Intratumoral Heterogeneity Promotes Collective Cancer Invasion through NOTCH1 Variation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Single Layer Invasion Model
2.2. Multilayer Invasion Model
2.3. Cell Culture
2.4. Cell Treatments
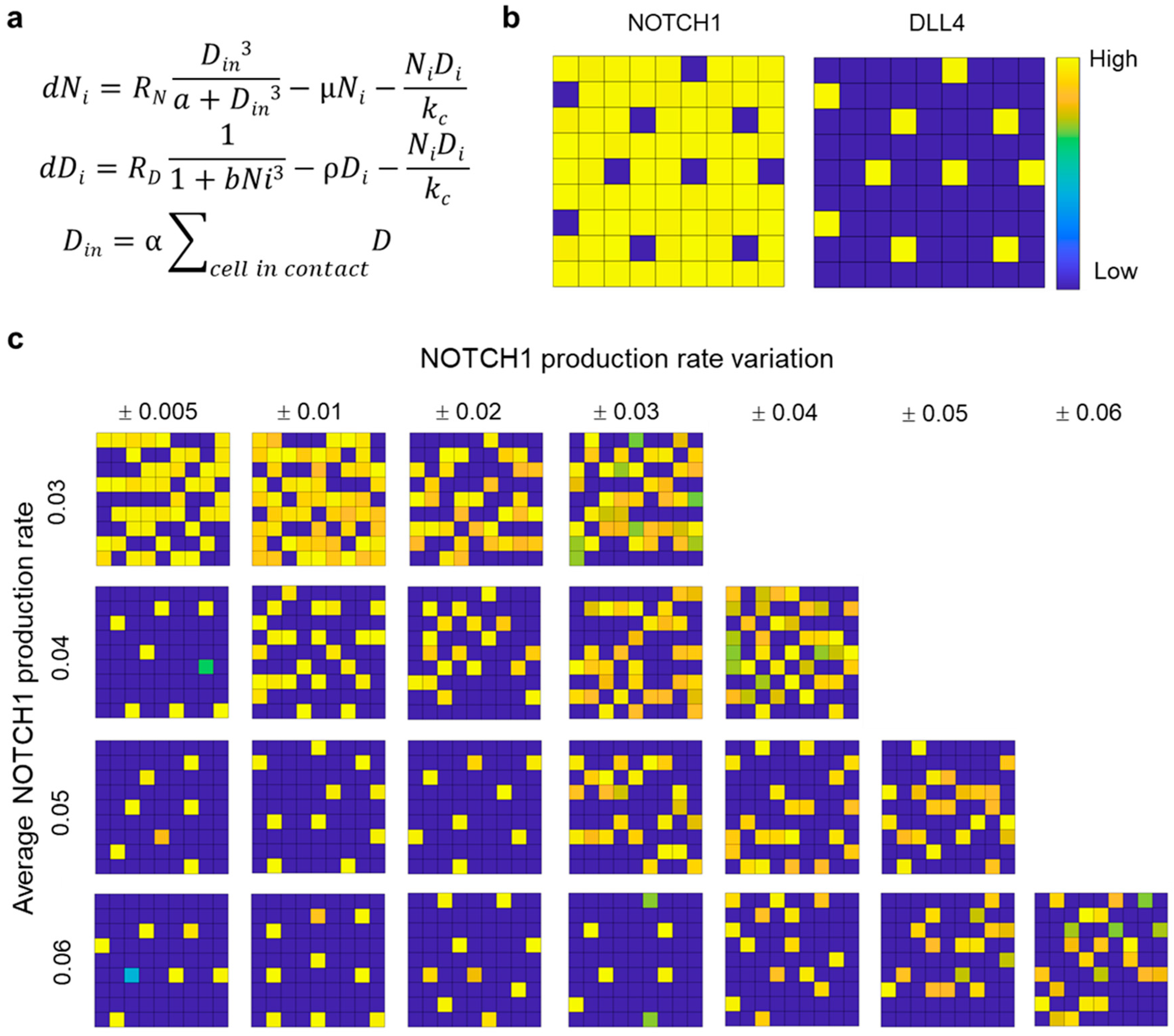
2.5. Computational Model
2.6. CRISPR Knockout
2.7. Statistics and Data Analysis
3. Results
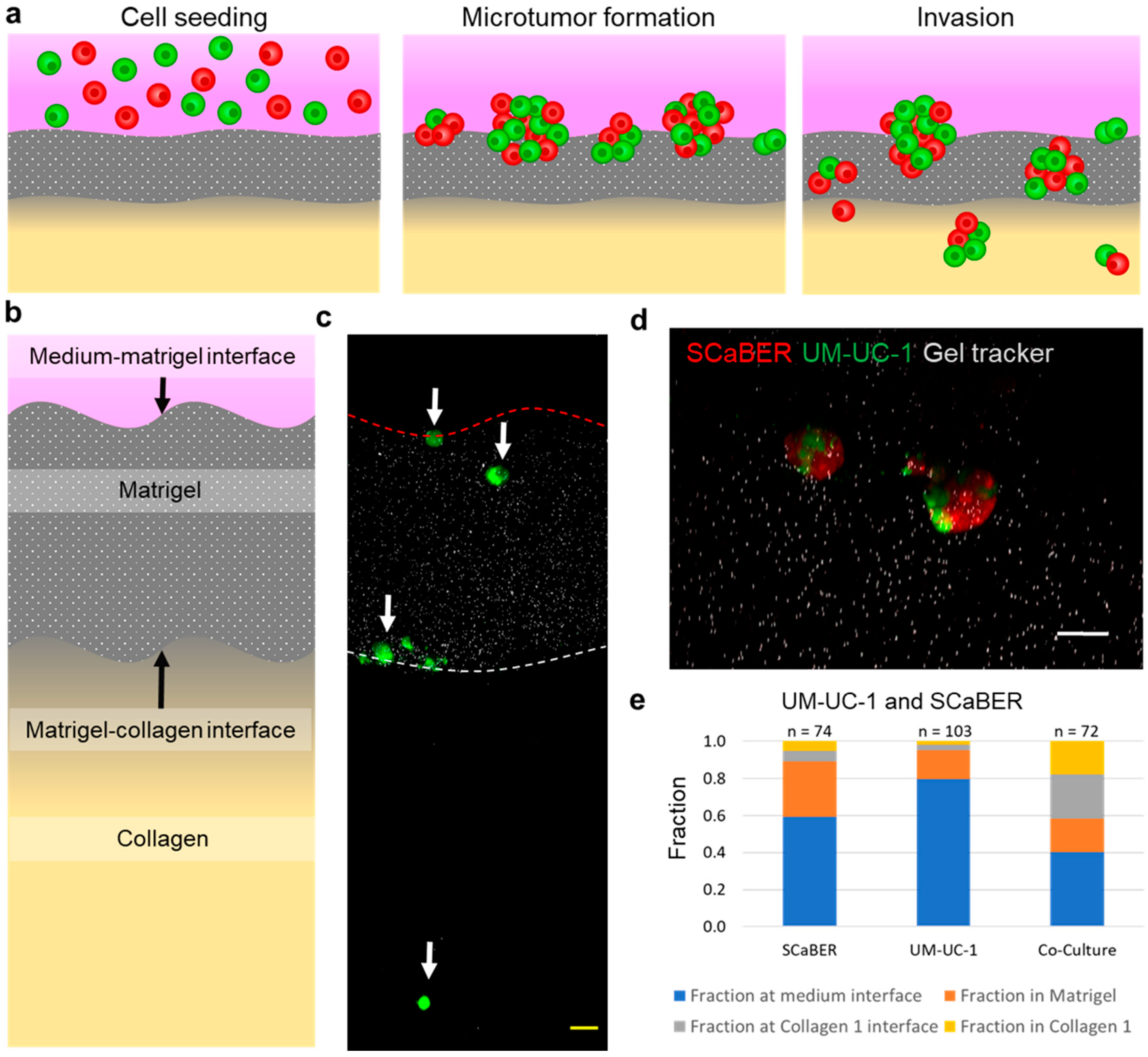
3.1. Cellular Heterogeneity Enhances Invasiveness of Bladder Microtumors
3.2. Heterogeneous Microtumors Efficiently Invade Matrices Found in the Bladder Wall
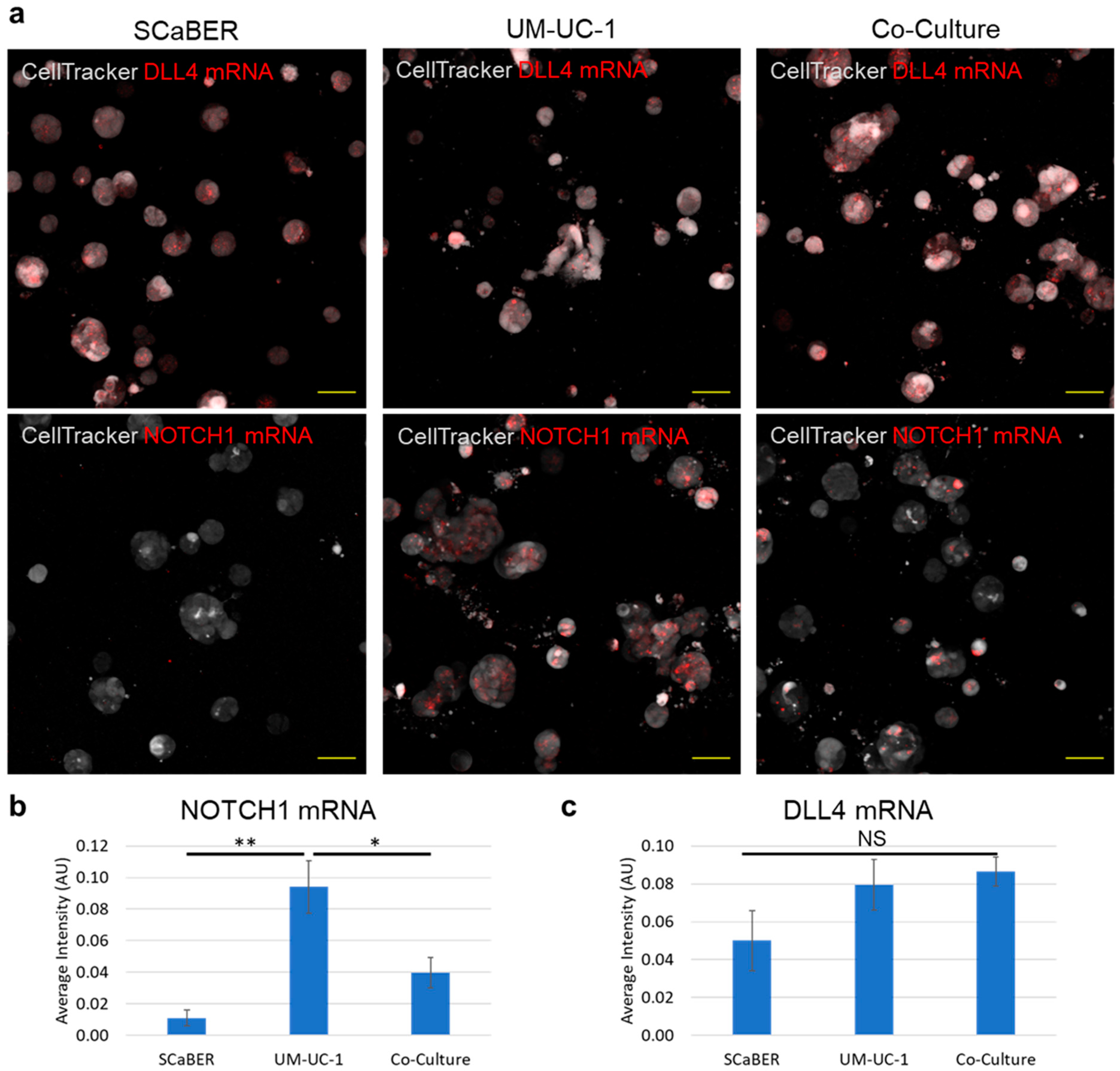
3.3. NOTCH1-DLL4 Signaling in Invasive Microtumors
3.4. UM-UC1- and UM-UC-1 FOXA1 KO Co-Culture Promotes Microtumor Invasion
3.5. Computational Modeling of Mixed Microtumor with Variations in NOTCH1 Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef] [Green Version]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.L.; et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damrauer, J.S.; Hoadley, K.A.; Chism, D.D.; Fan, C.; Tiganelli, C.J.; Wobker, S.E.; Yeh, J.J.; Milowsky, M.I.; Iyer, G.; Parker, J.S.; et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. USA 2014, 111, 3110–3115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamoun, A.; de Reynies, A.; Allory, Y.; Sjodahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef]
- Warrick, J.I.; Walter, V.; Yamashita, H.; Shuman, L.; Amponsa, V.O.; Zheng, Z.; Chan, W.; Whitcomb, T.L.; Yue, F.; Iyyanki, T.; et al. FOXA1, GATA3 and PPARɣ Cooperate to Drive Luminal Subtype in Bladder Cancer: A Molecular Analysis of Established Human Cell Lines. Sci. Rep. 2016, 6, 38531. [Google Scholar] [CrossRef]
- Yamashita, H.; Kawasawa, Y.I.; Shuman, L.; Zheng, Z.; Tran, T.; Walter, V.; Warrick, J.I.; Chen, G.; Al-Ahmadie, H.; Kaag, M.; et al. Repression of transcription factor AP-2 alpha by PPARgamma reveals a novel transcriptional circuit in basal-squamous bladder cancer. Oncogenesis 2019, 8, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015, 27, 15–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Bui, M.M.; Xu, B. Urothelial Carcinoma With Squamous Differentiation Is Associated With High Tumor Stage and Pelvic Lymph-Node Metastasis. Cancer Control 2017, 24, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Warrick, J.I.; Sjodahl, G.; Kaag, M.; Raman, J.D.; Merrill, S.; Shuman, L.; Chen, G.; Walter, V.; DeGraff, D.J. Intratumoral Heterogeneity of Bladder Cancer by Molecular Subtypes and Histologic Variants. Eur. Urol. 2019, 75, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Vilchez Mercedes, S.A.; Bocci, F.; Levine, H.; Onuchic, J.N.; Jolly, M.K.; Wong, P.K. Decoding leader cells in collective cancer invasion. Nat. Rev. Cancer 2021, 21, 592–604. [Google Scholar] [CrossRef]
- Carey, S.P.; Starchenko, A.; McGregor, A.L.; Reinhart-King, C.A. Leading malignant cells initiate collective epithelial cell invasion in a three-dimensional heterotypic tumor spheroid model. Clin. Exp. Metastasis 2013, 30, 615–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, K.J.; Gabrielson, E.; Werb, Z.; Ewald, A.J. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 2013, 155, 1639–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, P.Y.; Brenot, A.; King, A.C.; Longmore, G.D.; George, S.C. Randomly Distributed K14(+) Breast Tumor Cells Polarize to the Leading Edge and Guide Collective Migration in Response to Chemical and Mechanical Environmental Cues. Cancer Res. 2019, 79, 1899–1912. [Google Scholar] [CrossRef] [Green Version]
- Rampias, T.; Vgenopoulou, P.; Avgeris, M.; Polyzos, A.; Stravodimos, K.; Valavanis, C.; Scorilas, A.; Klinakis, A. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat. Med. 2014, 20, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Maraver, A.; Fernandez-Marcos, P.J.; Cash, T.P.; Mendez-Pertuz, M.; Duenas, M.; Maietta, P.; Martinelli, P.; Munoz-Martin, M.; Martinez-Fernandez, M.; Canamero, M.; et al. NOTCH pathway inactivation promotes bladder cancer progression. J. Clin. Investig. 2015, 125, 824–830. [Google Scholar] [CrossRef] [Green Version]
- Goriki, A.; Seiler, R.; Wyatt, A.W.; Contreras-Sanz, A.; Bhat, A.; Matsubara, A.; Hayashi, T.; Black, P.C. Unravelling disparate roles of NOTCH in bladder cancer. Nat. Rev. Urol. 2018, 15, 345–357. [Google Scholar] [CrossRef]
- Riahi, R.; Sun, J.; Wang, S.; Long, M.; Zhang, D.D.; Wong, P.K. Notch1-Dll4 signalling and mechanical force regulate leader cell formation during collective cell migration. Nat. Commun. 2015, 6, 6556. [Google Scholar] [CrossRef] [Green Version]
- Dean, Z.S.; Elias, P.; Jamilpour, N.; Utzinger, U.; Wong, P.K. Probing 3D Collective Cancer Invasion Using Double-Stranded Locked Nucleic Acid Biosensors. Anal. Chem. 2016, 88, 8902–8907. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Fan, H.Y.; Pang, X.; Zhang, M.; Yu, X.H.; Wu, J.S.; Chen, B.J.; Jiang, J.; Liang, X.H.; Tang, Y.L. Dll4/Notch1 signalling pathway is required in collective invasion of salivary adenoid cystic carcinoma. Oncol. Rep. 2021, 45, 1011–1022. [Google Scholar] [CrossRef]
- Torab, P.; Yan, Y.; Yamashita, H.; Warrick, J.I.; Raman, J.D.; DeGraff, D.J.; Wong, P.K. Three-Dimensional Microtumors for Probing Heterogeneity of Invasive Bladder Cancer. Anal. Chem. 2020, 92, 8768–8775. [Google Scholar] [CrossRef] [PubMed]
- Roeder, B.A.; Kokini, K.; Sturgis, J.E.; Robinson, J.P.; Voytik-Harbin, S.L. Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J. Biomech Eng. 2002, 124, 214–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, A.D. Generation of 3D Collagen Gels with Controlled Diverse Architectures. Curr. Protoc. Cell Biol. 2016, 72, 10.20.1–10.20.16. [Google Scholar] [CrossRef] [PubMed]
- Achilli, M.; Mantovani, D. Tailoring Mechanical Properties of Collagen-Based Scaffolds for Vascular Tissue Engineering: The Effects of pH, Temperature and Ionic Strength on Gelation. Polymers 2010, 2, 664–680. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Riahi, R.; Torab, P.; Zhang, D.D.; Wong, P.K. Collective Cell Migration in 3D Epithelial Wound Healing. ACS Nano 2019, 13, 1204–1212. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Zhu, N.; Lu, Y.; Wong, P.K. DNA Transformer for Visualizing Endogenous RNA Dynamics in Live Cells. Anal. Chem 2019, 91, 2626–2633. [Google Scholar] [CrossRef]
- Cohen, M.; Georgiou, M.; Stevenson, N.L.; Miodownik, M.; Baum, B. Dynamic Filopodia Transmit Intermittent Delta-Notch Signaling to Drive Pattern Refinement during Lateral Inhibition. Dev. Cell 2010, 19, 78–89. [Google Scholar] [CrossRef] [Green Version]
- Zuiverloon, T.C.M.; de Jong, F.C.; Costello, J.C.; Theodorescu, D. Systematic Review: Characteristics and Preclinical Uses of Bladder Cancer Cell Lines. Bladder Cancer 2018, 4, 169–183. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Zhu, Y.T.; Ma, L.L.; Pang, S.Y.; Wei, L.J.; Lei, C.Y.; He, C.W.; Tan, W.L. Characteristics of bladder transitional cell carcinoma with E-cadherin and N-cadherin double-negative expression. Oncol. Lett. 2016, 12, 530–536. [Google Scholar] [CrossRef] [Green Version]
- Fujiyama, C.; Jones, A.; Fuggle, S.; Bicknell, R.; Cranston, D.; Harris, A.L. Human bladder cancer invasion model using rat bladder in vitro and its use to test mechanisms and therapeutic inhibitors of invasion. Br. J. Cancer 2001, 84, 558–564. [Google Scholar] [CrossRef] [Green Version]
- Jager, W.; Moskalev, I.; Janssen, C.; Hayashi, T.; Gust, K.M.; Awrey, S.; Black, P.C. Minimally Invasive Establishment of Murine Orthotopic Bladder Xenografts. Jove J. Vis. Exp. 2014, 84, e51123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Style, R.W.; Hyland, C.; Boltyanskiy, R.; Wettlaufer, J.S.; Dufresne, E.R. Surface tension and contact with soft elastic solids. Nat. Commun. 2013, 4, 2728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, S.; Huss, W.; Foster, B.; Ohm, J.; Wang, J.; Azabdaftari, G.; Eng, K.H.; Woloszynska-Read, A. Transcriptional changes associated with in vivo growth of muscle-invasive bladder cancer cell lines in nude mice. Am. J. Clin. Exp. Urol. 2018, 6, 138–148. [Google Scholar]
- Padmanaban, V.; Tsehay, Y.; Cheung, K.J.; Ewald, A.J.; Bader, J.S. Between-tumor and within-tumor heterogeneity in invasive potential. PLoS Comput. Biol. 2020, 16, e1007464. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Riahi, R.; Li, N.; Zhang, D.D.; Wong, P.K. Single Cell Nanobiosensors for Dynamic Gene Expression Profiling in Native Tissue Microenvironments. Adv. Mater. 2015, 27, 6034–6038. [Google Scholar] [CrossRef]
- Tao, S.; Wang, S.; Moghaddam, S.J.; Ooi, A.; Chapman, E.; Wong, P.K.; Zhang, D.D. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res. 2014, 74, 7430–7441. [Google Scholar] [CrossRef] [Green Version]
- Warrick, J.I.; Kaag, M.; Raman, J.D.; Chan, W.; Tran, T.; Kunchala, S.; Shuman, L.; DeGraff, D.; Chen, G. FOXA1 and CK14 as markers of luminal and basal subtypes in histologic variants of bladder cancer and their associated conventional urothelial carcinoma. Virchows Arch. Int. J. Pathol. 2017, 471, 337–345. [Google Scholar] [CrossRef]
- Qiu, M.; Bao, W.; Wang, J.; Yang, T.; He, X.; Liao, Y.; Wan, X. FOXA1 promotes tumor cell proliferation through AR involving the Notch pathway in endometrial cancer. BMC Cancer 2014, 14, 78. [Google Scholar] [CrossRef]
- Fortini, M.E. Notch Signaling: The Core Pathway and Its Posttranslational Regulation. Dev. Cell 2009, 16, 633–647. [Google Scholar] [CrossRef] [Green Version]
- Shaya, O.; Sprinzak, D. From Notch signaling to fine-grained patterning: Modeling meets experiments. Curr. Opin. Genet. Dev. 2011, 21, 732–739. [Google Scholar] [CrossRef]
- Nguyen-Ngoc, K.V.; Cheung, K.J.; Brenot, A.; Shamir, E.R.; Gray, R.S.; Hines, W.C.; Yaswen, P.; Werb, Z.; Ewald, A.J. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc. Natl. Acad. Sci. USA 2012, 109, E2595–E2604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, A.G.; Vignjevic, D.M. Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 2015, 36, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Friedl, P.; Locker, J.; Sahai, E.; Segall, J.E. Classifying collective cancer cell invasion. Nat. Cell Biol. 2012, 14, 777–783. [Google Scholar] [CrossRef]
- Carey, S.P.; Martin, K.E.; Reinhart-King, C.A. Three-dimensional collagen matrix induces a mechanosensitive invasive epithelial phenotype. Sci. Rep. 2017, 7, 42088. [Google Scholar] [CrossRef] [Green Version]
- Anguiano, M.; Morales, X.; Castilla, C.; Pena, A.R.; Ederra, C.; Martinez, M.; Ariz, M.; Esparza, M.; Amaveda, H.; Mora, M.; et al. The use of mixed collagen-Matrigel matrices of increasing complexity recapitulates the biphasic role of cell adhesion in cancer cell migration: ECM sensing, remodeling and forces at the leading edge of cancer invasion. PLoS ONE 2020, 15, e0220019. [Google Scholar] [CrossRef]
- Summerbell, E.R.; Mouw, J.K.; Bell, J.S.K.; Knippler, C.M.; Pedro, B.; Arnst, J.L.; Khatib, T.O.; Commander, R.; Barwick, B.G.; Konen, J.; et al. Epigenetically heterogeneous tumor cells direct collective invasion through filopodia-driven fibronectin micropatterning. Sci. Adv. 2020, 6, eaaz6197. [Google Scholar] [CrossRef] [PubMed]
- Vilchez Mercedes, S.A.; Bocci, F.; Zhu, N.; Levine, H.; Onuchic, J.N.; Jolly, M.K.; Wong, P.K. Nrf2 modulates the hybrid epithelial/mesenchymal phenotype and Notch signaling during collective cancer migration. bioRxiv 2021, 2021-04. Available online: https://www.biorxiv.org/content/10.1101/2021.04.21.440858v2 (accessed on 10 September 2021). [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torab, P.; Yan, Y.; Ahmed, M.; Yamashita, H.; Warrick, J.I.; Raman, J.D.; DeGraff, D.J.; Wong, P.K. Intratumoral Heterogeneity Promotes Collective Cancer Invasion through NOTCH1 Variation. Cells 2021, 10, 3084. https://doi.org/10.3390/cells10113084
Torab P, Yan Y, Ahmed M, Yamashita H, Warrick JI, Raman JD, DeGraff DJ, Wong PK. Intratumoral Heterogeneity Promotes Collective Cancer Invasion through NOTCH1 Variation. Cells. 2021; 10(11):3084. https://doi.org/10.3390/cells10113084
Chicago/Turabian StyleTorab, Peter, Yue Yan, Mona Ahmed, Hironobu Yamashita, Joshua I. Warrick, Jay D. Raman, David J. DeGraff, and Pak Kin Wong. 2021. "Intratumoral Heterogeneity Promotes Collective Cancer Invasion through NOTCH1 Variation" Cells 10, no. 11: 3084. https://doi.org/10.3390/cells10113084





