miRNA-Mediated Fine Regulation of TLR-Induced M1 Polarization
Abstract
:1. Macrophages
2. Macrophage Polarization
2.1. Initiation of Macrophage Polarization
2.2. Regulation of Macrophage Polarization
3. Impact of Post-Transcriptional Regulatory Mechanisms
3.1. miRNAs
3.2. miRNA Expression Profiles of Macrophages from the M1 versus the M2 Activation Spectrum
3.3. TLR Signaling Cascade and miRNA Expression
3.4. Macrophage Polarization Influenced by miRNAs
4. Final Remarks
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Martinez, F.O.; Sica, A.; Mantovani, A.; Locati, M. Macrophage activation and polarization. Front. Biosci. Landmark 2008, 13, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Juhas, U.; Ryba-Stanislawowska, M.; Szargiej, P.; Mysliwska, J. Different pathways of macrophage activation and polarization. Postep. Hig. Med. Dosw. 2015, 69, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Abraham, E. MicroRNAs in immune response and macrophage polarization. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Malyshev, I.; Malyshev, Y. Current Concept and Update of the Macrophage Plasticity Concept: Intracellular Mechanisms of Reprogramming and M3 Macrophage “Switch” Phenotype. Biomed Res. Int. 2015, 2015, 341308. [Google Scholar] [CrossRef] [PubMed]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.-A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Q.; Dai, Y.; Yang, Y.; Huang, C.; Meng, X.M.; Wu, B.M.; Li, J. Emerging role of microRNAs in regulating macrophage activation and polarization in immune response and inflammation. Immunology 2016, 148, 237–248. [Google Scholar] [CrossRef]
- Zhou, D.; Huang, C.; Lin, Z.; Zhan, S.; Kong, L.; Fang, C.; Li, J. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell. Signal. 2014, 26, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS–) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Essandoh, K.; Li, Y.T.; Huo, J.Z.; Fan, G.C. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock 2016, 46, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Vergadi, E.; Ieronymaki, E.; Lyroni, K.; Vaporidi, K.; Tsatsanis, C. Akt signaling pathway in macrophage activation and M1/M2 polarization. J. Immunol. 2017, 198, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Paik, P.K.; Chen, J.; Yarilina, A.; Kockeritz, L.; Lu, T.T.; Woodgett, J.R.; Ivashkiv, L.B. IFN-γ Suppresses IL-10 Production and Synergizes with TLR2 by Regulating GSK3 and CREB/AP-1 Proteins. Immunity 2006, 24, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors the role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Rev. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Akira, S. Toll-like receptors. Curr. Protoc. Immunol. 2015, 109, 14.12.1–14.12.10. [Google Scholar] [CrossRef]
- Takeda, K.; Akira, S. TLR signaling pathways. Semin. Immunol. 2004, 16, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage M1-M2 polarization balance. Front. Immunol. 2014, 5, 113523. [Google Scholar] [CrossRef]
- Barton, G.M.; Medzhitov, R. Toll-like Receptor Signaling Pathways. Science 2003, 300, 1524–1525. [Google Scholar] [CrossRef]
- Bayraktar, R.; Bertilaccio, M.T.; Calin, G.A. The Interaction between Two Worlds: MicroRNAs and Toll-Like Receptors. Front. Immunol. 2019, 10, 437943. [Google Scholar] [CrossRef]
- Bazzoni, F.; Rossato, M.; Fabbri, M.; Gaudiosi, D.; Mirolo, M.; Mori, L.; Tamassia, N.; Mantovani, A.; Cassatella, M.A.; Locati, M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. USA 2009, 106, 5282–5287. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Abak, A.; Shoorei, H.; Talebi, S.F.; Mohaqiq, M.; Sarabi, P.; Taheri, M.; Mokhtari, M. Interaction between non-coding RNAs and Toll-like receptors. Biomed. Pharmacother. 2021, 140, 111784. [Google Scholar] [CrossRef]
- Lee, H.M.; Kim, T.S.; Jo, E.K. MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep. 2016, 49, 311–318. [Google Scholar] [CrossRef]
- Rothschild; McDaniel, D.K.; Ringel-Scaia, V.M.; Allen, I.C. Modulating inflammation through the negative regulation of NF-B signaling. J. Leukoc. Biol. 2018, 103, 1131–1150. [Google Scholar] [CrossRef] [PubMed]
- Virtue, A.; Wang, H.; Yang, X.F. MicroRNAs and Toll-like Receptor/Interleukin-1 Receptor Signaling. J. Hematol. Oncol. 2012, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Wang, X.; Zhu, X.; Zhang, J.; Zhu, Y.; Shao, X.; Zhou, X. JNK/AP1 Pathway Regulates MYC Expression and BCR Signaling through Ig Enhancers in Burkitt Lymphoma Cells. J. Cancer 2020, 11, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Vesely, P.W.; Staber, P.B.; Hoefler, G.; Kenner, L. Translational regulation mechanisms of AP-1 proteins. Mutat. Res. 2009, 682, 7–12. [Google Scholar] [CrossRef]
- Lee, H.; Kim, E.; Kim, S. miRNA-Induced Downregulation of IPMK in Macrophages Mediates Lipopolysaccharide-Triggered TLR4 Signaling. Biomolecules 2023, 13, 332. [Google Scholar] [CrossRef]
- Travassos, L.H.; Carneiro, L.A.M.; Girardin, S.E.; Boneca, I.G.; Lemos, R.; Bozza, M.T.; Domingues, R.C.P.; Coyle, A.J.; Bertin, H.; Philpott, D.J.; et al. Nod1 Participates in the Innate Immune Response to Pseudomonas aeruginosa. J. Biol. Chem. 2005, 280, 36714–36718. [Google Scholar] [CrossRef]
- McCormick, S.M.; Heller, N.M. Regulation of Macrophage, Dendritic Cell, and Microglial Phenotype and Function by the SOCS Proteins. Front. Immunol. 2015, 6, 167589. [Google Scholar] [CrossRef]
- Lawrence, T.; Natoli, G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 2011, 11, 750–761. [Google Scholar] [CrossRef]
- Ellis, T.N.; Leiman, S.A.; Kuehn, M.J. Naturally Produced Outer Membrane Vesicles from Pseudomonas aeruginosa Elicit a Potent Innate Immune Response via Combined Sensing of Both Lipopolysaccharide and Protein Components. Infect. Immun. 2010, 78, 3822–3831. [Google Scholar] [CrossRef]
- Gottschalk, R.A.; Dorrington, M.G.; Dutta, B.; Krauss, K.S.; Martins, A.J.; Uderhardt, S.; Chan, W.P.; Tsang, J.S.; Torabi-Parizi, P.; Fraser, I.D.; et al. IFN-mediated negative feedback supports bacteria class-specific macrophage inflammatory responses. Elife 2019, 8, e46836. [Google Scholar] [CrossRef] [PubMed]
- Androulidaki, A.; Iliopoulos, D.; Arranz, A.; Doxaki, C.; Schworer, S.; Zacharioudaki, V.; Margioris; Tsichlis, P.N.; Tsatsanis, C. The Kinase Akt1 Controls Macrophage Response to Lipopolysaccharide by Regulating MicroRNAs. Immunity 2009, 31, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.A.; Basith, S.; Choi, S. Negative regulatory approaches to the attenuation of Toll-like receptor signaling. Exp. Mol. Med. 2013, 45, e11. [Google Scholar] [CrossRef]
- Mellett, M.; Atzei, P.; Jackson, R.; O’Neill, L.A.; Moynagh, P.N. Mal Mediates TLR-Induced Activation of CREB and Expression of IL-10. J. Immunol. 2011, 186, 4925–4935. [Google Scholar] [CrossRef] [PubMed]
- Wen, A.Y.; Sakamoto, K.M.; Miller, L.S. The Role of the Transcription Factor CREB in Immune Function. J. Immunol. 2010, 185, 6413–6419. [Google Scholar] [CrossRef]
- Luan, B.; Yoon, Y.-S.; Le Lay, J.; Kaestner, K.H.; Hedrick, S.; Montminy, M. CREB pathway links PGE2 signaling with macrophage polarization. Proc. Natl. Acad. Sci. USA 2015, 112, 15642–15647. [Google Scholar] [CrossRef]
- Arranz, A.; Doxaki, C.; Vergadi, E.; Martinez de la Torre, Y.; Vaporidi, K.; Lagoudaki, E.D.; Ieronymaki, E.; Androulidaki, A.; Venihaki, M.; Margioris, A.N.; et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc. Natl. Acad. Sci. USA 2012, 109, 9517–9522. [Google Scholar] [CrossRef]
- Zhou, H.; Yu, M.J.; Fukuda, K.; Im, J.; Yao, P.; Cui, W.; Bulek, K.; Zepp, J.; Wan, Y.Z.; Kim, T.W.; et al. IRAK-M mediates Toll-like receptor/IL-1R-induced NF kappa B activation and cytokine production. EMBO J. 2013, 32, 583–596. [Google Scholar] [CrossRef]
- Wilson, H.M. SOCS Proteins in Macrophage Polarization and Function. Front. Immunol. 2014, 5, 104201. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Liu, W.C.; Sun, T.; Huang, Y.; Wang, Y.L.; Deb, D.K.; Yoon, D.; Kong, J.; Thadhani, R.; Li, Y.C. 1,25-Dihydroxyvitamin D Promotes Negative Feedback Regulation of TLR Signaling via Targeting MicroRNA-155-SOCS1 in Macrophages. J. Immunol. 2013, 190, 3687–3695. [Google Scholar] [CrossRef] [PubMed]
- Fric, J.; Zelante, T.; Wong, A.Y.; Mertes, A.; Yu, H.B.; Ricciardi-Castagnoli, P. NFAT control of innate immunity. Blood 2012, 120, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Coll, R.C.; O’Neill, L.A. New Insights into the Regulation of Signalling by Toll-Like Receptors and Nod-Like Receptors. J. Innate Immun. 2010, 2, 406–421. [Google Scholar] [CrossRef]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Müller-Esterl, W. (Ed.) Biochemie: Eine Einführung für Mediziner und Naturwissenschaftler.: Transkription—Umschrift Genetischer Information, 3rd ed.; Springer Spektrum: Berlin/Heidelberg, Germany, 2018; ISBN 978-3-662-54851-6. [Google Scholar]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Zhang, X.; Azhar, G.; Williams, E.D.; Rogers, S.C.; Wei, J.Y. MicroRNA Clusters in the Adult Mouse Heart: Age-Associated Changes. Biomed Res. Int. 2015, 2015, 732397. [Google Scholar] [CrossRef]
- Knippers, R. (Ed.) Molekulare Genetik, 9th ed.; Georg Thieme Verlag: New York, NY, USA, 2006; ISBN 978-3-13-477009-4. [Google Scholar]
- van Rooij, E.; Olson, E.N. MicroRNA therapeutics for cardiovascular disease: Opportunities and obstacles. Nat. Rev. Drug Discov. 2012, 11, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-S.; Phillips, M.D.; Betel, D.; Mu, P.; Ventura, A.; Siepel, A.C.; Chen, K.C.; Lai, E.C. Widespread regulatory activity of vertebrate microRNA* species. RNA 2011, 17, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Ipsaro, J.J.; Joshua-Tor, L. From Guide to Target: Molecular Insights into Eukaryotic RNAi Machinery. Nat. Struct. Mol. Biol. 2015, 22, 20–28. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Sheedy, F.J.; McCoy, C.E. MicroRNAs: The fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 2011, 11, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Thompson, W.E.; Chowdhury, I. Emerging roles of microRNAs in the regulation of Toll-like receptor (TLR)-signaling. Front. Biosci. Landmark 2021, 26, 771–796. [Google Scholar] [CrossRef] [PubMed]
- Dharap, A.; Pokrzywa, C.; Murali, S.; Pandi, G.; Vemuganti, R. MicroRNA miR-324-3p Induces Promoter-Mediated Expression of RelA Gene. PLoS ONE 2013, 8, e79467. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genes Dev. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.N.; Kausar, S.; Asma, B.; Ran, W.H.; Li, J.G.; Lin, Z.N.; Li, T.J.; Cui, H.J. MicroRNAs reshape the immunity of insects in response to bacterial infection. Front. Immunol. 2023, 14, 1176966. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; O’Neill, L.A. MicroRNAs and the resolution phase of inflammation in macrophages. Eur. J. Immunol. 2011, 41, 2482–2485. [Google Scholar] [CrossRef] [PubMed]
- Riechert, G.; Maucher, D.; Schmidt, B.; Schumann, J. miRNA-Mediated Priming of Macrophage M1 Differentiation Differs in Gram-Positive and Gram-Negative Settings. Genes 2022, 13, 211. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; McCurdy, S.; Huang, S.; Zhu, X.; Peplowska, K.; Tiirikainen, M.; Boisvert, W.A.; Garmire, L.X. Time Series miRNA-mRNA integrated analysis reveals critical miRNAs and targets in macrophage polarization. Sci. Rep. 2016, 6, 37446. [Google Scholar] [CrossRef] [PubMed]
- Cobos Jiménez, V.; Bradley, E.J.; Willemsen, A.M.; van Kampen, A.H.C.; Baas, F.; Kootstra, N.A. Next-generation sequencing of microRNAs uncovers expression signatures in polarized macrophages. Physiol. Genom. 2014, 46, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, M.; Zhong, M.; Suo, Q.; Lv, K. Expression profiles of miRNAs in polarized macrophages. Int. J. Mol. Med. 2013, 31, 797–802. [Google Scholar] [CrossRef]
- Verschoor, C.P.; Dorrington, M.G.; Novakowski, K.E.; Kaiser, J.; Radford, K.; Nair, P.; Anipindi, V.; Kaushic, C.; Surette, M.G.; Bowdisha, D.M.E. MicroRNA-155 is required for clearance of Streptococcus pneumoniae from the nasopharynx. Infect. Immun. 2014, 82, 4824–4833. [Google Scholar] [CrossRef]
- Ghorpade, D.S.; Leyland, R.; Kurowska-Stolarska, M.; Patil, S.A.; Balaji, K.N. MicroRNA-155 Is Required for Mycobacterium bovis BCG-Mediated Apoptosis of Macrophages. Mol. Cell. Biol. 2012, 32, 2239–2253. [Google Scholar] [CrossRef]
- Schnitger, A.K.D.; Machova, A.; Mueller, R.U.; Androulidaki, A.; Schermer, B.; Pasparakis, M.; Krönke, M.; Papadopoulou, N. Listeria monocytogenes Infection in Macrophages Induces Vacuolar-Dependent Host miRNA Response. PLoS ONE 2011, 6, e27435. [Google Scholar] [CrossRef]
- Cremer, T.J.; Ravneberg, D.H.; Clay, C.D.; Piper-Hunter, M.G.; Marsh, C.B.; Elton, T.S.; Gunn, J.S.; Amer, A.; Kanneganti, T.-D.; Schlesinger, L.S.; et al. MiR-155 Induction by F. novicida but Not the Virulent F. tularensis Results in SHIP Down-Regulation and Enhanced Pro-Inflammatory Cytokine Response. PLoS ONE 2009, 4, e8508. [Google Scholar] [CrossRef] [PubMed]
- Jennewein, C.; von Knethen, A.; Schmid, T.; Brüne, B. MicroRNA-27b Contributes to Lipopolysaccharide-mediated MicroRNA-27b Contributes to Lipopolysaccharide-mediated Peroxisome Proliferator-activated Receptor γ (PPARγ) mRNA Destabilization. J. Biol. Chem. 2010, 285, 11846–11853. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jin, H.; Yang, X.; Wang, L.; Su, L.; Liu, K.; Gu, Q.; Xu, X. MicroRNA-93 inhibits inflammatory cytokine production in LPS-stimulated murine macrophages by targeting IRAK4. FEBS Lett. 2014, 588, 1692–1698. [Google Scholar] [CrossRef] [PubMed]
- Arenas-Padilla, M.; Mata-Haro, V. Regulation of TLR signaling pathways by microRNAs: Implications in inflammatory diseases. Cent. Eur. J. Immunol. 2018, 43, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, B.; Zhu, J.; Huang, W.; Han, B.; Wang, Q.; Qi, C.; Wang, M.; Liu, F. miR-106b-5p Inhibits IRF1/IFN-β Signaling to miR-106b-5p Inhibits IRF1/IFN-β Signaling to Promote M2 Macrophage Polarization of Glioblastoma. OncoTargets Ther. 2020, 13, 7479–7492. [Google Scholar] [CrossRef]
- Ying, H.; Kang, Y.; Zhang, H.; Zhao, D.; Xia, J.; Lu, Z.; Wang, H.; Xu, F.; Shi, L. MiR-127 modulates macrophage polarization and promotes lung inflammation and injury by activating the JNK pathway. J. Immunol. 2015, 194, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zhao, J.-L.; Wang, L.; Gao, C.-C.; Liang, S.-Q.; An, D.-J.; Bai, J.; Chen, Y.; Han, H.; Qin, H.-Y. miR-148a-3p Mediates Notch Signaling to Promote the Differentiation and M1 Activation of Macrophages. Front. Immunol. 2017, 8, 1327. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Gao, Y.; Bi, D.; Xu, T. MicroRNA-148 as a negative regulator of the common TLR adaptor mediates inflammatory response in teleost fish. Sci. Rep. 2017, 7, 4124. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Liu, R.; Zheng, Y.; Liu, X.; Chang, L.; Xiong, S.; Chu, Y. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp. Cell Res. 2012, 318, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, Y.; Jiang, R.; Nie, C.; Zeng, Z.; Zhao, N.; Huang, C.; Shao, Q.; Ding, C.; Qing, C.; et al. miR-132 inhibits lipopolysaccharide-induced inflammation in alveolar macrophages by the cholinergic anti-inflammatory pathway. Exp. Lung Res. 2015, 41, 261–269. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.H.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef]
- Hop, H.T.; Arayan, L.T.; Huy, T.X.N.; Reyes, A.W.B.; Vu, S.H.; Min, W.; Lee, H.J.; Rhee, M.H.; Chang, H.H.; Kim, S. The Key Role of c-Fos for Immune Regulation and Bacterial Dissemination in Brucella Infected Macrophage. Front. Cell. Infect. Microbiol. 2018, 8, 287. [Google Scholar] [CrossRef]
- Macian, F.; Lopez-Rodriguez, C.; Rao, A. Partners in transcription: NFAT and AP-1. Oncogene 2001, 20, 2476–2489. [Google Scholar] [CrossRef]
- Rao, A.; Luo, C.; Hogan, P.G. Transcription factors of the NFAT family: Regulation and function. Annu. Review Immunol. 1997, 15, 707–747. [Google Scholar] [CrossRef]
- Hatzi, K.; Nance, J.P.; Kroenke, M.A.; Bothwell, M.; Haddad, E.K.; Melnick, A.; Crotty, S. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 2015, 212, 539–553. [Google Scholar] [CrossRef]
- Vartanian, R.; Masri, J.; Martin, J.; Cloninger, C.; Holmes, B.; Artinian, N.; Funk, A.; Ruegg, T.; Gera, J. AP-1 regulates cyclin D1 and c-MYC transcription in an AKT-dependent manner in response to mTOR inhibition: Role of AIP4/Itch-mediated JUNB degradation. Mol. Cancer Res. 2010, 9, 115–130. [Google Scholar] [CrossRef]
- Dueck, A.; Eichner, A.; Sixt, M.; Meister, G. A miR-155-dependent microRNA hierarchy in dendritic cell maturation and macrophage activation. FEBS Lett. 2014, 588, 632–640. [Google Scholar] [CrossRef]
- He, X.B.; Jing, Z.Z.; Cheng, G.F. MicroRNAs: New Regulators of Toll-Like Receptor Signalling Pathways. Biomed Res. Int. 2014, 2014, 945169. [Google Scholar] [CrossRef]
- Baer, P.C.; Neuhoff, A.K.; Schubert, R. microRNA Expression of Renal Proximal Tubular Epithelial Cells and Their Extracellular Vesicles in an Inflammatory Microenvironment In Vitro. Int. J. Mol. Sci. 2023, 24, 11069. [Google Scholar] [CrossRef]
- Li, Y.K.; Shi, X.Y. MicroRNAs in the regulation of TLR and RIG-I pathways. Cell. Mol. Immunol. 2013, 10, 65–71. [Google Scholar] [CrossRef]
- Yang, B.; Yang, R.C.; Xu, B.J.; Fu, J.Y.; Qu, X.Y.; Li, L.; Dai, M.H.; Tan, C.; Chen, H.C.; Wang, X.R. miR-155 and miR-146a collectively regulate meningitic Escherichia coli infection-mediated neuroinflammatory responses. J. Neuroinflamm. 2021, 18, 114. [Google Scholar] [CrossRef]
- Roessler, C.; Schumann, J. Transcriptom and miRNA data of PUFA-enriched stimulated murine macrophage and human endothelial cell lines. Sci. Data 2023, 10, 375. [Google Scholar] [CrossRef]
- Luo, X.; Zhang, X.; Wu, X.; Yang, X.; Han, C.; Wang, Z.; Du, Q.; Zhao, X.; Liu, S.-L.; Tong, D.; et al. Brucella Downregulates Tumor Necrosis Factor-α to Promote Intracellular Survival via Omp25 Regulation of Different MicroRNAs in Porcine and Murine Macrophages. Front. Immunol. 2018, 8, 2013. [Google Scholar] [CrossRef]
- Teixeira-Coelho, M.; Guedes, J.; Ferreirinha, P.; Howes, A.; Pedrosa, J.; Rodrigues, F.; Lai, W.S.; Blackshear, P.J.; O’Garra, A.; Castro, A.G.; et al. Differential post- transcriptional regulation of IL-10 by TLR2 and TLR4-activated macrophages. Eur. J. Immunol. 2014, 44, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Sticht, C.; de La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Aalaei-Andabili, S.H.; Rezaei, N. Toll like receptor (TLR)-induced differential expression of microRNAs (MiRs) promotes proper immune response against infections: A systematic review. J. Infect. 2013, 67, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.J.; Fatehchand, K.; Shah, P.; Gillette, D.; Patel, H.; Marsh, R.L.; Besecker, B.Y.; Rajaram, M.V.; Cormet-Boyaka, E.; Kanneganti, T.D.; et al. MiR-155 induction by microbes/microbial ligands requires NE-kappa B-dependent de novo protein synthesis. Front. Cell. Infect. Microbiol. 2012, 2, 73. [Google Scholar] [CrossRef]
- Nahid, M.A.; Satoh, M.; Chan, E.K.L. MicroRNA in TLR signaling and endotoxin tolerance. Cell. Mol. Immunol. 2011, 8, 388–403. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, I.; David, M. MicroRNAs in the immune response. Cytokine 2008, 43, 391–394. [Google Scholar] [CrossRef]
- Schulte, L.N.; Westermann, A.J.; Vogel, J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. 2013, 41, 542–553. [Google Scholar] [CrossRef]
- Wang, P.; Hou, J.; Lin, L.; Wang, C.M.; Liu, X.G.; Li, D.; Ma, F.; Wang, Z.G.; Cao, X.T. Inducible microRNA-155 Feedback Promotes Type I IFN Signaling in Antiviral Innate Immunity by Targeting Suppressor of Cytokine Signaling 1. J. Immunol. 2010, 185, 6226–6233. [Google Scholar] [CrossRef]
- Gao, H.; Xiao, D.Q.; Gao, L.B.; Li, X.H. MicroRNA-93 contributes to the suppression of lung inflammatory responses in LPS-induced acute lung injury in mice via the TLR4/MyD88/NF-kappa B signaling pathway. Int. J. Mol. Med. 2020, 46, 561–570. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, W.S.; Li, R.P.; Liu, Y. miRNA-93-5p in exosomes derived from M2 macrophages improves lipopolysaccharide-induced podocyte apoptosis by targeting Toll-like receptor 4. Bioengineered 2022, 13, 7683–7696. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, C.; Guo, M.; Tao, Y.; Cui, P.; Zhou, Y.; Qin, N.; Zheng, J.; Zhang, J.; Xu, L. MicroRNA-7 Deficiency Ameliorates the Pathologies of Acute Lung Injury through Elevating KlF4. Front. Immunol. 2016, 7, 389. [Google Scholar] [CrossRef]
- Wu, H.; Bao, Y.; Wang, L.; Li, X.; Sun, J. Mycobacterium marinum down-regulates miR-148a in macrophages in an EsxA-dependent manner. Int. Immunopharmacol. 2019, 73, 41–48. [Google Scholar] [CrossRef]
- Yin, M.; Lu, J.; Guo, Z.; Zhang, Y.; Liu, J.; Wu, T.; Guo, K.; Luo, T.; Guo, Z. Reduced SULT2B1b expression alleviates ox-LDL-induced inflammation by upregulating miR-148-3P via inhibiting the IKKβ/NF-κB pathway in macrophages. Aging 2021, 13, 3428–3442. [Google Scholar] [CrossRef]
- Arora, H.; Qureshi, R.; Jin, S.; Park, A.-K.; Park, W.-Y. miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFκB1. Exp. Mol. Med. 2011, 43, 298–304. [Google Scholar] [CrossRef]
- Qian, D.; Wei, G.; Xu, C.; He, Z.; Hua, J.; Li, J.; Hu, Q.; Lin, S.; Gong, J.; Meng, H.; et al. Bone marrow-derived mesenchymal stem cells (BMSCs) repair acute necrotized pancreatitis by secreting microRNA-9 to target the NF-κB1/p50 gene in rats. Sci. Rep. 2017, 7, 581. [Google Scholar] [CrossRef]
- Yue, P.; Jing, L.; Zhao, X.; Zhu, H.; Teng, J. Down-regulation of taurine-up-regulated gene 1 attenuates inflammation by sponging miR-9-5p via targeting NF-κB1/p50 in multiple sclerosis. Life Sci. 2019, 233, 116731. [Google Scholar] [CrossRef]
- Thulasingam, S.; Massilamany, C.; Gangaplara, A.; Dai, H.; Yarbaeva, S.; Subramaniam, S.; Riethoven, J.-J.; Eudy, J.; Lou, M.; Reddy, J. miR-27b*, an oxidative stress-responsive microRNA modulates nuclear factor-kB pathway in RAW 264.7 cells. Mol. Cell. Biochem. 2011, 352, 181–188. [Google Scholar] [CrossRef]
- Zhou, R.; O’Hara, S.P.; Chen, X.M. MicroRNA regulation of innate immune responses in epithelial cells. Cell. Mol. Immunol. 2011, 8, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; Wan, X.P.; Yuan, Y.; Huang, J.J.; Jiang, Y.J.; Zhao, K.Y.; Wang, Y.; Liu, Y.; Wang, Q.Q.; Jin, H.C. Opposite effects of miR-155 in the initial and later stages of lipopolysaccharide (LPS)-induced inflammatory response. J. Zhejiang Univ. Sci. B 2021, 22, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Tarassishin, L.; Loudig, O.; Bauman, A.; Shafit-Zagardo, B.; Suh, H.S.; Lee, S.C. Interferon Regulatory Factor 3 Inhibits Astrocyte Inflammatory Gene Expression Through Suppression of the Proinflammatory miR-155 and miR-155. Glia 2011, 59, 1911–1922. [Google Scholar] [CrossRef]
- Quinn; O’Neill, L.A. A trio of microRNAs that control Toll-like receptor signalling. Int. Immunol. 2011, 23, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Kirschner, M.B.; van Zandwijk, N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit. Rev. Oncol. Hematol. 2011, 80, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Link, F.; Krohn, K.; Schumann, J. Identification of stably expressed housekeeping miRNAs in endothelial cells and macrophages in an inflammatory setting. Sci. Rep. 2019, 9, 12786. [Google Scholar] [CrossRef]
- Hu, F.; Dong, X.; Li, W.; Lv, J.; Lin, F.; Song, G.; Hou, G.; Li, R. miR-351-5p aggravates lipopolysaccharide-induced acute lung injury via inhibiting AMPK. Mol. Med. Rep. 2021, 24, 689. [Google Scholar] [CrossRef]
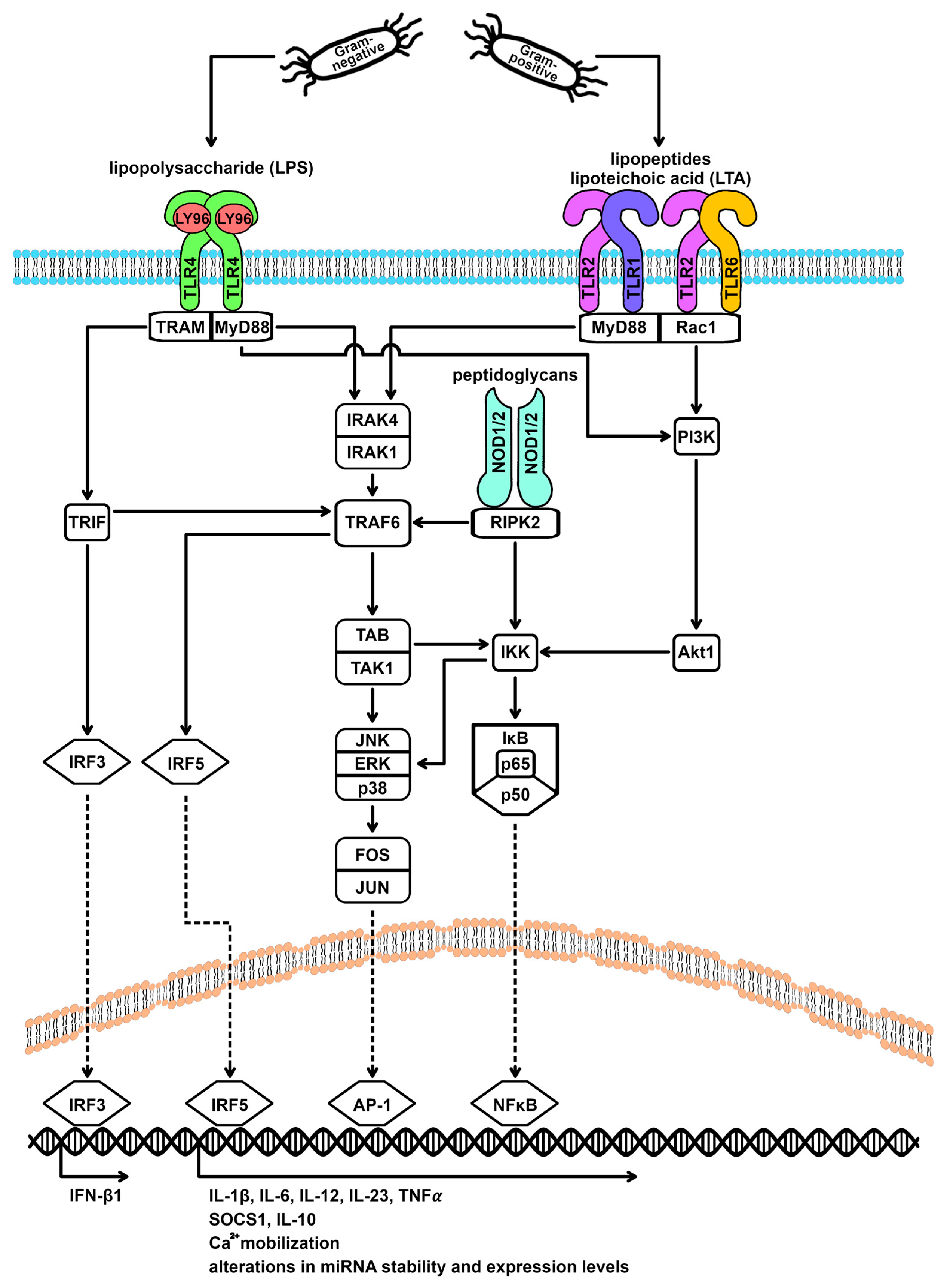

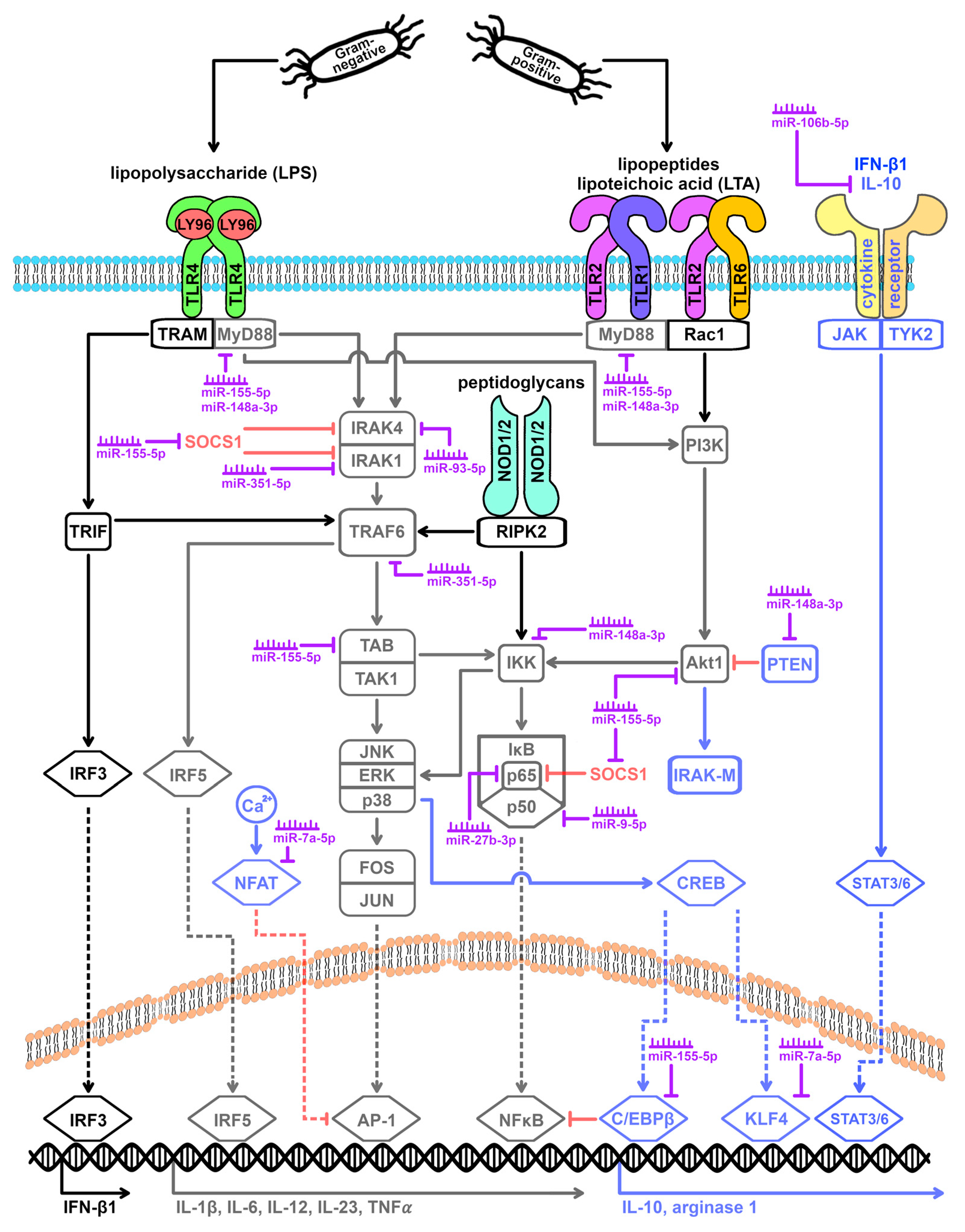
| TLR | Ligands |
|---|---|
| TLR1 | Triacyl lipopeptides |
| TLR2 | Peptidoglycan, lipopeptides, LTA, lipoarabinomannan, GPI anchors, phenol-soluble modulin, zymosan, glycolipids |
| TLR3 | Double-stranded RNA |
| TLR4 | LPS, Taxol, RSV fusion protein, MMTV envelope protein |
| TLR5 | Flagellin |
| TLR6 | Diacyl lipopeptides |
| TLR7 | Single-stranded RNA, imidazoquinolines |
| TLR8 | Single-stranded RNA, imidazoquinolines |
| TLR9 | CpG DNA |
| TLR10 | Unknown |
| TLR11 | Profilin, flagellin |
| TLR12 | Profilin |
| TLR13 | Bacterial 23S risbosomal RNA |
| miRNA | Associated Activation Spectrum | Direction of Expression Change | Reference(s) |
|---|---|---|---|
| miR-7a-5p | M1 | upregulation | [66] |
| miR-9-5p | M1 | downregulation | [66] |
| upregulation | [11,21,58,67] | ||
| miR-27b-3p | M1 | downregulation | [66] |
| upregulation | [74] | ||
| miR-29b-3p | M1 | upregulation | [68] |
| miR-93-5p | M1 | downregulation | [66,75,76] |
| miR-106b-5p | M1 | downregulation | [66,77] |
| miR-127-3p | M1 | upregulation | [11,78] |
| miR-143-3p | M1 | downregulation | [11,69] |
| miR-145-5p | M1 | downregulation | [11,69] |
| miR-147-3p | M1 | upregulation | [67] |
| miR-148a-3p | M1 | upregulation | [66,79,80] |
| miR-155-3p | M1 | upregulation | [11,67] |
| miR-155-5p | M1 | upregulation | [11,58,66,67,68,69,70,71,72,73] |
| miR-204-5p | M1 | upregulation | [11,69] |
| miR-351-5p | M1 | upregulation | [66] |
| miR-451 | M1 | upregulation | [11,69] |
| let-7c | M2 | upregulation | [67] |
| miR-23a-5p | M2 | downregulation | [11] |
| upregulation | [67] | ||
| miR-27a-3p | M2 | upregulation | [67] |
| miR-34a | M2 | downregulation | [11,81] |
| miR-99b | M2 | downregulation | [68] |
| miR-124 | M2 | downregulation | [11] |
| miR-132 | M2 | upregulation | [11,82] |
| miR-193b-3p | M2 | upregulation | [11,68] |
| miR-200a-3p | M2 | upregulation | [11] |
| miR-223 | M2 | downregulation | [11] |
| miR-511 | M2 | upregulation | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rumpel, N.; Riechert, G.; Schumann, J. miRNA-Mediated Fine Regulation of TLR-Induced M1 Polarization. Cells 2024, 13, 701. https://doi.org/10.3390/cells13080701
Rumpel N, Riechert G, Schumann J. miRNA-Mediated Fine Regulation of TLR-Induced M1 Polarization. Cells. 2024; 13(8):701. https://doi.org/10.3390/cells13080701
Chicago/Turabian StyleRumpel, Noah, Georg Riechert, and Julia Schumann. 2024. "miRNA-Mediated Fine Regulation of TLR-Induced M1 Polarization" Cells 13, no. 8: 701. https://doi.org/10.3390/cells13080701





