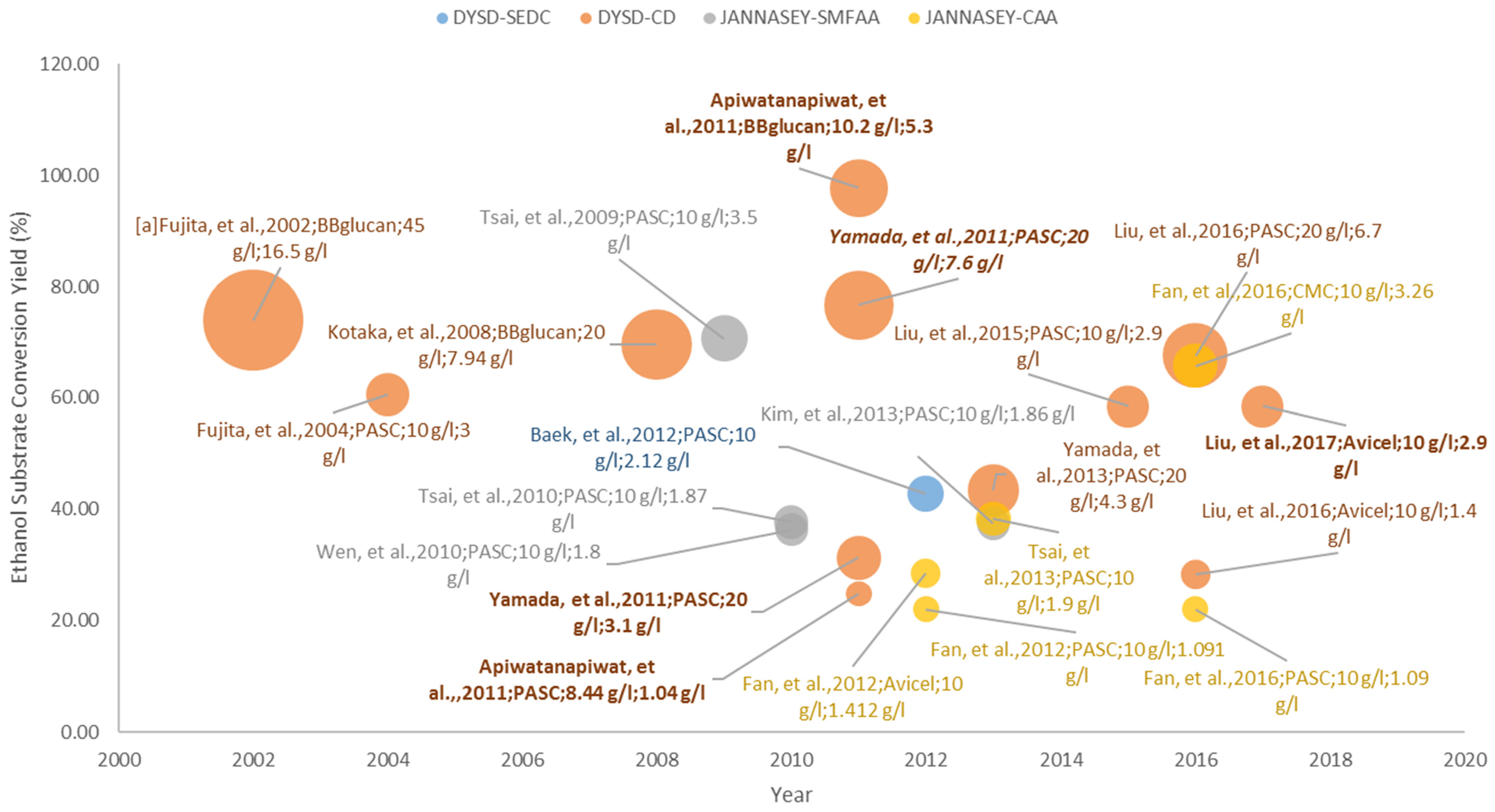
3.1. Pure Cellulose Substrates
3.1.1. DYSD Strategy for Cellulosic Bioethanol Fermentation
3.1.2. JANNASEY Strategy for Cellulosic Bioethanol Fermentation
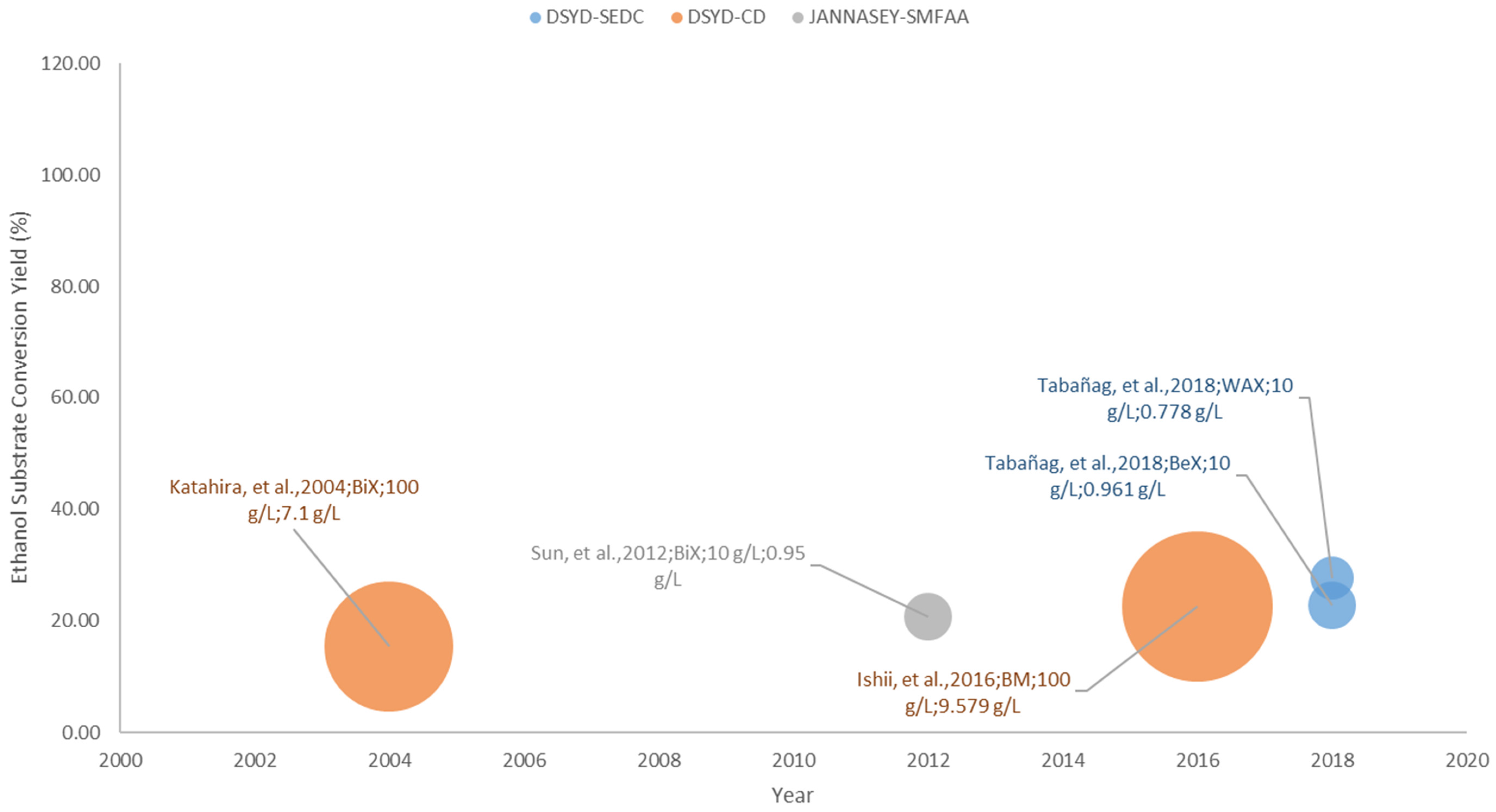
3.2. Pure Hemicellulose Substrates
3.2.1. DYSD Strategy for Hemicellulosic Bioethanol Fermentation
3.2.2. JANNASEY Strategy for Hemicellulosic Bioethanol Fermentation
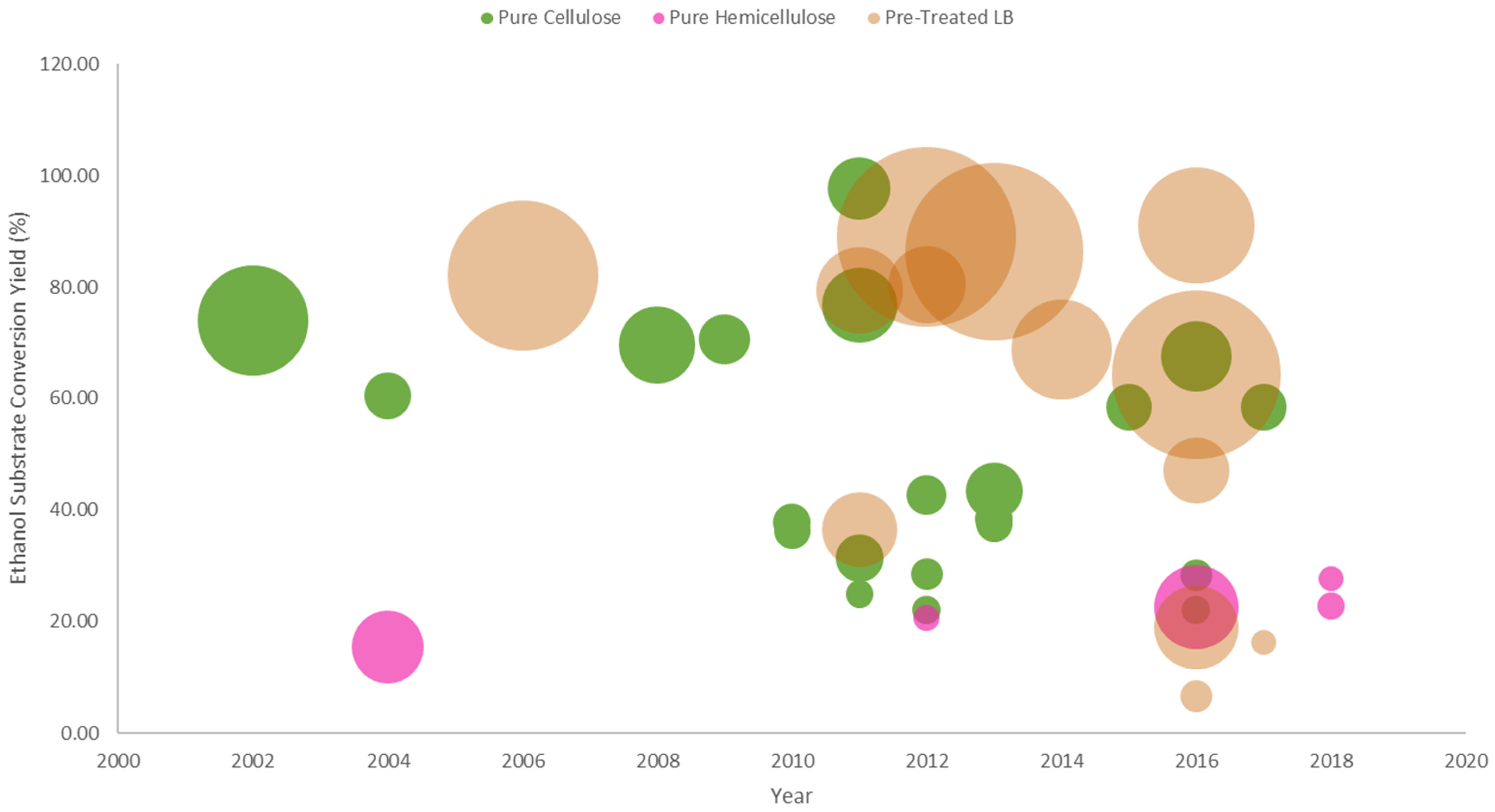
3.3. Pre-Treated Lignocellulosic Substrates
DYSD Strategy for Bioethanol Fermentation from Pre-Treated Lignocellulosic Biomass
3.4. Comparing the Various Yeast Surface Display Strategies for Bioethanol Fermentation from Lignocellulosic Biomass Substrates
3.4.1. Cellulosic Bioethanol Fermentation
3.4.2. Hemicelllulosic Bioethanol Fermentation
3.4.3. Bioethanol Fermentation from Pre-Treated Lignocellulosic Biomass
4. Avenues of Improvement for Bioethanol Production Using Yeast Surface Display
- Enhancement of Enzyme Functionality, and Display Efficiency—this area involves the following: nature of the enzyme being expressed in yeast that it may either retain or increase its activity upon heterologous expression (functionality), enhanced surface display efficiency for DYSD strategy, and adaptor assembly for JANNASEY strategy. The main goal of this area is to increase the hydrolysis efficiency of the resulting strain to optimally liberate the fermentable sugars from their lignocellulosic structure.
- Substrate Transport into the Yeast Cell—this area involves the efficient transport of the fermentable sugars into the cell. The goal is to provide efficient means transport for the different sugars (e.g., hexose, pentose, cellobiose) where the route is either specific to one type of sugar or a ubiquitous type for generic sugar transport.
- Metabolic Engineering for Efficient Ethanol Conversion—involves in the rewiring of the yeast itself to convert the sugars efficiently into ethanol by minimizing the generation of by-products.
- Optimization of Process Conditions for Enhanced Hydrolysis and Ethanol Production—optimization of the process conditions can either have an effect all the areas 1 to 3 or to one of them. This involves varying the process conditions (i.e., temperature, pH, media composition, induction) to achieve efficient enzymatic hydrolysis and cellular metabolism.
4.1. On the Enzyme Functionality and Display Efficiency
4.2. On Substrate Transport
4.3. On Metabolic Engineering for Improved Ethanol Prodution
4.4. On Process Parameters
4.4.1. Cultivation Conditions
4.4.2. Host Strain Type
4.4.3. Inhibitor Removal by Additional Process Treatment Steps
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
| Authors | Year | Refs. | Yeast Surface Display Strategy | Anchoring Protein | Subst. Abbrv | Substrate Conc. (g/L) | Ethanol Titer (g/L) | Ferm. Time. (h) | ESCY (%) i |
|---|---|---|---|---|---|---|---|---|---|
| PURE CELLULOSE SUBSTRATES | |||||||||
| a Fujita, et al. | 2002 | [58] | DYSD-CD | SAG1 (3′-half) | BBglucan | 45 | 16.5 | 48 | 73.97 |
| Fujita, et al. | 2004 | [59] | DYSD-CD | SAG1 (3′-half) | PASC | 10 | 3 | 40 | 60.52 |
| Kotaka, et al. | 2008 | [60] | DYSD-CD | SAG1 (3′-half) | BBglucan | 20 | 7.94 | 24 | 69.60 |
| Tsai, et al. | 2009 | [46] | JANNASEY-SMFAA | AGA1::AGA2 | PASC | 10 | 3.5 | 50 | 70.61 |
| Tsai, et al. | 2010 | [72] | JANNASEY-SMFAA | AGA1::AGA2 | PASC | 10 | 1.87 | 70 | 37.73 |
| Wen, et al. | 2010 | [73] | JANNASEY-SMFAA | AGA1::AGA2 | PASC | 10 | 1.8 | 70 | 36.31 |
| Apiwatanapiwat, et al. | 2011 | [67] | DYSD-CD | SAG1 (3′-half) | BBglucan | 10.2 | 5.3 | 36 | 97.70 |
| Apiwatanapiwat, et al. | 2011 | DYSD-CD | SAG1 (3′-half) | PASC | 8.44 | 1.04 | n.r. | 24.86 | |
| Yamada, et al. | 2011 | [66] | DYSD-CD | SAG1 (3′-half) | PASC | 20 | 3.1 | 72 | 31.27 |
| Yamada, et al. | 2011 | DYSD-CD | SAG1 (3′-half) | PASC | 20 | 7.6 | 72 | 76.66 | |
| Baek, et al. | 2012 | [45] | DYSD-SEDC | SAG1 (3′-half) | PASC | 10 | 2.12 | 85 | 42.77 |
| Fan, et al. | 2012 | [75] | JANNASEY-CAA | AGA1::AGA2 | PASC | 10 | 1.091 | 96 | 22.01 |
| Fan, et al. | 2012 | JANNASEY-CAA | AGA1::AGA2 | Avicel | 10 | 1.412 | 96 | 28.49 | |
| Tsai, et al. | 2013 | [76] | JANNASEY-CAA | AGA1::AGA2 | PASC | 10 | 1.9 | 72 | 38.33 |
| Yamada, et al. | 2013 | [68] | DYSD-CD | SAG1 (3′-half) | PASC | 20 | 4.3 | 72 | 43.38 |
| Kim, et al. | 2013 | [74] | JANNASEY-SMFAA | AGA1::AGA2 | PASC | 10 | 1.86 | 94 | 37.52 |
| Liu, et al. | 2015 | [61] | DYSD-CD | SED1 | PASC | 10 | 2.9 | 96 | 58.51 |
| Fan, et al. | 2016 | [77] | JANNASEY-CAA | AGA1::AGA2 | CMC | 10 | 3.26 | 60 | 65.77 |
| Fan, et al. | 2016 | JANNASEY-CAA | AGA1::AGA2 | PASC | 10 | 1.09 | 60 | 21.99 | |
| Liu, et al. | 2016 | [62] | DYSD-CD | SED1 | PASC | 20 | 6.7 | 96 | 67.59 |
| Liu, et al. | 2016 | DYSD-CD | SED1 | Avicel | 10 | 1.4 | 96 | 28.24 | |
| Liu, et al. | 2017 | [69] | DYSD-CD | SED1 | Avicel | 10 | 2.9 | 96 | 58.51 |
| PURE HEMICELLULOSE SUBSTRATES | |||||||||
| Fujita, et al. | 2002 | [96] | DYSD-CD | SAG1 (3′-half) | BiX | 10 | n.r. | n.r. | n.r. |
| Katahira, et al. | 2004 | [95] | DYSD-CD | SAG1 (3′-half) | BiX | 100 | 7.1 | 62 | 15.44 |
| Sun et al. | 2012 | [99] | JANNASEY-SMFAA | AGA1::AGA2 | BiX | 10 | 0.95 | 80 | 20.66 |
| Ishii, et al. | 2016 | [43] | DYSD-CD | Flo428p | BM | 100 | 9.579 | 312 | 22.59 |
| Tabañag, et al. | 2018 | [42] | DYSD-SEDC | SAG1 (3′-half) | BeX | 10 | 0.961 | 168 | 22.77 |
| Tabañag, et al. | 2018 | DYSD-SEDC | SAG1 (3′-half) | WAX | 10 | 0.778 | 168 | 27.66 | |
| PRE-TREATED LIGNOCELLULOSIC BIOMASS SUBSTRATES | |||||||||
| Murai, et al. | 1998 | [133] | DYSD-SEDC | SAG1 (3′-half) | RCSii | 625 | 23.25 | 168 | n.r. |
| Murai, et al. | 1998 | [134] | DYSD-SEDC | SAG1 (3′-half) | RCS+Term.ii | 626 | 53 | 169 | n.r. |
| Katahira, et al. | 2006 | [107] | DYSD-CD | SAG1 (3′-half) | WCH | 72 | 30.3 | 36 | 80.39 |
| Kotaka, et al. | 2008 | [135] | DYSD-CD | SAG1 (3′-half) | LCSii | 50 | 20 | 48 | n.r. |
| Apiwatanapiwat, et al. | 2011 | [67] | DYSD-CD | SAG1 (3′-half) | PT-CP | 50 | 10 | 48 | 79.40 |
| Sakamoto, et al. | 2012 | [105] | DYSD-CD | SAG1 (3′-half) | PT-RS | 800 | 8.2 | 72 | 80.39 |
| Matano, et al. | 2012 | [103] | DYSD-CD | SAG1 (3′-half) | PT-RS | 200 | 43.1 | 72 | 89.00 |
| Matano, et al. | 2013 | [104] | DYSD-CD | SAG1 (3′-half) | PT-RS | 200 | 42.2 | 360 | 86.30 |
| Yamada, et al. | 2011 | [66] | DYSD-CD | SAG1 (3′-half) | PT-RS | 100 | 7.5 | 72 | 33.00 |
| Inokuma, et al. | 2014 | [136] | DYSD-SEDC | SED1 | PT-RS-SF+CTec2 | 100 | 13.6 | 96 | 68.77 |
| Guirimand, et al. | 2016 | [106] | DYSD-CD | SED1 | PT-RSH | 500 | 5.8 | 96 | 79.48 |
| Guirimand, et al. | 2016 | DYSD-CD | SED1 | PT-RSH | 500 | 37.9 | 96 | 63.38 | |
| Ishii, et al. | 2016 | [43] | DYSD-CD | Flo428p | PT-INM | 100 | 9.632 | 216 | 18.89 |
| Liu, et al. | 2016 | [62] | DYSD-CD | SED1 | PT-RS-SF | 100 | 1.3 | 96 | 7.00 |
| Liu, et al. | 2016 | DYSD-CD | SED1 | PT-RS-SF+CTec2 | 100 | 18 | 96 | 80.00 | |
| Liu, et al. | 2017 | [69] | DYSD-CD | SED1 | PT-RS | 25 | 0.8 | 96 | n.r. |
| Other Studies | |||||||||
| Murai, et al. | 1997 | [137] | DYSD-CD | SAG1 (3′-half) | n.r. | n.r. | n.r. | n.r. | n.r. |
| Fukuda, et al. | 2007 | [111] | DYSD-SEDC | SAG1 (3′-half) | n.r. | n.r. | n.r. | n.r. | n.r. |
| Lilly, et al. | 2009 | [71] | JANNASEY-SMFAA | Cwp2 | n.r. | n.r. | n.r. | n.r. | n.r. |
| Kotaka, et al. | 2010 | [114] | DYSD-SEDC | SAG1 (3′-half) | n.r. | n.r. | n.r. | n.r. | n.r. |
| Yeasmin, et al. | 2011 | [110] | DYSD-SEDC | AGA1::AGA2 | n.r. | n.r. | n.r. | n.r. | n.r. |
| Gao, et al. | 2017 | [109] | DYSD-SEDC | AGA1::AGA2 | n.r. | n.r. | n.r. | n.r. | n.r. |
| Tang, et al. | 2017 | [113] | DYSD-CD | AGA1::AGA2 | n.r. | n.r. | n.r. | n.r. | n.r. |
References
- Stocker, T.F.; Qin, D.; Plattner, G.-K.; Tignor, M.M.B.; Allen, S.K.; Boschung, J.; Nauels, A.; Xia, Y.; Bex, V.; Midgley, P.M. Climate Change 2013: The Physical Science Basis: Working Group 1 Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Ronzon, T.; Lusser, M.; Klinkenberg, M.; Landa, L.; Lopez, J.S.; M’Barek, R.; Hadjamu, G.; Belward, A.; Camia, A.; Giuntoli, J.; et al. Bioeconomy Report 2016; European Commission: Brussels, Belgium, 2017. [Google Scholar]
- Langeveld, H.; Sanders, J.; Meeusen, M. The Biobased Economy: Biofuels, Materials and Chemicals in the Post-Oil Era; Earthscan LLC: London, UK, 2012. [Google Scholar]
- Commission, E. Innovating for Sustainable Growth: A Bioeconomy for Europe; European Commission: Brussels, Belgium, 2012. [Google Scholar]
- De Jong, E. Bio-Based Chemicals Value Added Products from Biorefineries; IEA Bioenergy—Task 42 Biorefinery: Wageningen, The Netherlands, 2012. [Google Scholar]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [PubMed]
- Kricka, W.; Fitzpatrick, J.; Bond, U. Metabolic engineering of yeasts by heterologous enzyme production for degradation of cellulose and hemicellulose from biomass: A perspective. Front. Microbiol. 2014, 5, 174. [Google Scholar] [CrossRef] [PubMed]
- Lynd, L.R.; van Zyl, W.H.; McBride, J.E.; Laser, M. Consolidated bioprocessing of cellulosic biomass: An update. Curr. Opin. Biotechnol. 2005, 16, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.G.; McBride, J.E.; Shaw, A.J.; Lynd, L.R. Recent progress in consolidated bioprocessing. Curr. Opin. Biotechnol. 2012, 23, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Van Zyl, W.; Lynd, L.; den Haan, R.; McBride, J. Consolidated bioprocessing for bioethanol production using Saccharomyces cerevisiae. In Biofuels; Olsson, L., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 108, pp. 205–235. [Google Scholar]
- Chiswell, D.J.; McCaffery, J. Phage antibodies: Will new ‘coliclonal’ antibodies replace monoclonal antibodies? Trends Biotechnol. 1992, 10, 80–84. [Google Scholar] [CrossRef]
- Scott, J.K.; Smith, G.P. Searching for peptide ligands with an epitope library. Science 1990, 249, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G.; Stathopoulos, C.; Daugherty, P.S.; Nayak, A.R.; Iverson, B.L.; Iii, R.C. Display of heterologous proteins on the surface of microorganisms: From the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 1997, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Little, M.; Fuchs, P.; Breitling, F.; Dübel, S. Bacterial surface presentation of proteins and peptides: An alternative to phage technology? Trends Biotechnol. 1993, 11, 3–5. [Google Scholar] [CrossRef]
- Georgiou, G.; Poetschke, H.L.; Stathopoulos, C.; Francisco, J.A. Practical applications of engineering gram-negative bacterial cell surfaces. Trends Biotechnol. 1993, 11, 6–10. [Google Scholar] [CrossRef]
- Michon, C.; Langella, P.; Eijsink, V.G.H.; Mathiesen, G.; Chatel, J.M. Display of recombinant proteins at the surface of lactic acid bacteria: Strategies and applications. Microb. Cell Fact. 2016, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Schüürmann, J.; Quehl, P.; Festel, G.; Jose, J. Bacterial whole-cell biocatalysts by surface display of enzymes: Toward industrial application. Appl. Microbiol. Biotechnol. 2014, 98, 8031–8046. [Google Scholar] [CrossRef] [PubMed]
- Desvaux, M.; Dumas, E.; Chafsey, I.; Hébraud, M. Protein cell surface display in gram-positive bacteria: From single protein to macromolecular protein structure. FEMS Microbiol. Lett. 2006, 256, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Choi, J.H.; Xu, Z. Microbial cell-surface display. Trends Biotechnol. 2003, 21, 45–52. [Google Scholar] [CrossRef]
- Ståhl, S.; Uhlén, M. Bacterial surface display: Trends and progress. Trends Biotechnol. 1997, 15, 185–192. [Google Scholar] [CrossRef]
- Van Bloois, E.; Winter, R.T.; Kolmar, H.; Fraaije, M.W. Decorating microbes: Surface display of proteins on escherichia coli. Trends Biotechnol. 2011, 29, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Wittrup, K.D. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 2001, 12, 395–399. [Google Scholar] [CrossRef]
- Murai, T.; Ueda, M.; Shibasaki, Y.; Kamasawa, N.; Osumi, M.; Imanaka, T.; Tanaka, A. Development of an arming yeast strain for efficient utilization of starch by co-display of sequential amylolytic enzymes on the cell surface. Appl. Microbiol. Biotechnol. 1999, 51, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Tanaka, A. Cell surface engineering of yeast: Construction of arming yeast with biocatalyst. J. Biosci. Bioeng. 2000, 90, 125–136. [Google Scholar] [CrossRef]
- Ueda, M.; Tanaka, A. Genetic immobilization of proteins on the yeast cell surface. Biotechnol. Adv. 2000, 18, 121–140. [Google Scholar] [CrossRef]
- Kondo, A.; Ueda, M. Yeast cell-surface display—Applications of molecular display. Appl. Microbiol. Biotechnol. 2004, 64, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Yamada, R.; Ogino, C.; Kondo, A. Recent developments in yeast cell surface display toward extended applications in biotechnology. Appl. Microbiol. Biotechnol. 2012, 95, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Kondo, A. Development of yeast cell factories for consolidated bioprocessing of lignocellulose to bioethanol through cell surface engineering. Biotechnol. Adv. 2012, 30, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Ueda, M. Arming technology in yeast—Novel strategy for whole-cell biocatalyst and protein engineering. Biomolecules 2013, 3, 632–650. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kondo, A. Cell-surface display of enzymes by the yeast Saccharomyces cerevisiae for synthetic biology. FEMS Yeast Res. 2015, 15, 1–9. [Google Scholar] [PubMed]
- Ueda, M. Establishment of cell surface engineering and its development. Biosci. Biotechnol. Biochem. 2016, 80, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces genome database: The genomics resource of budding yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mathias, A.; Stavrou, S.; Neville, J.D.M. A new yeast display vector permitting free SCFV amino termini can augment ligand binding affinities. Protein Eng. Des. Sel. 2005, 18, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Matsumoto, T.; Ueda, M.; Tanaka, A.; Fukuda, H.; Kondo, A. Long anchor using Flo1 protein enhances reactivity of cell surface-displayed glucoamylase to polymer substrates. Appl. Microbiol. Biotechnol. 2002, 60, 469–474. [Google Scholar] [PubMed]
- Matsumoto, T.; Fukuda, H.; Ueda, M.; Tanaka, A.; Kondo, A. Construction of yeast strains with high cell surface lipase activity by using novel display systems based on the Flo1p flocculation functional domain. Appl. Environ. Microbiol. 2002, 68, 4517–4522. [Google Scholar] [CrossRef] [PubMed]
- Lamed, R.; Setter, E.; Bayer, E.A. Characterization of a cellulose-binding, cellulase-containing complex in clostridium thermocellum. J. Bacteriol. 1983, 156, 828–836. [Google Scholar] [PubMed]
- Doi, R.H.; Kosugi, A. Cellulosomes: Plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2004, 2, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Artzi, L.; Bayer, E.A.; Morais, S. Cellulosomes: Bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 2017, 15, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Mitsuzawa, S.; Kagawa, H.; Li, Y.; Chan, S.L.; Paavola, C.D.; Trent, J.D. The rosettazyme: A synthetic cellulosome. J. Biotechnol. 2009, 143, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Nidetzky, B.; Steiner, W.; Hayn, M.; Claeyssens, M. Cellulose hydrolysis by the cellulases from trichoderma reesei: A new model for synergistic interaction. Biochem. J. 1994, 298, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Wyman, C.E. Effects of cellulase and xylanase enzymes on the deconstruction of solids from pretreatment of poplar by leading technologies. Biotechnol. Prog. 2009, 25, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Tabañag, I.D.; Tsai, S.-L. Hemicellulose degradation and utilization by a synthetic Saccharomyces cerevisiae consortium. bioRxiv 2018. [Google Scholar] [CrossRef]
- Ishii, J.; Okazaki, F.; Djohan, A.C.; Hara, K.Y.; Asai-Nakashima, N.; Teramura, H.; Andriani, A.; Tominaga, M.; Wakai, S.; Kahar, P.; et al. From mannan to bioethanol: Cell surface co-display of β-mannanase and β-mannosidase on yeast Saccharomyces cerevisiae. Biotechnol. Biofuels 2016, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Srikrishnan, S.; Chen, W.; Da Silva, N.A. Functional assembly and characterization of a modular xylanosome for hemicellulose hydrolysis in yeast. Biotechnol. Bioeng. 2013, 110, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.-H.; Kim, S.; Lee, K.; Lee, J.-K.; Hahn, J.-S. Cellulosic ethanol production by combination of cellulase-displaying yeast cells. Enzyme Microb. Technol. 2012, 51, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.-L.; Oh, J.; Singh, S.; Chen, R.; Chen, W. Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 2009, 75, 6087–6093. [Google Scholar] [CrossRef] [PubMed]
- Kondo, A.; Shigechi, H.; Abe, M.; Uyama, K.; Matsumoto, T.; Takahashi, S.; Ueda, M.; Tanaka, A.; Kishimoto, M.; Fukuda, H. High-level ethanol production from starch by a flocculent Saccharomyces cerevisiae strain displaying cell-surface glucoamylase. Appl. Microbiol. Biotechnol. 2002, 58, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Yan, R.; Liu, Y.; Jiang, W. In-depth investigation of enzymatic hydrolysis of biomass wastes based on three major components: Cellulose, hemicellulose and lignin. Bioresour. Technol. 2010, 101, 8217–8223. [Google Scholar] [CrossRef] [PubMed]
- Carere, C.R.; Sparling, R.; Cicek, N.; Levin, D.B. Third generation biofuels via direct cellulose fermentation. Int. J. Mol. Sci. 2008, 9, 1342–1360. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.; Muñoz-Dorado, J.; de la Rubia, T.; Martínez, J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: An overview. Int. Microbiol. 2002, 5, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Z.; Sathitsuksanoh, N.; Zhang, Y.H.P. Glycoside hydrolase family 9 processive endoglucanase from clostridium phytofermentans: Heterologous expression, characterization, and synergy with family 48 cellobiohydrolase. Bioresour. Technol. 2010, 101, 5534–5538. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Jun, H.-S.; Forsberg, C.W. Characterization and synergistic interactions of fibrobacter succinogenes glycoside hydrolases. Appl. Environ. Microbiol. 2007, 73, 6098–6105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-H.P.; Lynd, L.R. Toward an aggregated understanding of enzymatic hydrolysis of cellulose: Noncomplexed cellulase systems. Biotechnol. Bioeng. 2004, 88, 797–824. [Google Scholar] [CrossRef] [PubMed]
- Boisset, C.; Pétrequin, C.; Chanzy, H.; Henrissat, B.; Schülein, M. Optimized mixtures of recombinant humicola insolens cellulases for the biodegradation of crystalline cellulose. Biotechnol. Bioeng. 2001, 72, 339–345. [Google Scholar] [CrossRef]
- Zhou, S.; Ingram, L.O. Synergistic hydrolysis of carboxymethyl cellulose and acid-swollen cellulose by two endoglucanases (celz and cely) fromerwinia chrysanthemi. J. Bacteriol. 2000, 182, 5676–5682. [Google Scholar] [CrossRef] [PubMed]
- Boisset, C.; Fraschini, C.; Schülein, M.; Henrissat, B.; Chanzy, H. Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 2000, 66, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Väljamäe, P.; Sild, V.; Nutt, A.; Pettersson, G.; Johansson, G. Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur. J. Biochem. 1999, 266, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Takahashi, S.; Ueda, M.; Tanaka, A.; Okada, H.; Morikawa, Y.; Kawaguchi, T.; Arai, M.; Fukuda, H.; Kondo, A. Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes. Appl. Environ. Microbiol. 2002, 68, 5136–5141. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Ito, J.; Ueda, M.; Fukuda, H.; Kondo, A. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl. Environ. Microbiol. 2004, 70, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Kotaka, A.; Bando, H.; Kaya, M.; Kato-Murai, M.; Kuroda, K.; Sahara, H.; Hata, Y.; Kondo, A.; Ueda, M. Direct ethanol production from barley β-glucan by sake yeast displaying aspergillus oryzae β-glucosidase and endoglucanase. J. Biosci. Bioeng. 2008, 105, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Inokuma, K.; Ho, S.-H.; Haan, R.d.; Hasunuma, T.; Zyl, W.H.; Kondo, A. Combined cell-surface display- and secretion-based strategies for production of cellulosic ethanol with Saccharomyces cerevisiae. Biotechnol. Biofuels 2015, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ho, S.-H.; Sasaki, K.; den Haan, R.; Inokuma, K.; Ogino, C.; van Zyl, W.H.; Hasunuma, T.; Kondo, A. Engineering of a novel cellulose-adherent cellulolytic Saccharomyces cerevisiae for cellulosic biofuel production. Sci. Rep. 2016, 6, 24550. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ho, S.-H.; Hasunuma, T.; Chang, J.-S.; Ren, N.-Q.; Kondo, A. Recent advances in yeast cell-surface display technologies for waste biorefineries. Bioresour. Technol. 2016, 215, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Yamada, R.; Taniguchi, N.; Tanaka, T.; Ogino, C.; Fukuda, H.; Kondo, A. Cocktail delta-integration: A novel method to construct cellulolytic enzyme expression ratio-optimized yeast strains. Microb. Cell Fact. 2010, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Yamada, R.; Tanaka, T.; Ogino, C.; Fukuda, H.; Kondo, A. Novel strategy for yeast construction using δ-integration and cell fusion to efficiently produce ethanol from raw starch. Appl. Microbiol. Biotechnol. 2010, 85, 1491–1498. [Google Scholar] [CrossRef] [PubMed]
- Yamada, R.; Taniguchi, N.; Tanaka, T.; Ogino, C.; Fukuda, H.; Kondo, A. Direct ethanol production from cellulosic materials using a diploid strain of Saccharomyces cerevisiae with optimized cellulase expression. Biotechnol. Biofuels 2011, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Apiwatanapiwat, W.; Murata, Y.; Kosugi, A.; Yamada, R.; Kondo, A.; Arai, T.; Rugthaworn, P.; Mori, Y. Direct ethanol production from cassava pulp using a surface-engineered yeast strain co-displaying two amylases, two cellulases, and β-glucosidase. Appl. Microbiol. Biotechnol. 2011, 90, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Yamada, R.; Nakatani, Y.; Ogino, C.; Kondo, A. Efficient direct ethanol production from cellulose by cellulase-and cellodextrin transporter-co-expressing Saccharomyces cerevisiae. AMB Express 2013, 3, 34. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Inokuma, K.; Ho, S.-H.; den Haan, R.; van Zyl, W.H.; Hasunuma, T.; Kondo, A. Improvement of ethanol production from crystalline cellulose via optimizing cellulase ratios in cellulolytic Saccharomyces cerevisiae. Biotechnol. Bioeng. 2017, 114, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Kosugi, A.; Tanaka, T.; Kuroda, K.; Shibasaki, S.; Ogino, C.; Ueda, M.; Fukuda, H.; Doi, R.H.; Kondo, A. Regulation of the display ratio of enzymes on the Saccharomyces cerevisiae cell surface by the immunoglobulin g and cellulosomal enzyme binding domains. Appl. Environ. Microbiol. 2009, 75, 4149–4154. [Google Scholar] [CrossRef] [PubMed]
- Lilly, M.; Fierobe, H.-P.; Van Zyl, W.H.; Volschenk, H. Heterologous expression of a clostridium minicellulosome in Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 9, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.-L.; Goyal, G.; Chen, W. Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 2010, 76, 7514–7520. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Sun, J.; Zhao, H. Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl. Environ. Microbiol. 2010, 76, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Baek, S.-H.; Lee, K.; Hahn, J.-S. Cellulosic ethanol production using a yeast consortium displaying a minicellulosome and β-glucosidase. Microb. Cell Fact. 2013, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.H.; Zhang, Z.J.; Yu, X.Y.; Xue, Y.X.; Tan, T.W. Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc. Natl. Acad. Sci. USA 2012, 109, 13260–13265. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.L.; DaSilva, N.A.; Chen, W. Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth. Biol. 2013, 2, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.-H.; Zhang, Z.-J.; Mei, S.; Lu, Y.-Y.; Li, M.; Wang, Z.-Y.; Yang, J.-G.; Yang, S.-T.; Tan, T.-W. Engineering yeast with bifunctional minicellulosome and cellodextrin pathway for co-utilization of cellulose-mixed sugars. Biotechnol. Biofuels 2016, 9, 137. [Google Scholar] [CrossRef] [PubMed]
- Belgacem, M.N.; Gandini, A. Monomers, Polymers and Composites from Renewable Resources; Elsevier Science: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Akpinar, O.; Erdogan, K.; Bostanci, S. Production of xylooligosaccharides by controlled acid hydrolysis of lignocellulosic materials. Carbohydr. Res. 2009, 344, 660–666. [Google Scholar] [CrossRef] [PubMed]
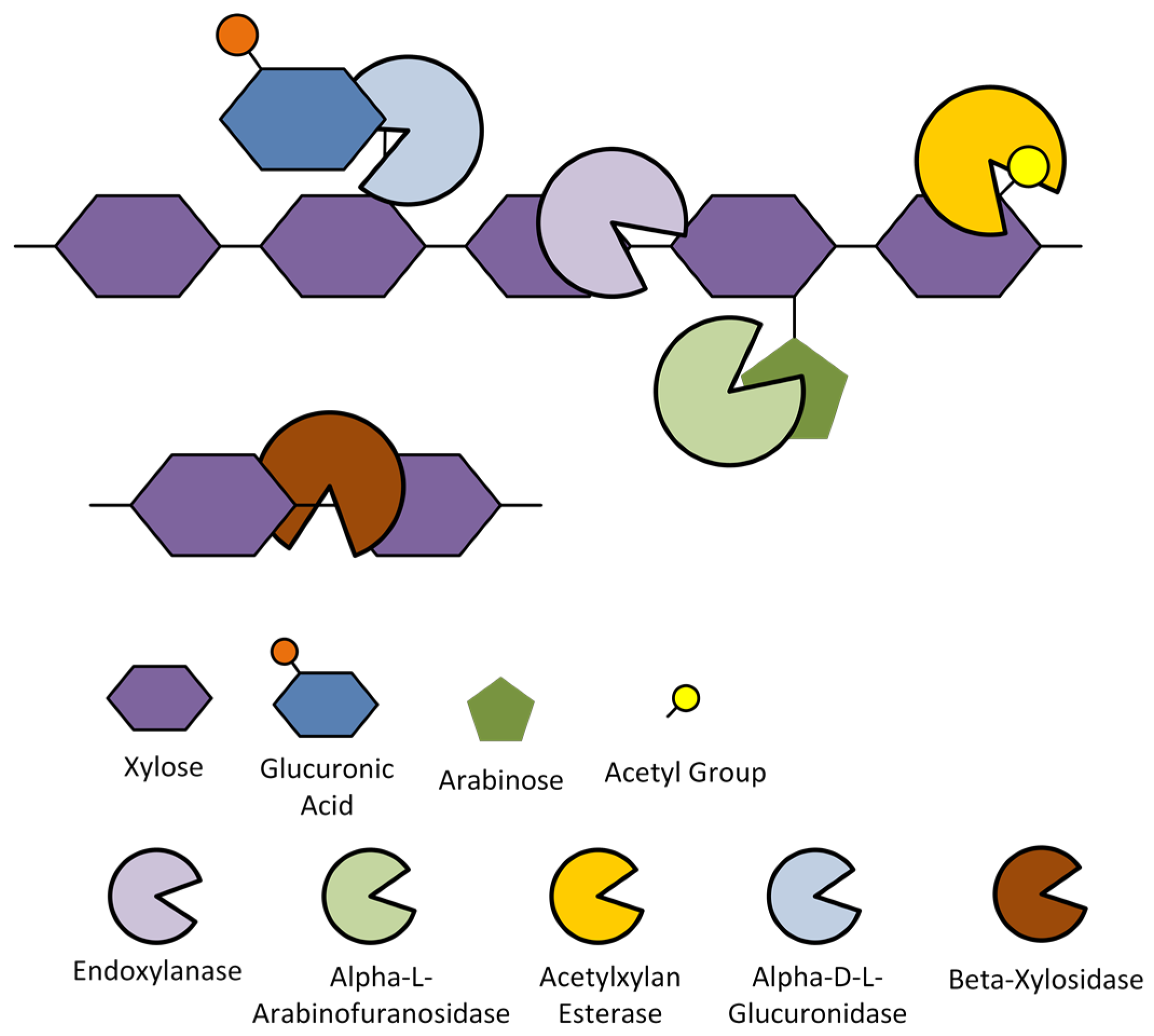
- Moreira, L.R.S.; Filho, E.X.F. Insights into the mechanism of enzymatic hydrolysis of xylan. Appl. Microbiol. Biotechnol. 2016, 100, 5205–5214. [Google Scholar] [CrossRef] [PubMed]
- Van Dyk, J.S.; Pletschke, B.I. Enzyme synergy for enhanced degradation of lignocellulosic waste. In Advances in Enzyme Biotechnology; Shukla, P., Pletschke, I.B., Eds.; Springer: New Delhi, India, 2013; pp. 57–65. [Google Scholar]
- Vardakou, M.; Katapodis, P.; Topakas, E.; Kekos, D.; Macris, B.J.; Christakopoulos, P. Synergy between enzymes involved in the degradation of insoluble wheat flour arabinoxylan. Innov. Food Sci. Emerg. Technol. 2004, 5, 107–112. [Google Scholar] [CrossRef]
- Sørensen, H.R.; Meyer, A.S.; Pedersen, S. Enzymatic hydrolysis of water-soluble wheat arabinoxylan. 1. Synergy between α-l-arabinofuranosidases, endo-1,4-β-xylanases, and β-xylosidase activities. Biotechnol. Bioeng. 2003, 81, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, H.R.; Pedersen, S.; Viksø-Nielsen, A.; Meyer, A.S. Efficiencies of designed enzyme combinations in releasing arabinose and xylose from wheat arabinoxylan in an industrial ethanol fermentation residue. Enzyme Microb. Technol. 2005, 36, 773–784. [Google Scholar] [CrossRef]
- Sørensen, H.R.; Pedersen, S.; Meyer, A.S. Synergistic enzyme mechanisms and effects of sequential enzyme additions on degradation of water insoluble wheat arabinoxylan. Enzyme Microb. Technol. 2007, 40, 908–918. [Google Scholar] [CrossRef]
- Kovacs, K. Production of Cellulolytic Enzymes with Trichoderma Atroviride Mutants for the Biomass-to-Bioethanol Process; Lund University: Lund, Sweden, 2009. [Google Scholar]
- Adelsberger, H.; Hertel, C.; Glawischnig, E.; Zverlov, V.V.; Schwarz, W.H. Enzyme system of clostridium stercorarium for hydrolysis of arabinoxylan: Reconstitution of the in vivo system from recombinant enzymes. Microbiology 2004, 150, 2257–2266. [Google Scholar] [CrossRef] [PubMed]
- Selig, M.J.; Adney, W.S.; Himmel, M.E.; Decker, S.R. The impact of cell wall acetylation on corn stover hydrolysis by cellulolytic and xylanolytic enzymes. Cellulose 2009, 16, 711–722. [Google Scholar] [CrossRef]
- De Vries, R.P.; Kester, H.C.M.; Poulsen, C.H.; Benen, J.A.E.; Visser, J. Synergy between enzymes from aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr. Res. 2000, 327, 401–410. [Google Scholar] [CrossRef]
- Ho, N.W.; Chen, Z.; Brainard, A.P. Genetically engineered saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 1998, 64, 1852–1859. [Google Scholar] [PubMed]
- Kuyper, M.; Winkler, A.A.; van Dijken, J.P.; Pronk, J.T. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: A proof of principle. FEMS Yeast Res. 2004, 4, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Kuyper, M.; Harhangi, H.; Stave, A.; Winkler, A.; Jetten, M.; De-Laat, W.; Den-Ridder, J.; Op den Camp, H.; van Dijken, J.; Pronk, J. High-level functional expression of a fungal xylose isomerase: The key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 2003, 4, 69–78. [Google Scholar] [CrossRef]
- Harhangi, H.; Akhmanova, A.; Emmens, R.; van der-Drift, C.; De-Laat, W.; Van-Dijken, J.; Jetten, M.; Pronk, J.; Op den Camp, H. Xylose metabolism in the anaerobic fungus Piromyces sp. Strain E2 follows the bacterial pathway. Arch. Microbiol. 2003, 180, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Karhumaa, K.; Garcia Sanchez, R.; Hahn-Hagerdal, B.; Gorwa-Grauslund, M. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 2007, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Katahira, S.; Fujita, Y.; Mizuike, A.; Fukuda, H.; Kondo, A. Construction of a xylan-fermenting yeast strain through codisplay of xylanolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 2004, 70, 5407–5414. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Katahira, S.; Ueda, M.; Tanaka, A.; Okada, H.; Morikawa, Y.; Fukuda, H.; Kondo, A. Construction of whole-cell biocatalyst for xylan degradation through cell-surface xylanase display in Saccharomyces cerevisiae. J. Mol. Catal. B Enzym. 2002, 17, 189–195. [Google Scholar] [CrossRef]
- Malgas, S.; van Dyk, J.S.; Pletschke, B.I. A review of the enzymatic hydrolysis of mannans and synergistic interactions between β-mannanase, β-mannosidase and α-galactosidase. World J. Microbiol. Biotechnol. 2015, 31, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Spivey, H.O.; Ovadi, J. Substrate channeling. Methods 1999, 19, 306–321. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wen, F.; Si, T.; Xu, J.-H.; Zhao, H. Direct conversion of xylan to ethanol by recombinant Saccharomyces cerevisiae strains displaying an engineered minihemicellulosome. Appl. Environ. Microbiol. 2012, 78, 3837–3845. [Google Scholar] [CrossRef] [PubMed]
- Wertz, J.L.; Bédué, O. Lignocellulosic Biorefineries; EFPL Press: Lausanne, Switzerland, 2013. [Google Scholar]
- Harmsen, P.; Huijgen, W.; Bermudez, L.; Bakker, R. Literature Review of Physical and Chemical Pretreatment Processes for Lignocellulosic Biomass; 9085857570; Wageningen UR Food & Biobased Research: Wageningen, The Netherlands, 2010. [Google Scholar]
- Mosier, N.; Wyman, C.; Dale, B.; Elander, R.; Lee, Y.Y.; Holtzapple, M.; Ladisch, M. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour. Technol. 2005, 96, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Matano, Y.; Hasunuma, T.; Kondo, A. Display of cellulases on the cell surface of Saccharomyces cerevisiae for high yield ethanol production from high-solid lignocellulosic biomass. Bioresour. Technol. 2012, 108, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Matano, Y.; Hasunuma, T.; Kondo, A. Cell recycle batch fermentation of high-solid lignocellulose using a recombinant cellulase-displaying yeast strain for high yield ethanol production in consolidated bioprocessing. Bioresour. Technol. 2013, 135, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Hasunuma, T.; Hori, Y.; Yamada, R.; Kondo, A. Direct ethanol production from hemicellulosic materials of rice straw by use of an engineered yeast strain codisplaying three types of hemicellulolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. J. Biotechnol. 2012, 158, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Guirimand, G.; Sasaki, K.; Inokuma, K.; Bamba, T.; Hasunuma, T.; Kondo, A. Cell surface engineering of Saccharomyces cerevisiae combined with membrane separation technology for xylitol production from rice straw hydrolysate. Appl. Microbiol. Biotechnol. 2016, 100, 3477–3487. [Google Scholar] [CrossRef] [PubMed]
- Katahira, S.; Mizuike, A.; Fukuda, H.; Kondo, A. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl. Microbiol. Biotechnol. 2006, 72, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Haan, R.; Rensburg, E.; Rose, S.H.; Görgens, J.F.; Zyl, W.H. Progress and challenges in the engineering of non-cellulolytic microorganisms for consolidated bioprocessing. Curr. Opin. Biotechnol. 2015, 33, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Mao, R.-Q.; Xiao, Y.; Zhou, J.; Liu, Y.-H.; Li, G. Efficient yeast cell-surface display of an endoglucanase of aspergillus flavus and functional characterization of the whole-cell enzyme. World J. Microbiol. Biotechnol. 2017, 33, 114. [Google Scholar] [CrossRef] [PubMed]
- Yeasmin, S.; Kim, C.; Park, H.; Sheikh, M.; Lee, J.; Kim, J.; Back, K.; Kim, S. Cell surface display of cellulase activity–free xylanase enzyme on Saccharomyces cerevisiae EBY100. Appl. Biochem. Biotechnol. 2011, 164, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Kato-Murai, M.; Kadonosono, T.; Sahara, H.; Hata, Y.; Suye, S.; Ueda, M. Enhancement of substrate recognition ability by combinatorial mutation of β-glucosidase displayed on the yeast cell surface. Appl. Microbiol. Biotechnol. 2007, 76, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Haan, R.; Kroukamp, H.; Zyl, J.H.-D.; Zyl, W.H. Cellobiohydrolase secretion by yeast: Current state and prospects for improvement. Process Biochem. 2013, 48, 1–12. [Google Scholar] [CrossRef]
- Tang, H.; Song, M.; He, Y.; Wang, J.; Wang, S.; Shen, Y.; Hou, J.; Bao, X. Engineering vesicle trafficking improves the extracellular activity and surface display efficiency of cellulases in Saccharomyces cerevisiae. Biotechnol. Biofuels 2017, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Kotaka, A.; Sahara, H.; Kuroda, K.; Kondo, A.; Ueda, M.; Hata, Y. Enhancement of β-glucosidase activity on the cell-surface of sake yeast by disruption of SED1. J. Biosci. Bioeng. 2010, 109, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, H.; Hashimoto, K.; Saijo, A.; Takada, Y.; Kondo, A.; Ueda, M.; Ooshima, H.; Tachibana, T.; Azuma, M. Cell wall structure suitable for surface display of proteins in Saccharomyces cerevisiae. Yeast 2014, 31, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Wentz, A.E.; Shusta, E.V. A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl. Environ. Microbiol. 2007, 73, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Shuler, M.L.; Kargi, F. Bioprocess Engineering: Basic Concepts; Prentice Hall: Upper Saddle River, NJ, USA, 2002. [Google Scholar]
- Özcan, S.; Johnston, M. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 1999, 63, 554–569. [Google Scholar] [PubMed]
- André, B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 1995, 11, 1575–1611. [Google Scholar] [CrossRef] [PubMed]
- Bisson, L.F.; Coons, D.M.; Kruckeberg, A.L.; Lewis, D.A. Yeast sugar transporters. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 259–308. [Google Scholar] [CrossRef] [PubMed]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Freidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999, 464, 123–128. [Google Scholar] [CrossRef]
- Solis-Escalante, D.; van den Broek, M.; Kuijpers, N.G.A.; Pronk, J.T.; Boles, E.; Daran, J.-M.; Daran-Lapujade, P. The genome sequence of the popular hexose-transport-deficient Saccharomyces cerevisiae strain EBY.Vw4000 reveals LoxP/Cre-induced translocations and gene loss. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.Y.; Kobayashi, J.; Yamada, R.; Sasaki, D.; Kuriya, Y.; Hirono-Hara, Y.; Ishii, J.; Araki, M.; Kondo, A. Transporter engineering in biomass utilization by yeast. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Kuijper, S.M. Engineering of Saccharomyces Cerevisiae for the Production of Fuel Ethanol from Xylose; Delft University of Technology: Delft, The Netherlands, 2006. [Google Scholar]
- Kuyper, M.; Hartog, M.; Toirkens, M.; Almering, M.; Winkler, A.; Van-Dijken, J.; Pronk, J. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005, 5, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-M.; Jellison, T.; Alper, H. Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol. Biofuels 2014, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Skerker, J.M.; Kang, W.; Lesmana, A.; Wei, N.; Arkin, A.P.; Jin, Y.-S. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e57048. [Google Scholar] [CrossRef] [PubMed]
- Demeke, M.; Dietz, H.; Li, Y.; Foulquie-Moreno, M.; Mutturi, S.; Deprez, S.; Den Abt, T.; Bonini, B.; Liden, G.; Dumortier, F.; et al. Development of a d-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol. Biofuels 2013, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Cheng, J.; Wang, B.; Fink, G.; Stephanopoulos, G. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab. Eng. 2012, 14, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-M.; Jellison, T.; Alper, H.S. Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 5708–5716. [Google Scholar] [CrossRef] [PubMed]
- Khatun, M.M.; Liu, C.-G.; Zhao, X.-Q.; Yuan, W.-J.; Bai, F.-W. Consolidated ethanol production from jerusalem artichoke tubers at elevated temperature by Saccharomyces cerevisiae engineered with inulinase expression through cell surface display. J. Ind. Microbiol. Biotechnol. 2017, 44, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Bae, J.G.; Fukai, K.; Tokumoto, N.; Kuroda, K.; Ogawa, J.; Nakatani, M.; Shimizu, S.; Ueda, M. Effect of pretreatment of hydrothermally processed rice straw with laccase-displaying yeast on ethanol fermentation. Appl. Microbiol. Biotechnol. 2012, 94, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Murai, T.; Ueda, M.; Kawaguchi, T.; Arai, M.; Tanaka, A. Assimilation of cellooligosaccharides by a cell surface-engineered yeast expressing β-glucosidase and carboxymethylcellulase from aspergillus aculeatus. Appl. Environ. Microbiol. 1998, 64, 4857–4861. [Google Scholar] [PubMed]
- Murai, T.; Yoshino, T.; Ueda, M.; Haranoya, I.; Ashikari, T.; Yoshizumi, H.; Tanaka, A. Evaluation of the function of arming yeast displaying glucoamylase on its cell surface by direct fermentation of corn to ethanol. J. Ferment. Bioeng. 1998, 86, 569–572. [Google Scholar] [CrossRef]
- Kotaka, A.; Sahara, H.; Hata, Y.; Abe, Y.; Kondo, A.; Kato-Murai, M.; Kuroda, K.; Ueda, M. Efficient and direct fermentation of starch to ethanol by sake yeast strains displaying fungal glucoamylases. Biosci. Biotechnol. Biochem. 2008, 72, 1376–1379. [Google Scholar] [CrossRef] [PubMed]
- Inokuma, K.; Hasunuma, T.; Kondo, A. Efficient yeast cell-surface display of exo- and endo-cellulase using the SED1 anchoring region and its original promoter. Biotechnol. Biofuels 2014, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Murai, T.; Ueda, M.; Yamamura, M.; Atomi, H.; Shibasaki, Y.; Kamasawa, N.; Osumi, M.; Amachi, T.; Tanaka, A. Construction of a starch-utilizing yeast by cell surface engineering. Appl. Environ. Microbiol. 1997, 63, 1362–1366. [Google Scholar] [PubMed]


| Lignocellulose Component | Composition (%) | Fermentable Sugars |
|---|---|---|
| Cellulose | ~35–50 | Glucose |
| Hemicellulose | ~20–35 | Xylose, Arabinose, Mannose, Galactose |
| Lignin | ~15–30 | - |
| Yeast Anchor Protein | Standard Name | Description |
|---|---|---|
| -agglutinin | SAG1 | Alpha-agglutinin of alpha-cells; binds to Aga1p during agglutination, N-terminal half is homologous to the immunoglobulin superfamily and contains binding site for a-agglutinin, C-terminal half is highly glycosylated and contains GPI anchor |
| SED | SED1 | Major stress-induced structural GPI-cell wall glycoprotein; associates with translating ribosomes, possible role in mitochondrial genome maintenance |
| Cwp2 | CWP2 | Covalently linked cell wall mannoprotein; major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
| Flo428p | FLO1 | Lectin-like protein involved in flocculation; cell wall protein that binds mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; important for co-flocculation with other yeasts, mediating interaction with specific species |
| a-agglutinin system | AGA1 | Anchorage subunit of a-agglutinin of a-cells; highly O-glycosylated protein with N-terminal secretion signal and C-terminal signal for addition of GPI anchor to cell wall, linked to adhesion subunit Aga2p via two disulfide bonds |
| AGA2 | Adhesion subunit of a-agglutinin of a-cells; C-terminal sequence acts as a ligand for alpha-agglutinin (Sag1p) during agglutination, modified with O-linked oligomannosyl chains, linked to anchorage subunit Aga1p via two disulfide bonds |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabañag, I.D.F.; Chu, I.-M.; Wei, Y.-H.; Tsai, S.-L. The Role of Yeast-Surface-Display Techniques in Creating Biocatalysts for Consolidated BioProcessing. Catalysts 2018, 8, 94. https://doi.org/10.3390/catal8030094
Tabañag IDF, Chu I-M, Wei Y-H, Tsai S-L. The Role of Yeast-Surface-Display Techniques in Creating Biocatalysts for Consolidated BioProcessing. Catalysts. 2018; 8(3):94. https://doi.org/10.3390/catal8030094
Chicago/Turabian StyleTabañag, Ian Dominic Flormata, I-Ming Chu, Yu-Hong Wei, and Shen-Long Tsai. 2018. "The Role of Yeast-Surface-Display Techniques in Creating Biocatalysts for Consolidated BioProcessing" Catalysts 8, no. 3: 94. https://doi.org/10.3390/catal8030094




