Flow Cytometry-Based Assay to Detect Alpha Galactosidase Enzymatic Activity at the Cellular Level
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culturing of Jurkat Cell Line
2.2. Mycoplasma Screening
2.3. Generation of GLA KO Jurkat Cells by CRISPR-Cas9 Genome Editing
2.4. Clone Selection and Validation of GLA KO Jurkat Cells
2.5. Flow Cytometric Monitoring of Alpha-Galactosidase A Enzyme Activity
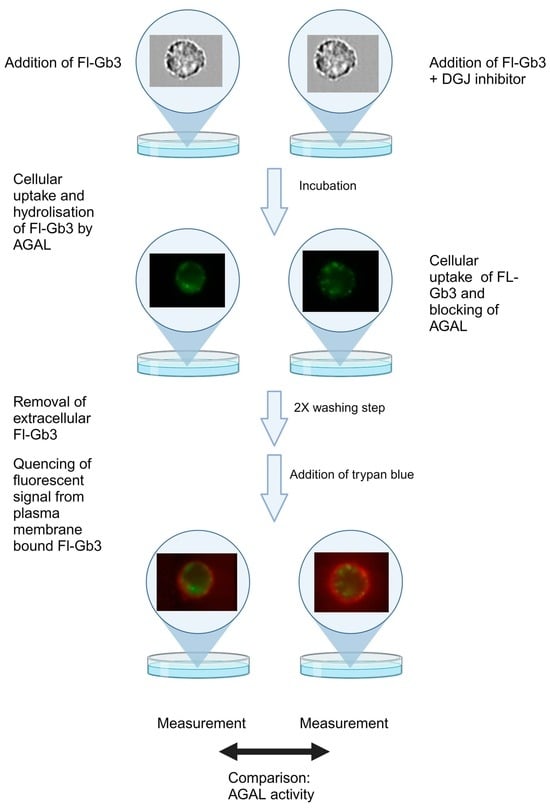
2.6. Imaging Cytometry for the Validation of AGAL Enzymatic Activity
2.7. Statistical Analysis
3. Results
3.1. Flow Cytometric Detection of Alpha-Galactosidase A Enzyme Activity
3.2. Generation of the GLA KO Cell Line
3.3. Validation of the Specificity of the AGAL Enzyme Activity Measurement with a Reversible AGAL Inhibitor (1-Deoxygalactonojirimycin)
3.4. Detection of Specific AGAL Activity in GLA KO Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peters, H.; Ellaway, C.; Nicholls, K.; Reardon, K.; Szer, J. Treatable Lysosomal Storage Diseases in the Advent of Disease-Specific Therapy. Intern. Med. J. 2020, 50, 5–27. [Google Scholar] [CrossRef] [PubMed]
- Platt, F.M.; d’Azzo, A.; Davidson, B.L.; Neufeld, E.F.; Tifft, C.J. Lysosomal Storage Diseases. Nat. Rev. Dis. Primers 2018, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Ellaway, C. Paediatric Fabry Disease. Transl. Pediatr. 2016, 5, 37–42. [Google Scholar] [PubMed]
- Li, Q.; Wang, J.; Tian, M.; Yang, Z.; Yu, L.; Liu, S.; Wang, C.; Wang, X.; Sun, S. Clinical Features and Enzyme Replacement Therapy in 10 Children with Fabry Disease. Front. Pediatr. 2023, 11, 1084336. [Google Scholar] [CrossRef] [PubMed]
- Kermond-Marino, A.; Weng, A.; Xi Zhang, S.K.; Tran, Z.; Huang, M.; Savige, J. Population Frequency of Undiagnosed Fabry Disease in the General Population. Kidney Int. Rep. 2023, 8, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Schiffmann, R.; Fuller, M.; Clarke, L.A.; Aerts, J.M.F.G. Is It Fabry Disease? Genet. Med. 2016, 18, 1181–1185. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.P.; Ferreira, S. Multiple Phenotypic Domains of Fabry Disease and Their Relevance for Establishing Genotype–Phenotype Correlations. Appl. Clin. Genet. 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Yogasundaram, H.; Nikhanj, A.; Putko, B.N.; Boutin, M.; Jain-Ghai, S.; Khan, A.; Auray-Blais, C.; West, M.L.; Oudit, G.Y. Elevated Inflammatory Plasma Biomarkers in Patients with Fabry Disease: A Critical Link to Heart Failure with Preserved Ejection Fraction. J. Am. Heart Assoc. Cardiovasc. Cerebrovasc. Dis. 2018, 7, e009098. [Google Scholar] [CrossRef] [PubMed]
- Weidemann, F.; Sanchez-Niño, M.D.; Politei, J.; Oliveira, J.P.; Wanner, C.; Warnock, D.G.; Ortiz, A. Fibrosis: A Key Feature of Fabry Disease with Potential Therapeutic Implications. Orphanet J. Rare Dis. 2013, 8, 116. [Google Scholar] [CrossRef]
- Kok, K.; Zwiers, K.C.; Boot, R.G.; Overkleeft, H.S.; Aerts, J.M.F.G.; Artola, M. Fabry Disease: Molecular Basis, Pathophysiology, Diagnostics and Potential Therapeutic Directions. Biomolecules 2021, 11, 271. [Google Scholar] [CrossRef]
- Hsu, M.J.; Chang, F.P.; Lu, Y.H.; Hung, S.C.; Wang, Y.C.; Yang, A.H.; Lee, H.J.; Sung, S.H.; Wang, Y.F.; Yu, W.C.; et al. Identification of Lysosomal and Extralysosomal Globotriaosylceramide (Gb3) Accumulations before the Occurrence of Typical Pathological Changes in the Endomyocardial Biopsies of Fabry Disease Patients. Genet. Med. 2019, 21, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Aerts, J.M.; Groener, J.E.; Kuiper, S.; Donker-Koopman, W.E.; Strijland, A.; Ottenhoff, R.; Van Roomen, C.; Mirzaian, M.; Wijburg, F.A.; Linthorst, G.E.; et al. Elevated Globotriaosylsphingosine Is a Hallmark of Fabry Disease. Proc. Natl. Acad. Sci. USA 2008, 105, 2812–2817. [Google Scholar] [CrossRef] [PubMed]
- Sirrs, S.; Bichet, D.G.; Iwanochko, M.R.; Khan, A.; Moore, D.; Oudit, G.; West, M.L. Canadian Fabry Disease Treatment Guidelines; 2019. [Google Scholar]
- Smid, B.E.; Van Der Tol, L.; Cecchi, F.; Elliott, P.M.; Hughes, D.A.; Linthorst, G.E.; Timmermans, J.; Weidemann, F.; West, M.L.; Biegstraaten, M.; et al. Uncertain Diagnosis of Fabry Disease: Consensus Recommendation on Diagnosis in Adults with Left Ventricular Hypertrophy and Genetic Variants of Unknown Significance. Int. J. Cardiol. 2014, 177, 400–408. [Google Scholar] [CrossRef] [PubMed]
- van der Tol, L.; Svarstad, E.; Ortiz, A.; Tøndel, C.; Oliveira, J.P.; Vogt, L.; Waldek, S.; Hughes, D.A.; Lachmann, R.H.; Terryn, W.; et al. Chronic Kidney Disease and an Uncertain Diagnosis of Fabry Disease: Approach to a Correct Diagnosis. Mol. Genet. Metab. 2015, 114, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Kovács, Á.F.; Ponyi, A.; Constantin, T.; Kovács, G.; Németh, K.; Varga, E.; Lengyel, A.; Pinti, É.; Bereczki, D.; Kádár, K.; et al. Fabry-Betegség: Diagnosztikai Útmutató. Gyermekgyógyászat 2021, 72, 73–84. [Google Scholar]
- Miller, D.T.; Lee, K.; Abul-Husn, N.S.; Amendola, L.M.; Brothers, K.; Chung, W.K.; Gollob, M.H.; Gordon, A.S.; Harrison, S.M.; Hershberger, R.E.; et al. ACMG SF v3.2 List for Reporting of Secondary Findings in Clinical Exome and Genome Sequencing: A Policy Statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2023, 25, 100866. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.A.; Aguiar, P.; Lidove, O.; Nicholls, K.; Nowak, A.; Thomas, M.; Torra, R.; Vujkovac, B.; West, M.L.; Feriozzi, S. Do Clinical Guidelines Facilitate or Impede Drivers of Treatment in Fabry Disease? Orphanet J. Rare Dis. 2022, 17, 42. [Google Scholar] [CrossRef] [PubMed]
- Linthorst, G.E.; Poorthuis, B.J.H.M.; Hollak, C.E.M. Enzyme Activity for Determination of Presence of Fabry Disease in Women Results in 40% False-Negative Results. J. Am. Coll. Cardiol. 2008, 51, 2082. [Google Scholar] [CrossRef] [PubMed]
- Germain, D.P.; Altarescu, G.; Barriales-Villa, R.; Mignani, R.; Pawlaczyk, K.; Pieruzzi, F.; Terryn, W.; Vujkovac, B.; Ortiz, A. An Expert Consensus on Practical Clinical Recommendations and Guidance for Patients with Classic Fabry Disease. Mol. Genet. Metab. 2022, 137, 49–61. [Google Scholar] [CrossRef]
- Andrade, J.; Waters, P.J.; Singh, R.S.; Levin, A.; Toh, B.C.; Vallance, H.D.; Sirrs, S. Screening for Fabry Disease in Patients with Chronic Kidney Disease: Limitations of Plasma α-Galactosidase Assay as a Screening Test. Clin. J. Am. Soc. Nephrol. 2008, 3, 139. [Google Scholar] [CrossRef]
- Daitx, V.V.; Mezzalira, J.; Goldim, M.P.d.S.; Coelho, J.C. Comparison between Alpha-Galactosidase A Activity in Blood Samples Collected on Filter Paper, Leukocytes and Plasma. Clin. Biochem. 2012, 45, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Jung, N.; Park, J.W.; Park, H.Y.; Jung, S.C. Epithelial–Mesenchymal Transition in Kidney Tubular Epithelial Cells Induced by Globotriaosylsphingosine and Globotriaosylceramide. PLoS ONE 2015, 10, e0136442. [Google Scholar] [CrossRef] [PubMed]
- Illien, F.; Rodriguez, N.; Amoura, M.; Joliot, A.; Pallerla, M.; Cribier, S.; Burlina, F.; Sagan, S. Quantitative Fluorescence Spectroscopy and Flow Cytometry Analyses of Cell-Penetrating Peptides Internalization Pathways: Optimization, Pitfalls, Comparison with Mass Spectrometry Quantification. Sci. Rep. 2016, 6, 36938. [Google Scholar] [CrossRef]
- Busetto, S.; Trevisan, E.; Patriarca, P.; Menegazzi, R. A Single-Step, Sensitive Flow Cytofluorometric Assay for the Simultaneous Assessment of Membrane-Bound and Ingested Candida Albicans in Phagocytosing Neutrophils. Cytom. Part. A 2004, 58A, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, L.C.S.; Rozenfeld, P.; Gatto, R.G.; Reisin, R.C.; Uchitel, O.D.; Weissmann, C. Upregulation of ASIC1a Channels in an in Vitro Model of Fabry Disease. Neurochem. Int. 2020, 140, 104824. [Google Scholar] [CrossRef]
- Fernández-Delgado, M.; Sendra, L.; Herrero, M.J.; Olivera-Pasquini, G.G.; Batista-Duharte, A.; Aliño, S.F. Study of Oligonucleotides Access and Distribution in Human Peripheral Blood Mononuclear Cells. Int. J. Mol. Sci. 2022, 23, 5839. [Google Scholar] [CrossRef] [PubMed]
- Hallows, W.C.; Skvorak, K.; Agard, N.; Kruse, N.; Zhang, X.; Zhu, Y.; Botham, R.C.; Chng, C.; Shukla, C.; Lao, J.; et al. Optimizing Human α-Galactosidase for Treatment of Fabry Disease. Sci. Rep. 2023, 13, 4748. [Google Scholar] [CrossRef]
- Kingsmore, S.F. Dispatches from Biotech Beginning BeginNGS: Rapid Newborn Genome Sequencing to End the Diagnostic and Therapeutic Odyssey. Am. J. Med. Genet. C Semin. Med. Genet. 2022, 190, 243–256. [Google Scholar] [CrossRef]
- Wojcik, M.H.; Zhang, T.; Ceyhan-Birsoy, O.; Genetti, C.A.; Lebo, M.S.; Yu, T.W.; Parad, R.B.; Holm, I.A.; Rehm, H.L.; Beggs, A.H.; et al. Discordant Results between Conventional Newborn Screening and Genomic Sequencing in the BabySeq Project. Genet. Med. 2021, 23, 1372–1375. [Google Scholar] [CrossRef]
- Spiekerkoetter, U.; Bick, D.; Scott, R.; Hopkins, H.; Krones, T.; Gross, E.S.; Bonham, J.R. Genomic Newborn Screening: Are We Entering a New Era of Screening? J. Inherit. Metab. Dis. 2023, 46, 778–795. [Google Scholar] [CrossRef]
- Kovács, Á.F.; Némethi, Z.; Abonyi, T.; Fekete, G.; Kovács, G.T. Enhancing Molecular Testing for Effective Delivery of Actionable Gene Diagnostics. Bioengineering 2022, 9, 745. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fekete, N.; Li, L.K.; Kozma, G.T.; Fekete, G.; Pállinger, É.; Kovács, Á.F. Flow Cytometry-Based Assay to Detect Alpha Galactosidase Enzymatic Activity at the Cellular Level. Cells 2024, 13, 706. https://doi.org/10.3390/cells13080706
Fekete N, Li LK, Kozma GT, Fekete G, Pállinger É, Kovács ÁF. Flow Cytometry-Based Assay to Detect Alpha Galactosidase Enzymatic Activity at the Cellular Level. Cells. 2024; 13(8):706. https://doi.org/10.3390/cells13080706
Chicago/Turabian StyleFekete, Nóra, Luca Kamilla Li, Gergely Tibor Kozma, György Fekete, Éva Pállinger, and Árpád Ferenc Kovács. 2024. "Flow Cytometry-Based Assay to Detect Alpha Galactosidase Enzymatic Activity at the Cellular Level" Cells 13, no. 8: 706. https://doi.org/10.3390/cells13080706