Identification and Characterization of HIRIP3 as a Histone H2A Chaperone
Abstract
:1. Introduction
2. Materials and Methods
2.1. Immunofluorescence
2.2. Protein Expression in Bacteria
2.3. Protein Expression in HeLa Cells
2.4. Tandem Affinity Purification from HeLa Cells
2.5. Protein Purification from Bacteria
2.6. SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.7. Staining of Protein Gels
2.8. Western Blotting
2.9. In-Gel Histone Deposition Assay
2.10. Mass Spectrometry
2.11. Structural Modelling
2.12. Kinase Assay
3. Results
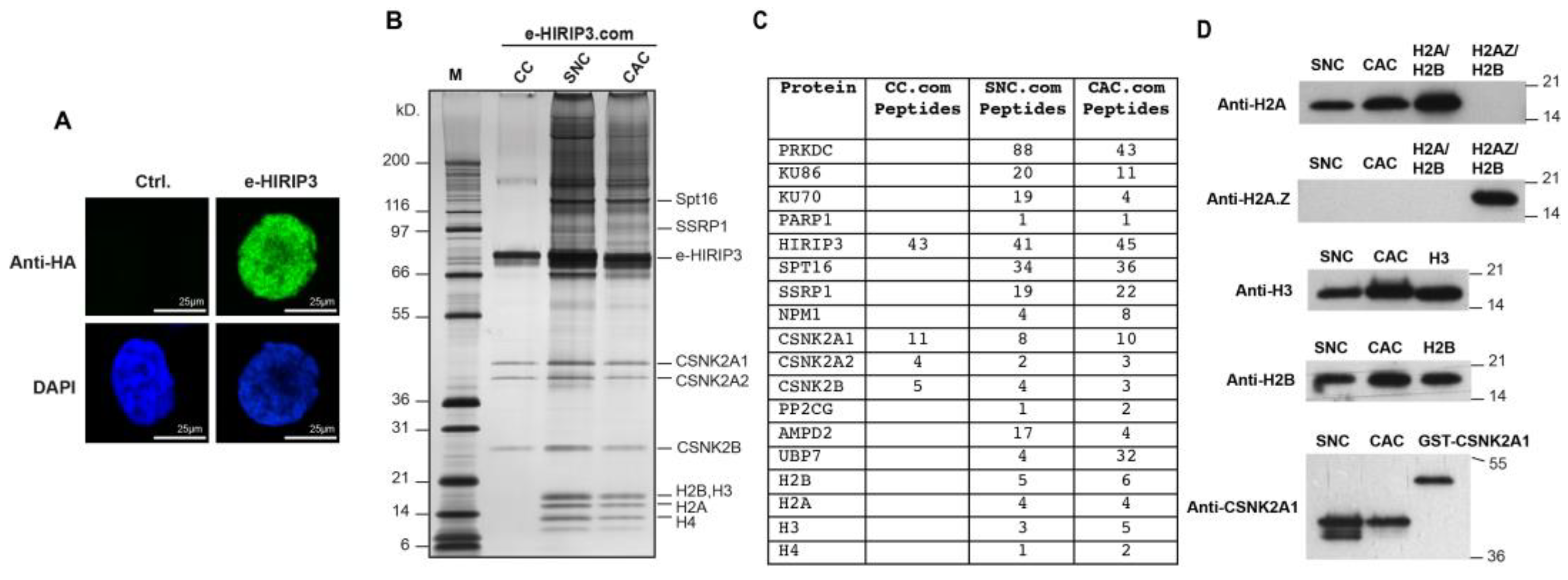
3.1. H2A/H2B Dimers Co-Purify with HIRIP3 In Vivo
3.2. HIRIP3 Physically Interacts with H2A/H2B and H2A.Z/H2B Dimers In Vitro through Its CHZ Motif
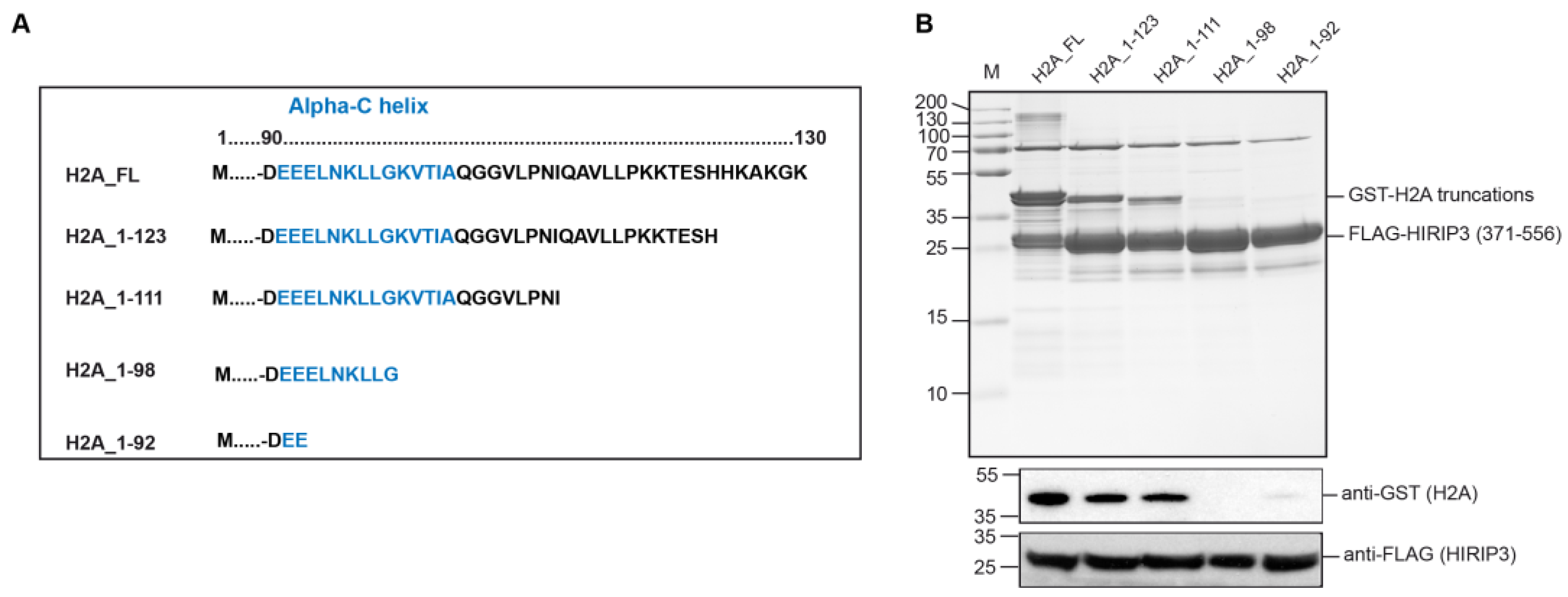
3.3. H2A Interacts with HIRIP3 through Its alphaC Domain
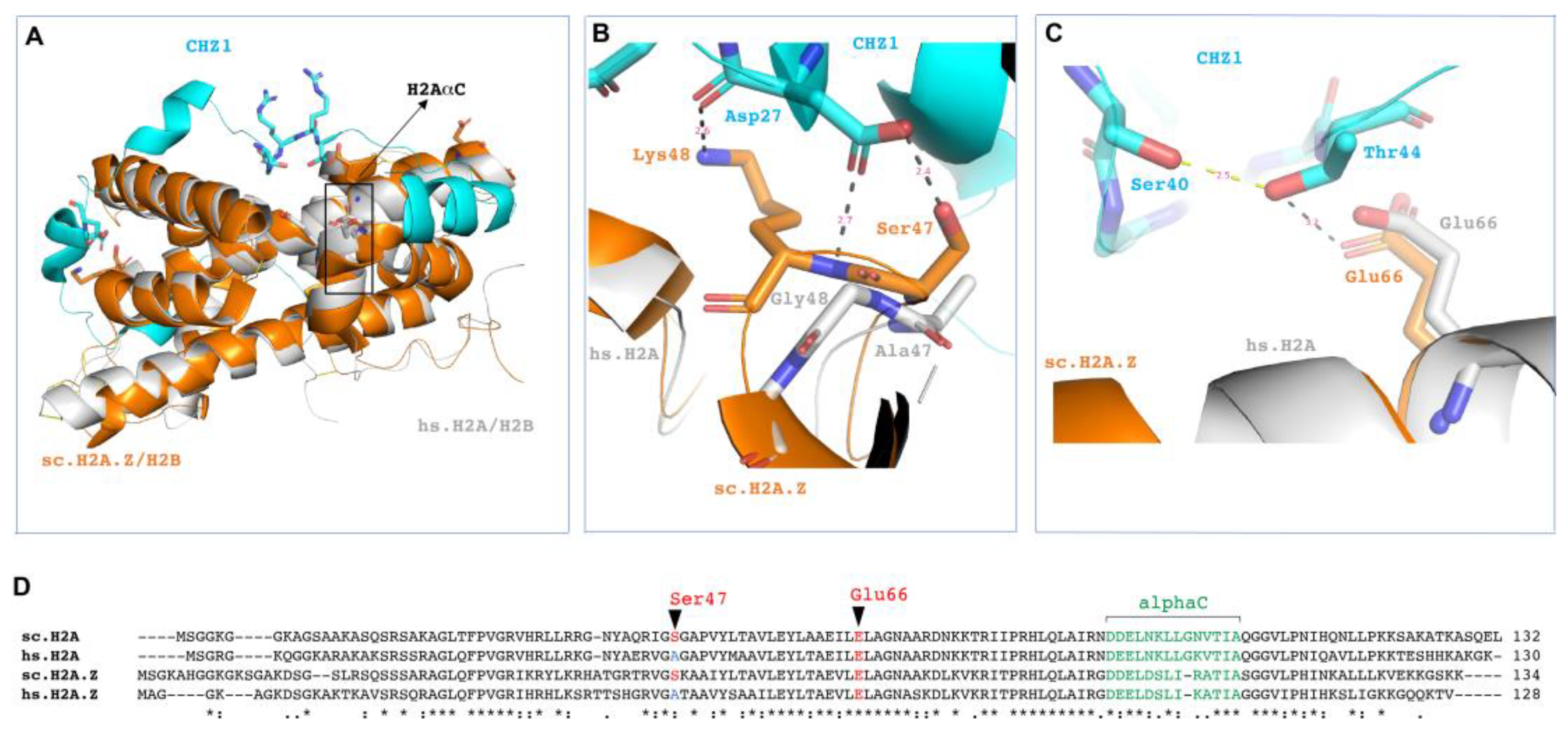
3.4. Structural Considerations of HIRIP3 and H2A/H2B Interaction
3.5. Recombinant CK2 Phosphorylates H2A at Serine 1
4. Discussion
4.1. HIRIP3 Interaction with H2A/H2B Dimers In Vivo and In Vitro
4.2. Structural Basis of HIRIP3 Interaction with H2A/H2B Dimer
4.3. CK2 Kinase Function within HIRIP3 Complex
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kornberg, R.D. Chromatin structure: A repeating unit of histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef]
- Polo, S.E.; Almouzni, G. Histone metabolic pathways and chromatin assembly factors as proliferation markers. Cancer Lett. 2005, 220, 1–9. [Google Scholar] [CrossRef]
- Filipescu, D.; Szenker, E.; Almouzni, G. Developmental roles of histone H3 variants and their chaperones. Trends Genet. 2013, 29, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Preis, J.; Morgan, H.D.; Whitelaw, E. The marks, mechanisms and memory of epigenetic states in mammals. Biochem. J. 2001, 356 Pt 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Skene, P.J.; Henikoff, S. Histone variants in pluripotency and disease. Development 2013, 140, 2513–2524. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Almouzni, G. Chromatin assembly: A basic recipe with various flavours. Curr. Opin. Genet. Dev. 2006, 16, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Elsasser, S.J.; D’arcy, S. Towards a mechanism for histone chaperones. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Tyler, J.K.; Kadonaga, J.T. Chromatin assembly factors: A dual function in nucleosome formation and mobilization? Genes Cells 1997, 2, 593–600. [Google Scholar] [CrossRef]
- Latrick, C.M.; Marek, M.; Ouararhni, K.; Papin, C.; Stoll, I.; Ignatyeva, M.; Obri, A.; Ennifar, E.; Dimitrov, S.; Romier, C.; et al. Molecular basis and specificity of H2A.Z-H2B recognition and deposition by the histone chaperone YL1. Nat. Struct. Mol. Biol. 2016, 23, 309–316. [Google Scholar] [CrossRef]
- Obri, A.; Ouararhni, K.; Papin, C.; Diebold, M.L.; Padmanabhan, K.; Marek, M.; Stoll, I.; Roy, L.; Reilly, P.T.; Mak, T.W.; et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 2014, 505, 648–653. [Google Scholar] [CrossRef]
- Wang, A.Y.; Aristizabal, M.J.; Ryan, C.; Krogan, N.J.; Kobor, M.S. Key functional regions in the histone variant H2A.Z C-terminal docking domain. Mol. Cell. Biol. 2011, 31, 3871–3884. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Sun, L.; Xu, N.; Shan, S.; Wu, F.; Liang, X.; Huang, Y.; Luk, E.; Wu, C.; et al. Structural insights into histone chaperone Chz1-mediated H2A.Z recognition and histone replacement. PLoS Biol. 2019, 17, e3000277. [Google Scholar] [CrossRef]
- Lorain, S.; Quivy, J.P.; Monier-Gavelle, F.; Scamps, C.; Lecluse, Y.; Almouzni, G.; Lipinski, M. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol. Cell. Biol. 1998, 18, 5546–5556. [Google Scholar] [CrossRef]
- Assrir, N.; Filhol, O.; Galisson, F.; Lipinski, M. HIRIP3 is a nuclear phosphoprotein interacting with and phosphorylated by the serine-threonine kinase CK2. Biol. Chem. 2007, 388, 391–398. [Google Scholar] [CrossRef]
- Luk, E.; Vu, N.D.; Patteson, K.; Mizuguchi, G.; Wu, W.H.; Ranjan, A.; Backus, J.; Sen, S.; Lewis, M.; Bai, Y.W.; et al. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell 2007, 25, 357–368. [Google Scholar] [CrossRef]
- Zhou, Z.; Feng, H.; Hansen, D.F.; Kato, H.; Luk, E.; Freedberg, D.I.; Kay, L.E.; Wu, C.; Bai, Y. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat. Struct. Mol. Biol. 2008, 15, 868–869. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.C.; Wang, A.H. Structural D/E-rich repeats play multiple roles especially in gene regulation through DNA/RNA mimicry. Mol. Biosyst. 2015, 11, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Ray-Gallet, D.; Quivy, J.P.; Scamps, C.; Martini, E.M.; Lipinski, M.; Almouzni, G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 2002, 9, 1091–1100. [Google Scholar] [CrossRef]
- Adam, S.; Polo, S.E.; Almouzni, G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 2013, 155, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Poustovoitov, M.V.; Ye, X.; Santos, H.A.; Chen, W.; Daganzo, S.M.; Erzberger, J.P.; Serebriiskii, I.G.; Canutescu, A.A.; Dunbrack, R.L.; et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 2005, 8, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Rai, T.S.; Puri, A.; McBryan, T.; Hoffman, J.; Tang, Y.; Pchelintsev, N.A.; van Tuyn, J.; Marmorstein, R.; Schultz, D.C.; Adams, P.D. Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex. Mol. Cell. Biol. 2011, 31, 4107–4118. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Yang, P.C. Construction of pET-32 α(+) Vector for Protein Expression and Purification. N. Am. J. Med. Sci. 2012, 4, 651–655. [Google Scholar] [CrossRef]
- Froger, A.; Hall, J.E. Transformation of plasmid DNA into E. coli using the heat shock method. J. Vis. Exp. 2007, 6, e253. [Google Scholar] [CrossRef]
- Serrano, M.; Lin, A.W.; McCurrach, M.E.; Beach, D.; Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.M.; Hanlon, C.D.; Andrew, D.J. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J. Cell Biol. 2010, 191, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Abmayr, S.M.; Yao, T.; Parmely, T.; Workman, J.L. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr. Protoc. Pharmacol. 2006, 12, 12.3.1–12.3.13. [Google Scholar] [CrossRef]
- Mahmood, T.; Yang, P.C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Drane, P.; Ouararhni, K.; Depaux, A.; Shuaib, M.; Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010, 24, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- White, C.L.; Suto, R.K.; Luger, K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001, 20, 5207–5218. [Google Scholar] [CrossRef]
- Shuaib, M.; Ouararhni, K.; Dimitrov, S.; Hamiche, A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. USA 2010, 107, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Straube, K.; Blackwell, J.S., Jr.; Pemberton, L.F. Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions. Traffic 2010, 11, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Soman, A.; Liew, C.W.; Teo, H.L.; Berezhnoy, N.V.; Olieric, V.; Korolev, N.; Rhodes, D.; Nordenskiold, L. The human telomeric nucleosome displays distinct structural and dynamic properties. Nucleic Acids Res. 2020, 48, 5383–5396. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Marin, O.; Pinna, L.A. Substrate specificity of protein kinase CK2. Cell. Mol. Biol. Res. 1994, 40, 401–409. [Google Scholar] [PubMed]
- Marin, O.; Bustos, V.H.; Cesaro, L.; Meggio, F.; Pagano, M.A.; Antonelli, M.; Allende, C.C.; Pinna, L.A.; Allende, J.E. A noncanonical sequence phosphorylated by casein kinase 1 in beta-catenin may play a role in casein kinase 1 targeting of important signaling proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 10193–10200. [Google Scholar] [CrossRef] [PubMed]
- Burgess, R.J.; Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 2013, 20, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Calvert, M.E.; Keck, K.M.; Ptak, C.; Shabanowitz, J.; Hunt, D.F.; Pemberton, L.F. Phosphorylation by casein kinase 2 regulates Nap1 localization and function. Mol. Cell. Biol. 2008, 28, 1313–1325. [Google Scholar] [CrossRef]
- Keller, D.M.; Zeng, X.; Wang, Y.; Zhang, Q.H.; Kapoor, M.; Shu, H.; Goodman, R.; Lozano, G.; Zhao, Y.; Lu, H. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 2001, 7, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Luo, Q.; Yin, L.; Wu, J.; Liu, Y.; Gan, J.; Dong, A.; Shen, W.H. OsChz1 acts as a histone chaperone in modulating chromatin organization and genome function in rice. Nat. Commun. 2020, 11, 5717. [Google Scholar] [CrossRef]
- Bonisch, C.; Hake, S.B. Histone H2A variants in nucleosomes and chromatin: More or less stable? Nucleic Acids Res. 2012, 40, 10719–10741. [Google Scholar] [CrossRef]
- Mao, P.; Kyriss, M.N.; Hodges, A.J.; Duan, M.; Morris, R.T.; Lavine, M.D.; Topping, T.B.; Gloss, L.M.; Wyrick, J.J. A basic domain in the histone H2B N-terminal tail is important for nucleosome assembly by FACT. Nucleic Acids Res. 2016, 44, 9142–9152. [Google Scholar] [CrossRef]
- Litchfield, D.W. Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003, 369, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Onikubo, T.; Takashi Onikubo, W.L.; Shechter, D. Histone H2A serine-1 phosphorylation is a chaperone dependent signal for dimerization with H2B and for enhanced deposition. bioRxiv 2022. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, A.T.; Seale, R.L. Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 1983, 258, 12675–12684. [Google Scholar] [CrossRef]
- Basnet, H.; Su, X.B.; Tan, Y.; Meisenhelder, J.; Merkurjev, D.; Ohgi, K.A.; Hunter, T.; Pillus, L.; Rosenfeld, M.G. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature 2014, 516, 267–271. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ignatyeva, M.; Patel, A.K.M.; Ibrahim, A.; Albiheyri, R.S.; Zari, A.T.; Bahieldin, A.; Bronner, C.; Sabir, J.S.M.; Hamiche, A. Identification and Characterization of HIRIP3 as a Histone H2A Chaperone. Cells 2024, 13, 273. https://doi.org/10.3390/cells13030273
Ignatyeva M, Patel AKM, Ibrahim A, Albiheyri RS, Zari AT, Bahieldin A, Bronner C, Sabir JSM, Hamiche A. Identification and Characterization of HIRIP3 as a Histone H2A Chaperone. Cells. 2024; 13(3):273. https://doi.org/10.3390/cells13030273
Chicago/Turabian StyleIgnatyeva, Maria, Abdul Kareem Mohideen Patel, Abdulkhaleg Ibrahim, Raed S. Albiheyri, Ali T. Zari, Ahmed Bahieldin, Christian Bronner, Jamal S. M. Sabir, and Ali Hamiche. 2024. "Identification and Characterization of HIRIP3 as a Histone H2A Chaperone" Cells 13, no. 3: 273. https://doi.org/10.3390/cells13030273





