Recognition of a Single β-D-Xylopyranose Molecule by Xylanase GH11 from Thermoanaerobacterium saccharolyticum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Preparation
2.2. Crystallization
2.3. X-ray Diffraction Data
2.4. Structure Determination
2.5. Bioinformatics
3. Results and Discussion
3.1. Structure Determination
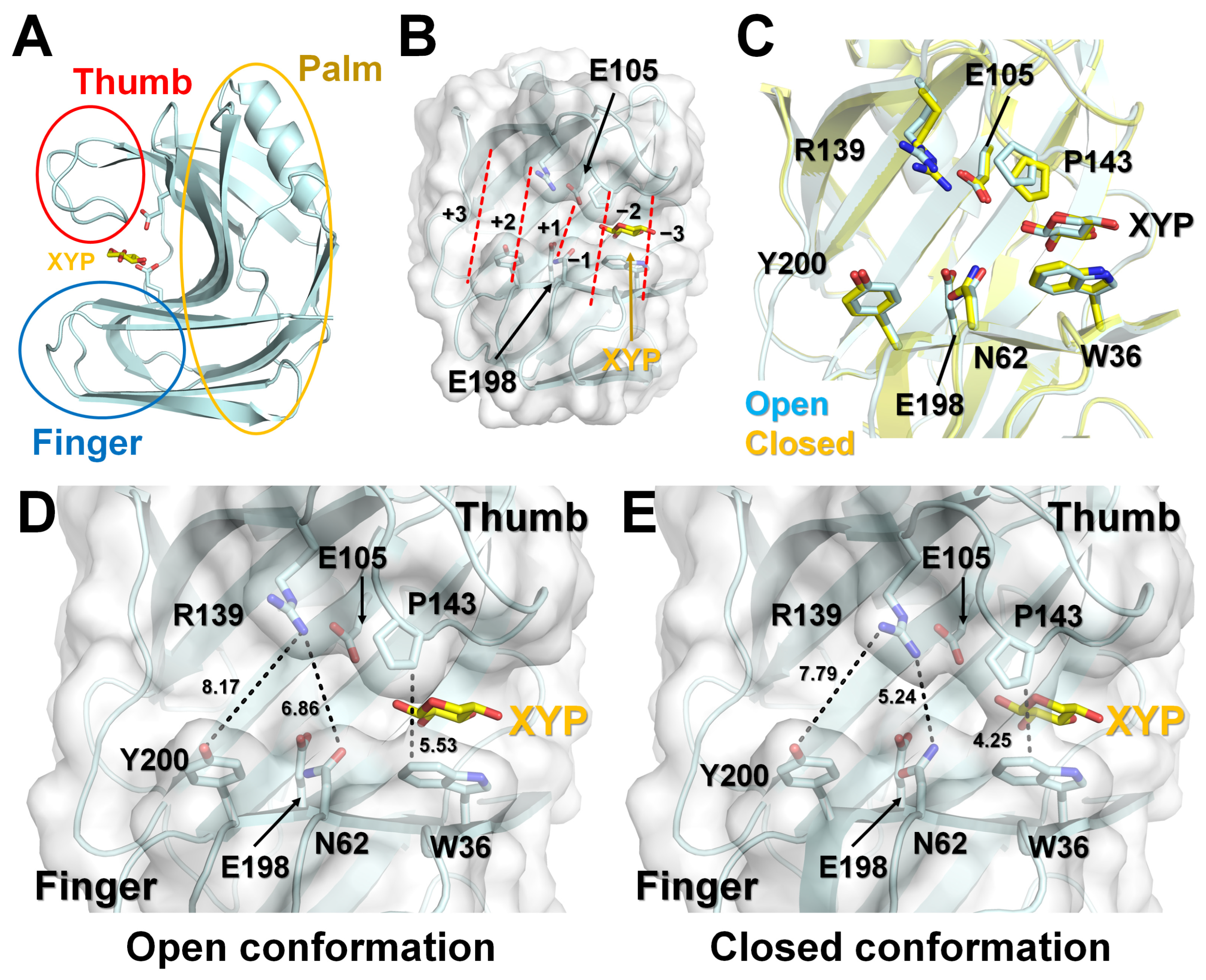
3.2. Analysis of XYP Binding in TsaGH11
3.3. Structure Comparison of Apo and XYP Binding in TsaGH11
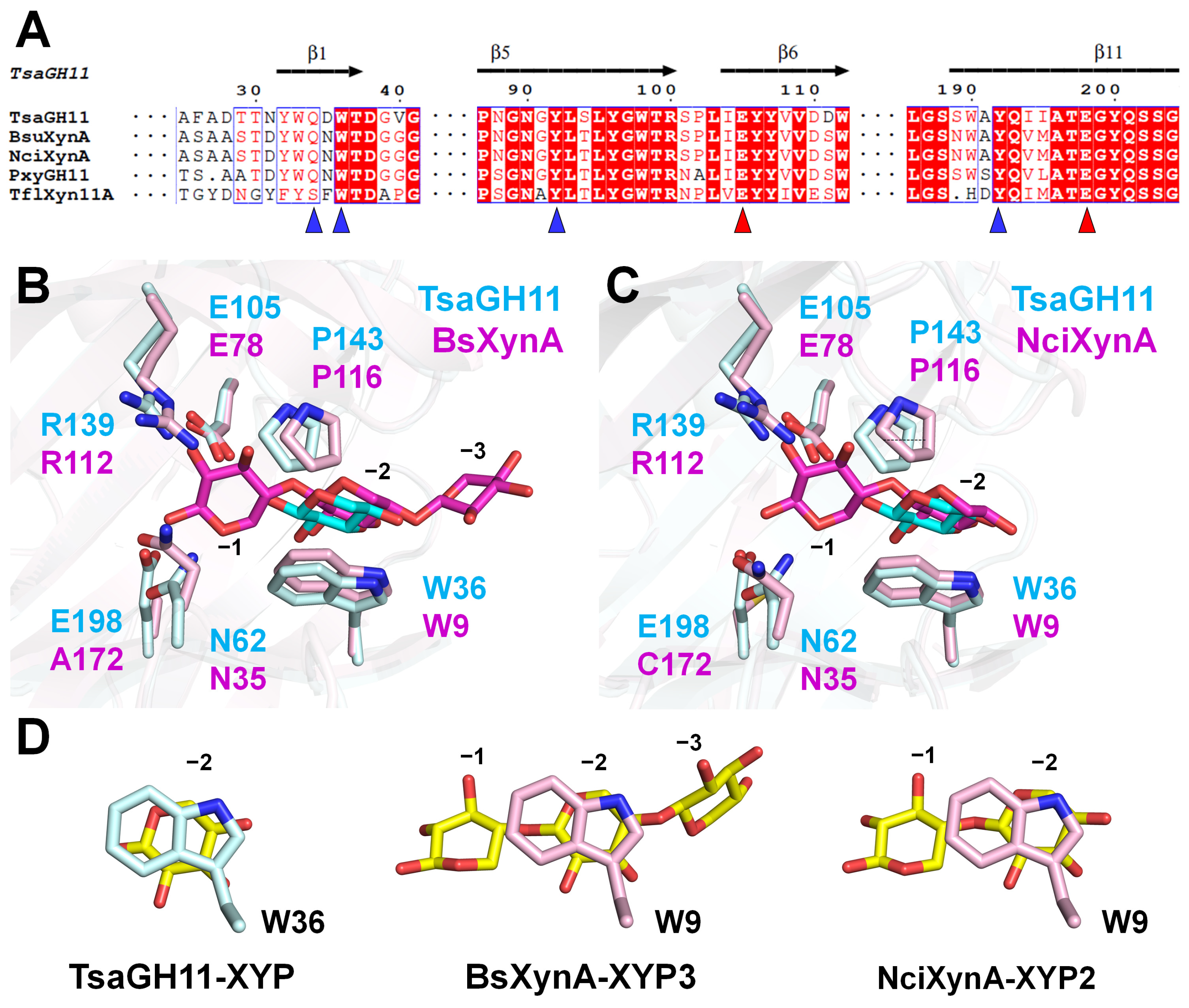
3.4. Structure Comparison of TsaGH11-XYP with Other GH11s
4. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Srivastava, N.; Rawat, R.; Singh Oberoi, H.; Ramteke, P.W. A Review on Fuel Ethanol Production from Lignocellulosic Biomass. Int. J. Green Energy 2014, 12, 949–960. [Google Scholar] [CrossRef]
- Srivastava, N.; Srivastava, M.; Mishra, P.K.; Gupta, V.K.; Molina, G.; Rodriguez-Couto, S.; Manikanta, A.; Ramteke, P.W. Applications of fungal cellulases in biofuel production: Advances and limitations. Renew. Sustain. Energy Rev. 2018, 82, 2379–2386. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef]
- Martínez-Abad, A.; Berglund, J.; Toriz, G.; Gatenholm, P.; Henriksson, G.; Lindström, M.; Wohlert, J.; Vilaplana, F. Regular Motifs in Xylan Modulate Molecular Flexibility and Interactions with Cellulose Surfaces. Plant Physiol. 2017, 175, 1579–1592. [Google Scholar] [CrossRef]
- Rennie, E.A.; Scheller, H.V. Xylan biosynthesis. Curr. Opin. Biotechnol. 2014, 26, 100–107. [Google Scholar] [CrossRef]
- Malgas, S.; Mafa, M.S.; Mkabayi, L.; Pletschke, B.I. A mini review of xylanolytic enzymes with regards to their synergistic interactions during hetero-xylan degradation. World J. Microbiol. Biotechnol. 2019, 35, 187. [Google Scholar] [CrossRef]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef]
- Marasinghe, S.D.; Jo, E.; Hettiarachchi, S.A.; Lee, Y.; Eom, T.-Y.; Gang, Y.; Kang, Y.-H.; Oh, C. Characterization of glycoside hydrolase family 11 xylanase from Streptomyces sp. strain J103; its synergetic effect with acetyl xylan esterase and enhancement of enzymatic hydrolysis of lignocellulosic biomass. Microb. Cell Fact. 2021, 20, 129. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Chakdar, H.; Kumar, M.; Pandiyan, K.; Singh, A.; Nanjappan, K.; Kashyap, P.L.; Srivastava, A.K. Bacterial xylanases: Biology to biotechnology. 3 Biotech 2016, 6, 150. [Google Scholar] [CrossRef] [PubMed]
- Paës, G.; Berrin, J.-G.; Beaugrand, J. GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol. Adv. 2012, 30, 564–592. [Google Scholar] [CrossRef]
- Mendis, M.; Simsek, S. Production of structurally diverse wheat arabinoxylan hydrolyzates using combinations of xylanase and arabinofuranosidase. Carbohydr. Polym. 2015, 132, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Uday, U.S.P.; Choudhury, P.; Bandyopadhyay, T.K.; Bhunia, B. Classification, mode of action and production strategy of xylanase and its application for biofuel production from water hyacinth. Int. J. Biol. Macromol. 2016, 82, 1041–1054. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, R.; Kuthiala, T.; Singh, G.; Rarotra, S.; Kaur, A.; Arya, S.K.; Kumar, P. Current status of xylanase for biofuel production: A review on classification and characterization. Biomass Convers. Biorefin. 2021, 13, 8773–8791. [Google Scholar] [CrossRef]
- Dodd, D.; Cann, I.K.O. Enzymatic deconstruction of xylan for biofuel production. GCB Bioenergy 2009, 1, 2–17. [Google Scholar] [CrossRef]
- Kim, I.J.; Kim, S.R.; Bornscheuer, U.T.; Nam, K.H. Engineering of GH11 Xylanases for Optimal pH Shifting for Industrial Applications. Catalysts 2023, 13, 1405. [Google Scholar] [CrossRef]
- Ravn, J.L.; Thøgersen, J.C.; Eklöf, J.; Pettersson, D.; Ducatelle, R.; van Immerseel, F.; Pedersen, N.R. GH11 xylanase increases prebiotic oligosaccharides from wheat bran favouring butyrate-producing bacteria in vitro. Anim. Feed Sci. Technol. 2017, 226, 113–123. [Google Scholar] [CrossRef]
- Walia, A.; Guleria, S.; Mehta, P.; Chauhan, A.; Parkash, J. Microbial xylanases and their industrial application in pulp and paper biobleaching: A review. 3 Biotech 2017, 7, 11. [Google Scholar] [CrossRef]
- Törrönen, A.; Harkki, A.; Rouvinen, J. Three-dimensional structure of endo-1,4-beta-xylanase II from Trichoderma reesei: Two conformational states in the active site. EMBO J. 1994, 13, 2493–2501. [Google Scholar] [CrossRef]
- Quiocho, F.A. Molecular Features and Basic Understanding of Protein-Carbohydrate Interactions: The Arabinose-Binding Protein-Sugar Complex. In Carbohydrate-Protein Interaction; Springer: Berlin/Heidelberg, Germany, 1988; pp. 135–148. [Google Scholar]
- Davies, G.J.; Wilson, K.S.; Henrissat, B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 1997, 321, 557–559. [Google Scholar] [CrossRef] [PubMed]
- Sabini, E.; Sulzenbacher, G.; Dauter, M.; Dauter, Z.; Jørgensen, P.L.; Schülein, M.; Dupont, C.; Davies, G.J.; Wilson, K.S. Catalysis and specificity in enzymatic glycoside hydrolysis: A 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase. Chem. Biol. 1999, 6, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, X.; Li, C.; Kovalevsky, A.; Wan, Q. Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography. Protein J. 2020, 39, 671–680. [Google Scholar] [CrossRef]
- Cheng, Y.-S.; Chen, C.-C.; Huang, C.-H.; Ko, T.-P.; Luo, W.; Huang, J.-W.; Liu, J.-R.; Guo, R.-T. Structural Analysis of a Glycoside Hydrolase Family 11 Xylanase from Neocallimastix patriciarum. J. Biol. Chem. 2014, 289, 11020–11028. [Google Scholar] [CrossRef]
- Wan, Q.; Zhang, Q.; Hamilton-Brehm, S.; Weiss, K.; Mustyakimov, M.; Coates, L.; Langan, P.; Graham, D.; Kovalevsky, A. X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: Implications for the catalytic mechanism. Acta Crystallogr. D Biol. 2013, 70, 11–23. [Google Scholar] [CrossRef]
- Shaw, A.J.; Podkaminer, K.K.; Desai, S.G.; Bardsley, J.S.; Rogers, S.R.; Thorne, P.G.; Hogsett, D.A.; Lynd, L.R. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc. Natl. Acad. Sci. USA 2008, 105, 13769–13774. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.E.; Jain, M.K.; Lee, C.Y.; Lowe, S.E.; Zeikus, J.G. Taxonomic Distinction of Saccharolytic Thermophilic Anaerobes: Description of Thermoanaerobacterium xylanolyticum gen. nov., sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov.; Reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and Clostridium thermohydrosulfuricum E100-69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov., and Thermoanaerobacter thermohydrosulfuricus comb. nov., Respectively; and Transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus. Int. J. Syst. Bacteriol. 1993, 43, 41–51. [Google Scholar] [CrossRef]
- Kim, I.J.; Kim, S.R.; Kim, K.H.; Bornscheuer, U.T.; Nam, K.H. Characterization and structural analysis of the endo-1,4-β-xylanase GH11 from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum useful for lignocellulose saccharification. Sci. Rep. 2023, 13, 17332. [Google Scholar] [CrossRef]
- Nam, K.H. Data of serial synchrotron crystallography of xylanase GH11 from Thermoanaerobacterium saccharolyticum. Data Brief 2024, 52, 110055. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Comparative Analysis of Room Temperature Structures Determined by Macromolecular and Serial Crystallography. Crystals 2024, 14, 276. [Google Scholar] [CrossRef]
- Nam, K.H. pH-Induced structural changes in xylanase GH11 from Thermoanaerobacterium saccharolyticum. F1000Research 2024, 13, 242. [Google Scholar] [CrossRef]
- Gu, D.H.; Eo, C.; Hwangbo, S.A.; Ha, S.C.; Kim, J.H.; Kim, H.; Lee, C.S.; Seo, I.D.; Yun, Y.D.; Lee, W.; et al. BL-11C Micro-MX: A high-flux microfocus macromolecular-crystallography beamline for micrometre-sized protein crystals at Pohang Light Source II. J. Synchrotron Radiat. 2021, 28, 1210–1215. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Casañal, A.; Lohkamp, B.; Emsley, P. Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 2020, 29, 1055–1064. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Metoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef]


| Data Collection | Data I | Data II |
|---|---|---|
| X-ray source | Beamline 11C, PLS-II | |
| Wavelength (Å) | 0.9794 Å | |
| Space group | P43212 | P43212 |
| Cell dimension | ||
| a, b, c (Å) | 73.10, 73.10, 165.10 | 73.23, 73.23, 164.78 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50.0–1.90 (1.93–1.90) | 50.0–1.7 (1.73–1.70) |
| Unique reflections | 35,309 (1721) | 49,993 (2406) |
| Completeness (%) | 96.9 (95.7) | 99.7 (99.4) |
| Redundancy | 6.0 (4.3) | 18.7 (11.6) |
| I/σ | 21.20 (2.04) | 9.98 (2.73) |
| Rmerge | 0.109 (0.445) | 0.150 (0.552) |
| CC1/2 | 0.989 (0.579) | 0.996 (0.623) |
| CC* | 0.997 (0.856) | 0.999 (0.876) |
| Refinement | ||
| Resolution (Å) | 49.33–1.90 | 49.40–1.70 |
| Rwork a | 0.1721 | 0.1652 |
| Rfree b | 0.2067 | 0.1936 |
| RMS deviations | ||
| Bonds (Å) | 0.007 | 0.006 |
| Angles (°) | 0.900 | 0.853 |
| B factors (Å2) | ||
| Protein (Chain A/B) | 24.59/34.26 | 16.55/24.22 |
| XYP (Chain A/B) | 39.30/41.73 | 26.34/33.41 |
| Water | 40.44 | 33.32 |
| Ramachandran plot | ||
| Favored (%) | 97.51 | 97.79 |
| Allowed (%) | 2.49 | 2.21 |
| Disallowed (%) | 0.00 | 0.00 |
| PDB code | 8YYN | 8YYO |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, K.H. Recognition of a Single β-D-Xylopyranose Molecule by Xylanase GH11 from Thermoanaerobacterium saccharolyticum. Crystals 2024, 14, 402. https://doi.org/10.3390/cryst14050402
Nam KH. Recognition of a Single β-D-Xylopyranose Molecule by Xylanase GH11 from Thermoanaerobacterium saccharolyticum. Crystals. 2024; 14(5):402. https://doi.org/10.3390/cryst14050402
Chicago/Turabian StyleNam, Ki Hyun. 2024. "Recognition of a Single β-D-Xylopyranose Molecule by Xylanase GH11 from Thermoanaerobacterium saccharolyticum" Crystals 14, no. 5: 402. https://doi.org/10.3390/cryst14050402





