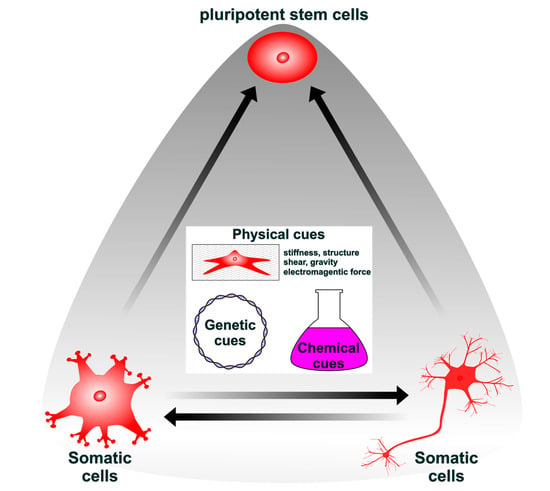
Lineage Reprogramming: Genetic, Chemical, and Physical Cues for Cell Fate Conversion with a Focus on Neuronal Direct Reprogramming and Pluripotency Reprogramming
Abstract
:1. Introduction
2. Genetic, Chemical, and Physical Cues Affect the Induction and Maintenance of Pluripotency
2.1. The Modulation of Signaling Pathways and Epigenetic Statuses Influences the Induction and Maintenance of Pluripotency
2.2. Genetic Cues Phase-Dependently Affect the Induction of Pluripotency
2.3. The Phase-Dependent Effect of Chemical Cues Affects Pluripotency
2.4. Maximum Efficacy of Pluripotency Induction by Chemical Cues Is Obtained by Optimizing the Timing of Treatment
2.5. Physical Cues Generated by the Substrate Surface, Electromagnetic Fields, and Confined Space Affect the Induction of Pluripotency
3. Direct Reprogramming by Genetic, Chemical, and Physical Cues
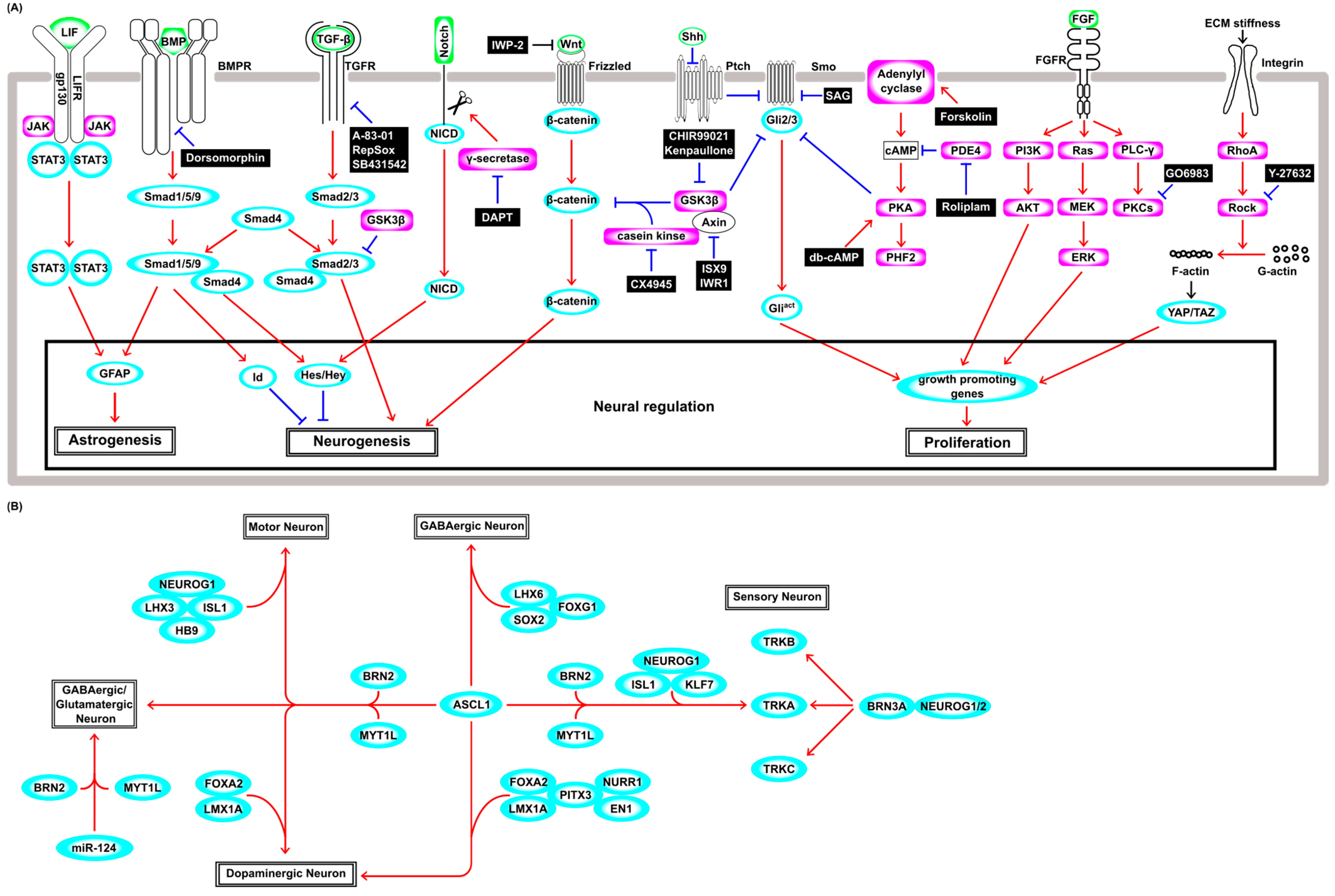
3.1. Direct Reprogramming by Lineage-Specific Factors
3.2. Neuronal Differentiation from Neural Stem Cells in a Physiological Condition
3.3. Direct Neuronal Reprogramming from Fibroblasts by Genetic Cues
3.4. Chemical Cues Boost the Efficiency of Direct Neuronal Reprogramming
3.5. Chemical Direct Reprogramming without Genetic Cues
3.6. Direct Reprogramming by Physical Cues
3.7. Direct Neuronal Reprogramming from Glia
3.8. Direct In Vivo Reprogramming for Treatment of Neural Injury
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
| AAV | Adeno-associated virus |
| AuNGs | Gold nanoparticles |
| bHLH | basic helix–loop–helix |
| BMP | Bone morphogenetic protein |
| C/EBP-α | CCAAT/Enhancer-binding protein-α |
| CiPSCs | Chemically induced pluripotent stem cells |
| DOT1L | Disruptor of telomeric silencing 1-like |
| EL-EMFs | Extremely low-frequency electromagnetic fields |
| Eras | ESC-specific Ras isoform |
| ERK1/2 | Extracellular signal-regulated kinase 1/2 |
| ERVs | Endogenous retroviruses |
| ESCs | Embryonic stem cells |
| FGF | Fibroblast growth factor |
| GABA | Gamma aminobutyric acid |
| GFAP | Glial fibrillary acidic protein |
| Gli | Glioblastoma-associated oncogene |
| GSK3β | Glycogen synthase kinase-3β |
| HDAC | Histone deacetylase |
| Hes | Hairy and enhancer of split |
| hiMNs | human induced motor neurons |
| Id | Inhibitor of differentiation or inhibitor of DNA binding |
| iPSCs | induced pluripotent stem cells |
| JAK-STAT3 | Janus kinase-signal transducers and activators of transcription 3 |
| LATS1/2 | Large tumor suppressor 1/2 |
| LIF | Leukemia inhibitory factor |
| LSD1 | Lysine-specific demethylase 1 |
| Mll2 | Myeloid/mixed-lineage leukemia 2 |
| MST1/2 | Mammalian Ste20-like serine/threonine kinase 1/2 |
| NG2 | Polydendrocytes |
| NICD | Intracellular domain of NOTCH |
| NSC | Neural stem cell |
| OAC1/2 | Oct4-activating compounds 1/2 |
| PDMS | Polydimethylsiloxane |
| PI3K-AKT | Phosphatidyl inositol 3-kinase-protein kinase B |
| PKA | Protein kinase A |
| PKC | Protein kinase C |
| PLCγ | Phospholibase C γ isoform |
| Ptch | Patched |
| REST | Repressor element 1-silencing transcription factor |
| ROCK | Rho kinase |
| SCI | Spinal cord injury |
| Shh | Sonic hedgehog |
| Smad | Small mothers against decapentaplegic |
| Smo | Smoothened |
| Tcf3 | T cell factor 3 |
| TEAD | Transcriptional enhanced associated domain |
| TF | Transcription factor |
| TGF-β | Transforming growth factor-β |
| TSS | Transcription start site |
| vGLUT1 | vecicular glutamate transporter 1 |
| VPA | Valproic acid |
| Wnt | Wingless/Int |
| YAP/TAZ | Yes-associated protein/transcriptional coactivator with PDZ binding motif |
References
- Gurdon, J.B. Adult Frogs Derived from the Nuclei of Single Somatic Cells. Dev. Biol. 1962, 4, 256–273. [Google Scholar] [CrossRef] [PubMed]
- Blau, H.M.; Chiu, C.-P.; Webster, C. Cytoplasmic Activation of Human Nuclear Genes in Stable Heterocaryons. Cell 1983, 32, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Felsenfeld, G. A Brief History of Epigenetics. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. The Yin and Yang of Histone Marks in Transcription. Annu. Rev. Genom. Hum. Genet. 2021, 22, 2021. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; David, B.T.; Trawczynski, M.; Fessler, R.G. Advances in Pluripotent Stem Cells: History, Mechanisms, Technologies, and Applications. Stem Cell Rev. Rep. 2020, 16, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Breunig, J.J.; Haydar, T.F.; Rakic, P. Neural Stem Cells: Historical Perspective and Future Prospects. Neuron 2011, 70, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Blessing, D.; Déglon, N. Adeno-Associated Virus and Lentivirus Vectors: A Refined Toolkit for the Central Nervous System. Curr. Opin. Virol. 2016, 21, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Ogawa, K.; Shimosato, D.; Adachi, K. A Parallel Circuit of LIF Signalling Pathways Maintains Pluripotency of Mouse ES Cells. Nature 2009, 460, 118–122. [Google Scholar] [CrossRef]
- Ohtsuka, S.; Nakai-Futatsugi, Y.; Niwa, H. LIF Signal in Mouse Embryonic Stem Cells. JAKSTAT 2015, 4, e1086520. [Google Scholar] [CrossRef]
- Dahéron, L.; Opitz, S.L.; Zaehres, H.; Lensch, W.M.; Andrews, P.W.; Itskovitz-Eldor, J.; Daley, G.Q. LIF/STAT3 Signaling Fails to Maintain Self-Renewal of Human Embryonic Stem Cells. Stem Cells 2004, 22, 770–778. [Google Scholar] [CrossRef]
- Humphrey, R.K.; Beattie, G.M.; Lopez, A.D.; Bucay, N.; King, C.C.; Firpo, M.T.; Rose-John, S.; Hayek, A. Maintenance of Pluripotency in Human Embryonic Stem Cells Is STAT3 Independent. Stem Cells 2004, 22, 522–530. [Google Scholar] [CrossRef]
- Hollnagel, A.; Oehlmann, V.; Heymer, J.; Rüther, U.; Nordheim, A. Id Genes Are Direct Targets of Bone Morphogenetic Protein Induction in Embryonic Stem Cells. J. Biol. Chem. 1999, 274, 19838–19845. [Google Scholar] [CrossRef]
- Ying, Q.L.; Nichols, J.; Chambers, I.; Smith, A. BMP Induction of Id Proteins Suppresses Differentiation and Sustains Embryonic Stem Cell Self-Renewal in Collaboration with STAT3. Cell 2003, 115, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Meijer, L.; Skaltsounis, L.; Greengard, P.; Brivanlou, A.H. Maintenance of Pluripotency in Human and Mouse Embryonic Stem Cells through Activation of Wnt Signaling by a Pharmacological GSK-3-Specific Inhibitor. Nat. Med. 2004, 10, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Atlasi, Y.; Noori, R.; Gaspar, C.; Franken, P.; Sacchetti, A.; Rafati, H.; Mahmoudi, T.; Decraene, C.; Calin, G.A.; Merrill, B.J.; et al. Wnt Signaling Regulates the Lineage Differentiation Potential of Mouse Embryonic Stem Cells through Tcf3 Down-Regulation. PLoS Genet. 2013, 9, e1003424. [Google Scholar] [CrossRef] [PubMed]
- Blair, K.; Wray, J.; Smith, A. The Liberation of Embryonic Stem Cells. PLoS Genet. 2011, 7, e1002019. [Google Scholar] [CrossRef]
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell 2014, 158, 1254–1269. [Google Scholar] [CrossRef] [PubMed]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of Novel Human Ground State Naive Pluripotent Stem Cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef]
- Boyer, L.A.; Tong, I.L.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef]
- Driskill, J.H.; Pan, D. Control of Stem Cell Renewal and Fate by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2023, 24, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Azzolin, L.; Panciera, T.; Soligo, S.; Enzo, E.; Bicciato, S.; Dupont, S.; Bresolin, S.; Frasson, C.; Basso, G.; Guzzardo, V.; et al. YAP/TAZ Incorporation in the β-Catenin Destruction Complex Orchestrates the Wnt Response. Cell 2014, 158, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Morey, R.; O’Neil, R.C.; He, Y.; Daughtry, B.; Schultz, M.D.; Hariharan, M.; Nery, J.R.; Castanon, R.; Sabatini, K.; et al. Abnormalities in Human Pluripotent Cells Due to Reprogramming Mechanisms. Nature 2014, 511, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Shin, J.Y.; Tonge, P.D.; Puri, M.C.; Lee, S.; Park, H.; Lee, W.C.; Hussein, S.M.I.; Bleazard, T.; Yun, J.Y.; et al. An Epigenomic Roadmap to Induced Pluripotency Reveals DNA Methylation as a Reprogramming Modulator. Nat. Commun. 2014, 5, 5619. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Anokye-Danso, F.; Trivedi, C.M.; Juhr, D.; Gupta, M.; Cui, Z.; Tian, Y.; Zhang, Y.; Yang, W.; Gruber, P.J.; Epstein, J.A.; et al. Highly Efficient MiRNA-Mediated Reprogramming of Mouse and Human Somatic Cells to Pluripotency. Cell Stem Cell 2011, 8, 376–388. [Google Scholar] [CrossRef]
- Polo, J.M.; Anderssen, E.; Walsh, R.M.; Schwarz, B.A.; Nefzger, C.M.; Lim, S.M.; Borkent, M.; Apostolou, E.; Alaei, S.; Cloutier, J.; et al. A Molecular Roadmap of Reprogramming Somatic Cells into IPS Cells. Cell 2012, 151, 1617–1632. [Google Scholar] [CrossRef]
- Rais, Y.; Zviran, A.; Geula, S.; Gafni, O.; Chomsky, E.; Viukov, S.; Mansour, A.A.; Caspi, I.; Krupalnik, V.; Zerbib, M.; et al. Deterministic Direct Reprogramming of Somatic Cells to Pluripotency. Nature 2013, 502, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, B.; Sardina, J.L.; Van Oevelen, C.; Collombet, S.; Kallin, E.M.; Vicent, G.P.; Lu, J.; Thieffry, D.; Beato, M.; Graf, T. C/EBPα Poises B Cells for Rapid Reprogramming into Induced Pluripotent Stem Cells. Nature 2014, 506, 235–239. [Google Scholar] [CrossRef]
- Tan, D.S.; Chen, Y.; Gao, Y.; Bednarz, A.; Wei, Y.; Malik, V.; Ho, D.H.H.; Weng, M.; Ho, S.Y.; Srivastava, Y.; et al. Directed Evolution of an Enhanced POU Reprogramming Factor for Cell Fate Engineering. Mol. Biol. Evol. 2021, 38, 2854–2868. [Google Scholar] [CrossRef]
- MacCarthy, C.M.; Wu, G.; Malik, V.; Menuchin-Lasowski, Y.; Velychko, T.; Keshet, G.; Fan, R.; Bedzhov, I.; Church, G.M.; Jauch, R.; et al. Highly Cooperative Chimeric Super-SOX Induces Naive Pluripotency across Species. Cell Stem Cell 2024, 31, 127–147.e9. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, D.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Snitow, M.; Chen, A.E.; Melton, D.A. Induction of Pluripotent Stem Cells by Defined Factors Is Greatly Improved by Small-Molecule Compounds. Nat. Biotechnol. 2008, 26, 795–797. [Google Scholar] [CrossRef] [PubMed]
- Krämer, O.H.; Zhu, P.; Ostendorff, H.P.; Golebiewski, M.; Tiefenbach, J.; Peters, M.A.; Brill, B.; Groner, B.; Bach, I.; Heinzel, T.; et al. The Histone Deacetylase Inhibitor Valproic Acid Selectively Induces Proteasomal Degradation of HDAC2. EMBO J. 2003, 22, 3411–3420. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tian, E.; Chen, Z.X.; Sun, G.Q.; Ye, P.; Yang, S.; Lu, D.; Xie, J.; Ho, T.V.; Tsark, W.M.; et al. Identification of Oct4-Activating Compounds That Enhance Reprogramming Efficiency. Proc. Natl. Acad. Sci. USA 2012, 109, 20853–20858. [Google Scholar] [CrossRef] [PubMed]
- Ichida, J.K.; Blanchard, J.; Lam, K.; Son, E.Y.; Chung, J.E.; Egli, D.; Loh, K.M.; Carter, A.C.; Di Giorgio, F.P.; Koszka, K.; et al. A Small-Molecule Inhibitor of Tgf-β Signaling Replaces Sox2 in Reprogramming by Inducing Nanog. Cell Stem Cell 2009, 5, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Daigle, S.R.; Olhava, E.J.; Therkelsen, C.A.; Majer, C.R.; Sneeringer, C.J.; Song, J.; Johnston, L.D.; Scott, M.P.; Smith, J.J.; Xiao, Y.; et al. Selective Killing of Mixed Lineage Leukemia Cells by a Potent Small-Molecule DOT1L Inhibitor. Cancer Cell 2011, 20, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Onder, T.T.; Kara, N.; Cherry, A.; Sinha, A.U.; Zhu, N.; Bernt, K.M.; Cahan, P.; Marcarci, B.O.; Unternaehrer, J.; Gupta, P.B.; et al. Chromatin-Modifying Enzymes as Modulators of Reprogramming. Nature 2012, 483, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.P.; Choi, J.; Yoon, J.; Bruder, J.M.; Shin, B.; Kim, J.; Arauzo-Bravo, M.J.; Han, D.; Wu, G.; Han, D.W.; et al. Permissive Epigenomes Endow Reprogramming Competence to Transcriptional Regulators. Nat. Chem. Biol. 2021, 17, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wan, H.; Zhao, X.; Zhu, S.; Zhou, Q.; Ding, S. Brief Report: Combined Chemical Treatment Enables Oct4-Induced Reprogramming from Mouse Embryonic Fibroblasts. Stem Cells 2011, 29, 549–553. [Google Scholar] [CrossRef]
- Chen, J.; Liu, J.; Yang, J.; Chen, Y.; Chen, J.; Ni, S.; Song, H.; Zeng, L.; Ding, K.; Pei, D. BMPs Functionally Replace Klf4 and Support Efficient Reprogramming of Mouse Fibroblasts by Oct4 Alone. Cell Res. 2011, 21, 205–212. [Google Scholar] [CrossRef]
- Moon, J.H.; Heo, J.S.; Kim, J.S.; Jun, E.K.; Lee, J.H.; Kim, A.; Kim, J.; Whang, K.Y.; Kang, Y.K.; Yeo, S.; et al. Reprogramming Fibroblasts into Induced Pluripotent Stem Cells with Bmi1. Cell Res. 2011, 21, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Yin, X.; Yang, W.; Du, Y.; Hou, P.; Ge, J.; Liu, C.; Zhang, W.; Zhang, X.; et al. Generation of IPSCs from Mouse Fibroblasts with a Single Gene, Oct4, and Small Molecules. Cell Res. 2011, 21, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Li, Y.; Zhang, X.; Liu, C.; Guan, J.; Li, H.; Zhao, T.; Ye, J.; Yang, W.; Liu, K.; et al. Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Wang, G.; Wang, J.; Zhang, Z.; Fu, Y.; Cheng, L.; Meng, G.; Lyu, Y.; Zhu, J.; Li, Y.; et al. Chemical Reprogramming of Human Somatic Cells to Pluripotent Stem Cells. Nature 2022, 605, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Liuyang, S.; Wang, G.; Wang, Y.; He, H.; Lyu, Y.; Cheng, L.; Yang, Z.; Guan, J.; Fu, Y.; Zhu, J.; et al. Highly Efficient and Rapid Generation of Human Pluripotent Stem Cells by Chemical Reprogramming. Cell Stem Cell 2023, 30, 450–459.e9. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, Y.; Wang, L.; Ma, X.; Pu, J.; Lin, L.; Deng, Q.; Li, Y.; Wang, W.; Jin, Y.; et al. A Fast Chemical Reprogramming System Promotes Cell Identity Transition through a Diapause-like State. Nat. Cell Biol. 2023, 25, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Panciera, T.; Azzolin, L.; Cordenonsi, M.; Piccolo, S. Mechanobiology of YAP and TAZ in Physiology and Disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Downing, T.L.; Soto, J.; Morez, C.; Houssin, T.; Fritz, A.; Yuan, F.; Chu, J.; Patel, S.; Schaffer, D.V.; Li, S. Biophysical Regulation of Epigenetic State and Cell Reprogramming. Nat. Mater. 2013, 12, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Caiazzo, M.; Okawa, Y.; Ranga, A.; Piersigilli, A.; Tabata, Y.; Lutolf, M.P. Defined Three-Dimensional Microenvironments Boost Induction of Pluripotency. Nat. Mater. 2016, 15, 344–352. [Google Scholar] [CrossRef]
- Baek, S.; Quan, X.; Kim, S.; Lengner, C.; Park, J.K.; Kim, J. Electromagnetic Fields Mediate Efficient Cell Reprogramming into a Pluripotent State. ACS Nano 2014, 8, 10125–10138. [Google Scholar] [CrossRef]
- Roy, B.; Venkatachalapathy, S.; Ratna, P.; Wang, Y.; Jokhun, D.S.; Nagarajan, M.; Shivashankar, G.V. Laterally Confined Growth of Cells Induces Nuclear Reprogramming in the Absence of Exogenous Biochemical Factors. Proc. Natl. Acad. Sci. USA 2018, 115, E4741–E4750. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Yuan, L.; Lee, Y.; Bharti, A.; Mitra, A.; Shivashankar, G.V. Fibroblast Rejuvenation by Mechanical Reprogramming and Redifferentiation. Proc. Natl. Acad. Sci. USA 2020, 117, 10131–10141. [Google Scholar] [CrossRef] [PubMed]
- Mertens, J.; Paquola, A.C.M.; Ku, M.; Hatch, E.; Böhnke, L.; Ladjevardi, S.; McGrath, S.; Campbell, B.; Lee, H.; Herdy, J.R.; et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 2015, 17, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a Single Transfected CDNA Converts Fibmblasts to Myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Dall’Agnese, A.; Caputo, L.; Nicoletti, C.; di Iulio, J.; Schmitt, A.; Gatto, S.; Diao, Y.; Ye, Z.; Forcato, M.; Perera, R.; et al. Transcription Factor-Directed Re-Wiring of Chromatin Architecture for Somatic Cell Nuclear Reprogramming toward Trans-Differentiation. Mol. Cell 2019, 76, 453–472.e8. [Google Scholar] [CrossRef] [PubMed]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Akiyama, M.; Tamura, F.; Isomi, M.; Yamakawa, H.; Sadahiro, T.; Muraoka, N.; Kojima, H.; Haginiwa, S.; Kurotsu, S.; et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell 2018, 22, 91–103.e5. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.R.; Gifford, C.A.; Thomas, R.; Pratt, K.J.B.; Samse-Knapp, K.; Mohamed, T.M.A.; Radzinsky, E.M.; Schricker, A.; Ye, L.; Yu, P.; et al. Context-Specific Transcription Factor Functions Regulate Epigenomic and Transcriptional Dynamics during Cardiac Reprogramming. Cell Stem Cell 2019, 25, 87–102.e9. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Zhang, L.; Gao, Y.; He, Z.; Yao, D.; Wu, Z.; Cen, J.; Chen, X.; Liu, C.; Hu, Y.; et al. Direct Reprogramming of Human Fibroblasts to Functional and Expandable Hepatocytes. Cell Stem Cell 2014, 14, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Suzuki, A. Direct Conversion of Mouse Fibroblasts to Hepatocyte-like Cells by Defined Factors. Nature 2011, 475, 390–395. [Google Scholar] [CrossRef]
- Huang, P.; He, Z.; Ji, S.; Sun, H.; Xiang, D.; Liu, C.; Hu, Y.; Wang, X.; Hui, L. Induction of Functional Hepatocyte-like Cells from Mouse Fibroblasts by Defined Factors. Nature 2011, 475, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Park, C.; Han, J.W.; Kim, J.Y.; Cho, K.; Kim, E.J.; Kim, S.; Lee, S.J.; Oh, S.Y.; Tanaka, Y.; et al. Direct Reprogramming of Human Dermal Fibroblasts into Endothelial Cells Using ER71/ETV2. Circ. Res. 2017, 120, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Desbordes, S.C.; Xie, H.; Sanchez Tillo, E.; Pixley, F.; Stanley, E.R.; Graf, T. PU. 1 and C/EBP/Convert Fibroblasts into Macrophage-like Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 6057–6062. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, S.; Seale, P.; Kubota, K.; Lunsford, E.; Frangioni, J.V.; Gygi, S.P.; Spiegelman, B.M. Initiation of Myoblast to Brown Fat Switch by a PRDM16-C/EBP-β Transcriptional Complex. Nature 2009, 460, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Buganim, Y.; Itskovich, E.; Hu, Y.C.; Cheng, A.W.; Ganz, K.; Sarkar, S.; Fu, D.; Welstead, G.G.; Page, D.C.; Jaenisch, R. Direct Reprogramming of Fibroblasts into Embryonic Sertoli-like Cells by Defined Factors. Cell Stem Cell 2012, 11, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Batta, K.; Florkowska, M.; Kouskoff, V.; Lacaud, G. Direct Reprogramming of Murine Fibroblasts to Hematopoietic Progenitor Cells. Cell Rep. 2014, 9, 1871–1884. [Google Scholar] [CrossRef] [PubMed]
- Szabo, E.; Rampalli, S.; Risueño, R.M.; Schnerch, A.; Mitchell, R.; Fiebig-Comyn, A.; Levadoux-Martin, M.; Bhatia, M. Direct Conversion of Human Fibroblasts to Multilineage Blood Progenitors. Nature 2010, 468, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Ang, H.Y.K.; El Farran, C.A.; Li, P.; Fang, H.T.; Liu, T.M.; Kong, S.L.; Chin, M.L.; Ling, W.Y.; Lim, E.K.H.; et al. Reprogramming Mouse Fibroblasts into Engraftable Myeloerythroid and Lymphoid Progenitors. Nat. Commun. 2016, 7, 13396. [Google Scholar] [CrossRef]
- Yu, B.; He, Z.Y.; You, P.; Han, Q.W.; Xiang, D.; Chen, F.; Wang, M.J.; Liu, C.C.; Lin, X.W.; Borjigin, U.; et al. Reprogramming Fibroblasts into Bipotential Hepatic Stem Cells by Defined Factors. Cell Stem Cell 2013, 13, 328–340. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, N.; Huang, Y.; Spencer, C.I.; Fu, J.D.; Yu, C.; Liu, K.; Nie, B.; Xu, T.; Li, K.; et al. Expandable Cardiovascular Progenitor Cells Reprogrammed from Fibroblasts. Cell Stem Cell 2016, 18, 368–381. [Google Scholar] [CrossRef]
- Ito, N.; Kii, I.; Shimizu, N.; Tanaka, H.; Shin’Ichi, T. Direct Reprogramming of Fibroblasts into Skeletal Muscle Progenitor Cells by Transcription Factors Enriched in Undifferentiated Subpopulation of Satellite Cells. Sci. Rep. 2017, 7, 8097. [Google Scholar] [CrossRef]
- Miura, S.; Suzuki, A. Generation of Mouse and Human Organoid-Forming Intestinal Progenitor Cells by Direct Lineage Reprogramming. Cell Stem Cell 2017, 21, 456–471.e5. [Google Scholar] [CrossRef]
- Najm, F.J.; Lager, A.M.; Zaremba, A.; Wyatt, K.; Caprariello, A.V.; Factor, D.C.; Karl, R.T.; Maeda, T.; Miller, R.H.; Tesar, P.J. Transcription Factor-Mediated Reprogramming of Fibroblasts to Expandable, Myelinogenic Oligodendrocyte Progenitor Cells. Nat. Biotechnol. 2013, 31, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Zuchero, J.B.; Ahlenius, H.; Marro, S.; Ng, Y.H.; Vierbuchen, T.; Hawkins, J.S.; Geissler, R.; Barres, B.A.; Wernig, M. Generation of Oligodendroglial Cells by Direct Lineage Conversion. Nat. Biotechnol. 2013, 31, 434–439. [Google Scholar] [CrossRef]
- Lujan, E.; Chanda, S.; Ahlenius, H.; Südhof, T.C.; Wernig, M. Direct Conversion of Mouse Fibroblasts to Self-Renewing, Tripotent Neural Precursor Cells. Proc. Natl. Acad. Sci. USA 2012, 109, 2527–2532. [Google Scholar] [CrossRef] [PubMed]
- Ring, K.L.; Tong, L.M.; Balestra, M.E.; Javier, R.; Andrews-Zwilling, Y.; Li, G.; Walker, D.; Zhang, W.R.; Kreitzer, A.C.; Huang, Y. Direct Reprogramming of Mouse and Human Fibroblasts into Multipotent Neural Stem Cells with a Single Factor. Cell Stem Cell 2012, 11, 100–109. [Google Scholar] [CrossRef]
- Bonni, A.; Sun, Y.; Nadal-Vicens, M.; Bhatt, A.; Frank, D.A.; Rozovsky, I.; Stahl, N.; Yancopoulos, G.D.; Greenberg, M.E. Regulation of Gliogenesis in the Central Nervous System by the JAK-STAT Signaling Pathway. Science 1997, 278, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yanagisawa, M.; Arakawa, H.; Kimura, N.; Hisatsune, T.; Kawabata, M.; Miyazono, K.; Taga, T. Synergistic Signaling in Fetal Brain by STAT3-Smad1 Complex Brideged by p300. Science 1999, 284, 479–482. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Takizawa, T.; Ochiai, W.; Yanagisawa, M.; Hisatsune, T.; Nakafuku, M.; Miyazono, K.; Kishimoto, T.; Kageyama, R.; Taga, T. BMP2-Mediated Alteration in the Developmental Pathway of Fetal Mouse Brain Cells from Neurogenesis to Astrocytogenesis. Proc. Natl. Acad. Sci. USA 2001, 98, 5868–5873. [Google Scholar] [CrossRef]
- Sueda, R.; Kageyama, R. Regulation of Active and Quiescent Somatic Stem Cells by Notch Signaling. Dev. Growth Differ. 2020, 62, 59–66. [Google Scholar] [CrossRef]
- Takizawa, T.; Ochiai, W.; Nakashima, K.; Taga, T. Enhanced Gene Activation by Notch and BMP Signaling Cross-Talk. Nucleic Acids Res. 2003, 31, 5723–5731. [Google Scholar] [CrossRef] [PubMed]
- Hiew, L.-F.; Poon, C.-H.; You, H.-Z.; Lim, L.-W. TGF-β/Smad Signalling in Neurogenesis: Implications for Neuropsychiatric Diseases. Cells 2021, 10, 1382. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, Y.; Itoh, Y.; Tabata, H.; Nakajima, K.; Akiyama, T.; Masuyama, N.; Gotoh, Y. The Wnt/β-Catenin Pathway Directs Neuronal Differentation of Cortical Neural Precursor Cells. Development 2004, 131, 2791–2801. [Google Scholar] [CrossRef] [PubMed]
- Inestrosa, N.C.; Varela-Nallar, L. Wnt Signalling in Neuronal Differentiation and Development. Cell Tissue Res. 2015, 359, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, A.; Sakai, H.; Xu, Y.; Itoh, Y.; Hirabayashi, Y.; Gotoh, Y. Tcf3 Represses Wnt-β-Catenin Signaling and Maintains Neural Stem Cell Population during Neocortical Development. PLoS ONE 2014, 9, e94408. [Google Scholar] [CrossRef] [PubMed]
- Pietrobono, S.; Gagliardi, S.; Stecca, B. Non-Canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors beyond Smoothened. Front. Genet. 2019, 10, 556. [Google Scholar] [CrossRef] [PubMed]
- Diez del Corral, R.; Morales, A.V. The Multiple Roles of FGF Signaling in the Developing Spinal Cord. Front. Cell Dev. Biol. 2017, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.S.; Sideris, A.; Sutachan, J.J.; Montoya, G.J.V.; Blanck, T.J.J.; Recio-Pinto, E. Differential Regulation of Proliferation and Neuronal Differentiation in Adult Rat Spinal Cord Neural Stem/Progenitors by ERK1/2, Akt, and PLCγ. Front. Mol. Neurosci. 2013, 6, 23. [Google Scholar] [CrossRef]
- Ma, D.K.; Ponnusamy, K.; Song, M.R.; Ming, G.L.; Song, H. Molecular Genetic Analysis of FGFR1 Signalling Reveals Distinct Roles of MAPK and PLC1 Activation for Self-Renewal of Adult Neural Stem Cells. Mol. Brain 2009, 2, 16. [Google Scholar] [CrossRef]
- Corbit, K.C.; Soh, J.-W.; Yoshida, K.; Eves, E.M.; Weinstein, I.B.; Rosner, M.R. Different Protein Kinase C Isoforms Determine Growth Factor Specificity in Neuronal Cells. Mol. Cell Biol. 2000, 20, 5392–5403. [Google Scholar] [CrossRef]
- Geribaldi-Doldán, N.; Flores-Giubi, E.; Murillo-Carretero, M.; García-Bernal, F.; Carrasco, M.; Macías-Sánchez, A.J.; Domínguez-Riscart, J.; Verástegui, C.; Hernández-Galán, R.; Castro, C. 12-Deoxyphorbols Promote Adult Neurogenesis by Inducing Neural Progenitor Cell Proliferation via PKC Activation. Int. J. Neuropsychopharmacol. 2016, 19, pyv085. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Xiao, C.; Shu, Q.; Cheng, B.; Wang, Z.; Xue, R.; Wen, Z.; Wang, J.; Shi, H.; Fan, D.; et al. Cell Response to Mechanical Microenvironment Cues via Rho Signaling: From Mechanobiology to Mechanomedicine. Acta Biomater. 2023, 159, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Chanda, S.; Ang, C.E.; Davila, J.; Pak, C.; Mall, M.; Lee, Q.Y.; Ahlenius, H.; Jung, S.W.; Südhof, T.C.; Wernig, M. Generation of Induced Neuronal Cells by the Single Reprogramming Factor ASCL1. Stem Cell Rep. 2014, 3, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct Conversion of Fibroblasts to Functional Neurons by Defined Factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Marro, S.; Pang, Z.P.; Yang, N.; Tsai, M.C.; Qu, K.; Chang, H.Y.; Südhof, T.C.; Wernig, M. Direct Lineage Conversion of Terminally Differentiated Hepatocytes to Functional Neurons. Cell Stem Cell 2011, 9, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.L.; Vierbuchen, T.; Qu, K.; Lee, Q.Y.; Chanda, S.; Fuentes, D.R.; Giresi, P.G.; Ng, Y.H.; Marro, S.; Neff, N.F.; et al. Hierarchical Mechanisms for Direct Reprogramming of Fibroblasts to Neurons. Cell 2013, 155, 621. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.L.; Lee, Q.Y.; Chen, A.C.; Li, R.; Corces, M.R.; Ang, C.E.; Treutlein, B.; Xiang, C.; Baubet, V.; Suchy, F.P.; et al. Rapid Chromatin Switch in the Direct Reprogramming of Fibroblasts to Neurons. Cell Rep. 2017, 20, 3236–3247. [Google Scholar] [CrossRef]
- Pang, Z.P.; Yang, N.; Vierbuchen, T.; Ostermeier, A.; Fuentes, D.R.; Yang, T.Q.; Citri, A.; Sebastiano, V.; Marro, S.; Südhof, T.C.; et al. Induction of Human Neuronal Cells by Defined Transcription Factors. Nature 2011, 476, 220–223. [Google Scholar] [CrossRef]
- Son, E.Y.; Ichida, J.K.; Wainger, B.J.; Toma, J.S.; Rafuse, V.F.; Woolf, C.J.; Eggan, K. Conversion of Mouse and Human Fibroblasts into Functional Spinal Motor Neurons. Cell Stem Cell 2011, 9, 205–218. [Google Scholar] [CrossRef]
- Pfisterer, U.; Kirkeby, A.; Torper, O.; Wood, J.; Nelander, J.; Dufour, A.; Björklund, A.; Lindvall, O.; Jakobsson, J.; Parmar, M. Direct Conversion of Human Fibroblasts to Dopaminergic Neurons. Proc. Natl. Acad. Sci. USA 2011, 108, 10343–10348. [Google Scholar] [CrossRef]
- Torper, O.; Pfisterer, U.; Wolf, D.A.; Pereira, M.; Lau, S.; Jakobsson, J.; Björklund, A.; Grealish, S.; Parmar, M. Generation of Induced Neurons via Direct Conversion in Vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 7038–7043. [Google Scholar] [CrossRef] [PubMed]
- Colasante, G.; Lignani, G.; Rubio, A.; Medrihan, L.; Yekhlef, L.; Sessa, A.; Massimino, L.; Giannelli, S.G.; Sacchetti, S.; Caiazzo, M.; et al. Rapid Conversion of Fibroblasts into Functional Forebrain GABAergic Interneurons by Direct Genetic Reprogramming. Cell Stem Cell 2015, 17, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Martinat, C.; Bacci, J.-J.; Leete, T.; Kim, J.; Vanti, W.B.; Newman, A.H.; Cha, J.H.; Gether, U.; Wang, H.; Abeliovich, A. Cooperative Transcription Activation by Nurr1 and Pitx3 Induces Embryonic Stem Cell Maturation to the Midbrain Dopamine Neuron Phenotype. Proc. Natl. Acad. Sci. USA 2006, 103, 2874–2879. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Su, S.C.; Wang, H.; Cheng, A.W.; Cassady, J.P.; Lodato, M.A.; Lengner, C.J.; Chung, C.Y.; Dawlaty, M.M.; Tsai, L.H.; et al. Functional Integration of Dopaminergic Neurons Directly Converted from Mouse Fibroblasts. Cell Stem Cell 2011, 9, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Leung, A.; Han, B.S.; Chang, M.Y.; Moon, J., II; Kim, C.H.; Hong, S.; Pruszak, J.; Isacson, O.; Kim, K.S. Wnt1-Lmx1a Forms a Novel Autoregulatory Loop and Controls Midbrain Dopaminergic Differentiation Synergistically with the SHH-FoxA2 Pathway. Cell Stem Cell 2009, 5, 646–658. [Google Scholar] [CrossRef] [PubMed]
- Caiazzo, M.; Dell’Anno, M.T.; Dvoretskova, E.; Lazarevic, D.; Taverna, S.; Leo, D.; Sotnikova, T.D.; Menegon, A.; Roncaglia, P.; Colciago, G.; et al. Direct Generation of Functional Dopaminergic Neurons from Mouse and Human Fibroblasts. Nature 2011, 476, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Dell’Anno, M.T.; Caiazzo, M.; Leo, D.; Dvoretskova, E.; Medrihan, L.; Colasante, G.; Giannelli, S.; Theka, I.; Russo, G.; Mus, L.; et al. Remote Control of Induced Dopaminergic Neurons in Parkinsonian Rats. J. Clin. Investig. 2014, 124, 3215–3229. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, J.W.; Eade, K.T.; Szůcs, A.; Lo Sardo, V.; Tsunemoto, R.K.; Williams, D.; Sanna, P.P.; Baldwin, K.K. Selective Conversion of Fibroblasts into Peripheral Sensory Neurons. Nat. Neurosci. 2015, 18, 25–35. [Google Scholar] [CrossRef]
- Wainger, B.J.; Buttermore, E.D.; Oliveira, J.T.; Mellin, C.; Lee, S.; Saber, W.A.; Wang, A.J.; Ichida, J.K.; Chiu, I.M.; Barrett, L.; et al. Modeling Pain in Vitro Using Nociceptor Neurons Reprogrammed from Fibroblasts. Nat. Neurosci. 2015, 18, 17–24. [Google Scholar] [CrossRef]
- Yoo, A.S.; Sun, A.X.; Li, L.; Shcheglovitov, A.; Portmann, T.; Li, Y.; Lee-Messer, C.; Dolmetsch, R.E.; Tsien, R.W.; Crabtree, G.R. MicroRNA-Mediated Conversion of Human Fibroblasts to Neurons. Nature 2011, 476, 228–231. [Google Scholar] [CrossRef]
- Ambasudhan, R.; Talantova, M.; Coleman, R.; Yuan, X.; Zhu, S.; Lipton, S.A.; Ding, S. Direct Reprogramming of Adult Human Fibroblasts to Functional Neurons under Defined Conditions. Cell Stem Cell 2011, 9, 113–118. [Google Scholar] [CrossRef]
- Victor, M.B.; Richner, M.; Hermanstyne, T.O.; Ransdell, J.L.; Sobieski, C.; Deng, P.Y.; Klyachko, V.A.; Nerbonne, J.M.; Yoo, A.S. Generation of Human Striatal Neurons by MicroRNA-Dependent Direct Conversion of Fibroblasts. Neuron 2014, 84, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cao, N.; Spencer, C.I.; Nie, B.; Ma, T.; Xu, T.; Zhang, Y.; Wang, X.; Srivastava, D.; Ding, S. Small Molecules Enable Cardiac Reprogramming of Mouse Fibroblasts with a Single Factor, Oct4. Cell Rep. 2014, 6, 951–960. [Google Scholar] [CrossRef]
- Zhu, S.; Ambasudhan, R.; Sun, W.; Kim, H.J.; Talantova, M.; Wang, X.; Zhang, M.; Zhang, Y.; Laurent, T.; Parker, J.; et al. Small Molecules Enable OCT4-Mediated Direct Reprogramming into Expandable Human Neural Stem Cells. Cell Res. 2014, 24, 126–129. [Google Scholar] [CrossRef]
- Liu, M.L.; Zang, T.; Zou, Y.; Chang, J.C.; Gibson, J.R.; Huber, K.M.; Zhang, C.L. Small Molecules Enable Neurogenin 2 to Efficiently Convert Human Fibroblasts into Cholinergic Neurons. Nat. Commun. 2013, 4, 2183. [Google Scholar] [CrossRef]
- Liu, M.L.; Zang, T.; Zhang, C.L. Direct Lineage Reprogramming Reveals Disease-Specific Phenotypes of Motor Neurons from Human ALS Patients. Cell Rep. 2016, 14, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Karow, M.; Sánchez, R.; Schichor, C.; Masserdotti, G.; Ortega, F.; Heinrich, C.; Gascón, S.; Khan, M.A.; Lie, D.C.; Dellavalle, A.; et al. Reprogramming of Pericyte-Derived Cells of the Adult Human Brain into Induced Neuronal Cells. Cell Stem Cell 2012, 11, 471–476. [Google Scholar] [CrossRef]
- Karow, M.; Gray Camp, J.; Falk, S.; Gerber, T.; Pataskar, A.; Gac-Santel, M.; Kageyama, J.; Brazovskaja, A.; Garding, A.; Fan, W.; et al. Direct Pericyte-to-Neuron Reprogramming via Unfolding of a Neural Stem Cell-like Program. Nat. Neurosci. 2018, 21, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Huang, C.; Xu, X.; Gu, H.; Ye, Y.; Jiang, C.; Qiu, Z.; Xie, X. Direct Reprogramming of Mouse Fibroblasts into Cardiomyocytes with Chemical Cocktails. Cell Res. 2015, 25, 1013–1024. [Google Scholar] [CrossRef]
- Cao, N.; Huang, Y.; Zheng, J.; Spencer, C.I.; Zhang, Y.; Fu, J.; Nie, B.; Xie, M.; Zhang, M.; Wang, H.; et al. Conversion of Human Fibroblasts into Functional Cardiomyocytes by Small Molecules. Science 2016, 352, 1216–1220. [Google Scholar] [CrossRef]
- Tian, E.; Sun, G.; Sun, G.; Chao, J.; Ye, P.; Warden, C.; Riggs, A.D.; Shi, Y. Small-Molecule-Based Lineage Reprogramming Creates Functional Astrocytes. Cell Rep. 2016, 16, 781–792. [Google Scholar] [CrossRef]
- Hu, W.; Qiu, B.; Guan, W.; Wang, Q.; Wang, M.; Li, W.; Gao, L.; Shen, L.; Huang, Y.; Xie, G.; et al. Direct Conversion of Normal and Alzheimer’s Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell 2015, 17, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zuo, X.; Jing, J.; Ma, Y.; Wang, J.; Liu, D.; Zhu, J.; Du, X.; Xiong, L.; Du, Y.; et al. Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 2015, 17, 195–203. [Google Scholar] [CrossRef]
- Mahato, B.; Kaya, K.D.; Fan, Y.; Sumien, N.; Shetty, R.A.; Zhang, W.; Davis, D.; Mock, T.; Batabyal, S.; Ni, A.; et al. Pharmacologic Fibroblast Reprogramming into Photoreceptors Restores Vision. Nature 2020, 581, 83–88. [Google Scholar] [CrossRef]
- Sahu, M.R.; Mondal, A.C. Neuronal Hippo Signaling: From Development to Diseases. Dev. Neurobiol. 2021, 81, 92–109. [Google Scholar] [CrossRef] [PubMed]
- Meli, V.S.; Veerasubramanian, K.; Downing, T.L.; Wang, W.; Liu, W.F. Mechanosensation to Inflammation: Roles for YAP/TAZ in Innate Immune Cells. Sci. Signal 2023, 16, eadc9656. [Google Scholar] [CrossRef]
- Della Chiara, G.; Gervasoni, F.; Fakiola, M.; Godano, C.; D’Oria, C.; Azzolin, L.; Bonnal, R.J.P.; Moreni, G.; Drufuca, L.; Rossetti, G.; et al. Epigenomic Landscape of Human Colorectal Cancer Unveils an Aberrant Core of Pan-Cancer Enhancers Orchestrated by YAP/TAZ. Nat. Commun. 2021, 12, 2340. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.K.; Mei, S.C.; Chen, E.H.; Zheng, Y.; Pan, D. YAP Induces an Oncogenic Transcriptional Program through TET1-Mediated Epigenetic Remodeling in Liver Growth and Tumorigenesis. Nat. Genet. 2022, 54, 1202–1213. [Google Scholar] [CrossRef]
- Li, H.; Wu, B.K.; Kanchwala, M.; Cai, J.; Wang, L.; Xing, C.; Zheng, Y.; Pan, D. YAP/TAZ Drives Cell Proliferation and Tumour Growth via a Polyamine–EIF5A Hypusination–LSD1 Axis. Nat. Cell Biol. 2022, 24, 373–383. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in Mechanotransduction. Nature 2011, 474, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, Y.; Li, P.; Sun, Y.; Lv, S.; Wang, Y.; He, X.; Xu, J.; Xu, Z.; Li, L.; et al. Soft Substrates Promote Direct Chemical Reprogramming of Fibroblasts into Neurons. Acta Biomater. 2022, 152, 255–272. [Google Scholar] [CrossRef]
- Kurotsu, S.; Sadahiro, T.; Fujita, R.; Tani, H.; Yamakawa, H.; Tamura, F.; Isomi, M.; Kojima, H.; Yamada, Y.; Abe, Y.; et al. Soft Matrix Promotes Cardiac Reprogramming via Inhibition of YAP/TAZ and Suppression of Fibroblast Signatures. Stem Cell Rep. 2020, 15, 612–628. [Google Scholar] [CrossRef]
- Yoo, J.; Noh, M.; Kim, H.; Jeon, N.L.; Kim, B.S.; Kim, J. Nanogrooved Substrate Promotes Direct Lineage Reprogramming Offibroblasts to Functional Induced Dopaminergic Neurons. Biomaterials 2015, 45, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Sia, J.; Yu, P.; Srivastava, D.; Li, S. Effect of Biophysical Cues on Reprogramming to Cardiomyocytes. Biomaterials 2016, 103, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Conley, B.M.; Rathnam, C.; Cho, H.Y.; Pongkulapa, T.; Conklin, B.; Lee, K.B. Predictive Biophysical Cue Mapping for Direct Cell Reprogramming Using Combinatorial Nanoarrays. ACS Nano 2022, 16, 5577–5586. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Lee, E.; Kim, H.Y.; Youn, D.H.; Jung, J.; Kim, H.; Chang, Y.; Lee, W.; Shin, J.; Baek, S.; et al. Electromagnetized Gold Nanoparticles Mediate Direct Lineage Reprogramming into Induced Dopamine Neurons in Vivo for Parkinson’s Disease Therapy. Nat. Nanotechnol. 2017, 12, 1006–1014. [Google Scholar] [CrossRef]
- Song, Y.; Soto, J.; Chen, B.; Hoffman, T.; Zhao, W.; Zhu, N.; Peng, Q.; Liu, L.; Ly, C.; Wong, P.K.; et al. Transient Nuclear Deformation Primes Epigenetic State and Promotes Cell Reprogramming. Nat. Mater. 2022, 21, 1191–1199. [Google Scholar] [CrossRef]
- Clifford, T.; Finkel, Z.; Rodriguez, B.; Joseph, A.; Cai, L. Current Advancements in Spinal Cord Injury Research—Glial Scar Formation and Neural Regeneration. Cells 2023, 12, 853. [Google Scholar] [CrossRef]
- Heins, N.; Malatesta, P.; Cecconi, F.; Nakafuku, M.; Tucker, K.L.; Hack, M.A.; Chapouton, P.; Barde, Y.A.; Goötz, M. Glial Cells Generate Neurons: The Role of the Transcription Factor Pax6. Nat. Neurosci. 2002, 5, 308–315. [Google Scholar] [CrossRef]
- Berninger, B.; Costa, M.R.; Koch, U.; Schroeder, T.; Sutor, B.; Grothe, B.; Götz, M. Functional Properties of Neurons Derived from in Vitro Reprogrammed Postnatal Astroglia. J. Neurosci. 2007, 27, 8654–8664. [Google Scholar] [CrossRef] [PubMed]
- Masserdotti, G.; Gillotin, S.; Sutor, B.; Drechsel, D.; Irmler, M.; Jørgensen, H.F.; Sass, S.; Theis, F.J.; Beckers, J.; Berninger, B.; et al. Transcriptional Mechanisms of Proneural Factors and REST in Regulating Neuronal Reprogramming of Astrocytes. Cell Stem Cell 2015, 17, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Petryniak, M.A.; Potter, G.B.; Rowitch, D.H.; Rubenstein, J.L.R. Dlx1 and Dlx2 Control Neuronal versus Oligodendroglial Cell Fate Acquisition in the Developing Forebrain. Neuron 2007, 55, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, C.; Blum, R.; Gascón, S.; Masserdotti, G.; Tripathi, P.; Sánchez, R.; Tiedt, S.; Schroeder, T.; Götz, M.; Berninger, B. Directing Astroglia from the Cerebral Cortex into Subtype Specific Functional Neurons. PLoS Biol. 2010, 8, e1000373. [Google Scholar] [CrossRef] [PubMed]
- Kadkhodaei, B.; Ito, T.; Joodmardi, E.; Mattsson, B.; Rouillard, C.; Carta, M.; Muramatsu, S.I.; Sumi-Ichinose, C.; Nomura, T.; Metzger, D.; et al. Nurr1 Is Required for Maintenance of Maturing and Adult Midbrain Dopamine Neurons. J. Neurosci. 2009, 29, 15923–15932. [Google Scholar] [CrossRef] [PubMed]
- Addis, R.C.; Hsu, F.C.; Wright, R.L.; Dichter, M.A.; Coulter, D.A.; Gearhart, J.D. Efficient Conversion of Astrocytes to Functional Midbrain Dopaminergic Neurons Using a Single Polycistronic Vector. PLoS ONE 2011, 6, e28719. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhang, L.; Wu, Z.; Chen, Y.; Wang, F.; Chen, G. In Vivo Direct Reprogramming of Reactive Glial Cells into Functional Neurons after Brain Injury and in an Alzheimer’s Disease Model. Cell Stem Cell 2014, 14, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, T.; Irie, T.; Katsurabayashi, S.; Hayashi, Y.; Nagai, T.; Hamazaki, N.; Adefuin, A.M.D.; Miura, F.; Ito, T.; Kimura, H.; et al. Pioneer Factor NeuroD1 Rearranges Transcriptional and Epigenetic Profiles to Execute Microglia-Neuron Conversion. Neuron 2019, 101, 472–485.e7. [Google Scholar] [CrossRef] [PubMed]
- Matsuda-Ito, K.; Matsuda, T.; Nakashima, K. Expression Level of the Reprogramming Factor NeuroD1 Is Critical for Neuronal Conversion Efficiency from Different Cell Types. Sci. Rep. 2022, 12, 17980. [Google Scholar] [CrossRef]
- Niu, W.; Zang, T.; Zou, Y.; Fang, S.; Smith, D.K.; Bachoo, R.; Zhang, C.L. In Vivo Reprogramming of Astrocytes to Neuroblasts in the Adult Brain. Nat. Cell Biol. 2013, 15, 1164–1175. [Google Scholar] [CrossRef]
- Su, Z.; Niu, W.; Liu, M.L.; Zou, Y.; Zhang, C.L. In Vivo Conversion of Astrocytes to Neurons in the Injured Adult Spinal Cord. Nat. Commun. 2014, 5, 3338. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Su, Z.; Tai, W.; Zou, Y.; Xu, X.M.; Zhang, C.L. The P53 Pathway Controls SOX2-Mediated Reprogramming in the Adult Mouse Spinal Cord. Cell Rep. 2016, 17, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, B.; Cananzi, S.; Han, C.; Wang, L.-L.; Zou, Y.; Fu, Y.-X.; Hon, G.C.; Zhang, C.-L. A Single Factor Elicits Multilineage Reprogramming of Astrocytes in the Adult Mouse Striatum. Proc. Natl. Acad. Sci. USA 2022, 119, e2107339119. [Google Scholar] [CrossRef]
- Chen, Y.C.; Ma, N.X.; Pei, Z.F.; Wu, Z.; Do-Monte, F.H.; Keefe, S.; Yellin, E.; Chen, M.S.; Yin, J.C.; Lee, G.; et al. A NeuroD1 AAV-Based Gene Therapy for Functional Brain Repair after Ischemic Injury through In Vivo Astrocyte-to-Neuron Conversion. Mol. Ther. 2020, 28, 217–234. [Google Scholar] [CrossRef]
- Torper, O.; Ottosson, D.R.; Pereira, M.; Lau, S.; Cardoso, T.; Grealish, S.; Parmar, M. InVivo Reprogramming of Striatal NG2 Glia into Functional Neurons That Integrate into Local Host Circuitry. Cell Rep. 2015, 12, 474–481. [Google Scholar] [CrossRef]
- Tai, W.; Wu, W.; Wang, L.L.; Ni, H.; Chen, C.; Yang, J.; Zang, T.; Zou, Y.; Xu, X.M.; Zhang, C.L. In Vivo Reprogramming of NG2 Glia Enables Adult Neurogenesis and Functional Recovery Following Spinal Cord Injury. Cell Stem Cell 2021, 28, 923–937.e4. [Google Scholar] [CrossRef]
- Biddy, B.A.; Kong, W.; Kamimoto, K.; Guo, C.; Waye, S.E.; Sun, T.; Morris, S.A. Single-Cell Mapping of Lineage and Identity in Direct Reprogramming. Nature 2018, 564, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Li, B.; Bhakta, M.; Xie, S.; Zhou, P.; Munshi, N.V.; Hon, G.C. Rational Reprogramming of Cellular States by Combinatorial Perturbation. Cell Rep. 2019, 27, 3486–3499.e6. [Google Scholar] [CrossRef]
- Masaki, T.; Qu, J.; Cholewa-Waclaw, J.; Burr, K.; Raaum, R.; Rambukkana, A. Reprogramming Adult Schwann Cells to Stem Cell-like Cells by Leprosy Bacilli Promotes Dissemination of Infection. Cell 2013, 152, 51–67. [Google Scholar] [CrossRef]
- Ohta, K.; Kawano, R.; Ito, N. Lactic Acid Bacteria Convert Human Fibroblasts to Multipotent Cells. PLoS ONE 2012, 7, e51866. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umeyama, T.; Matsuda, T.; Nakashima, K. Lineage Reprogramming: Genetic, Chemical, and Physical Cues for Cell Fate Conversion with a Focus on Neuronal Direct Reprogramming and Pluripotency Reprogramming. Cells 2024, 13, 707. https://doi.org/10.3390/cells13080707
Umeyama T, Matsuda T, Nakashima K. Lineage Reprogramming: Genetic, Chemical, and Physical Cues for Cell Fate Conversion with a Focus on Neuronal Direct Reprogramming and Pluripotency Reprogramming. Cells. 2024; 13(8):707. https://doi.org/10.3390/cells13080707
Chicago/Turabian StyleUmeyama, Taichi, Taito Matsuda, and Kinichi Nakashima. 2024. "Lineage Reprogramming: Genetic, Chemical, and Physical Cues for Cell Fate Conversion with a Focus on Neuronal Direct Reprogramming and Pluripotency Reprogramming" Cells 13, no. 8: 707. https://doi.org/10.3390/cells13080707