Microbial Genes for a Circular and Sustainable Bio-PET Economy
Abstract
:1. Introduction
2. PET Metabolism
2.1. Enzymatic Hydrolysis of PET
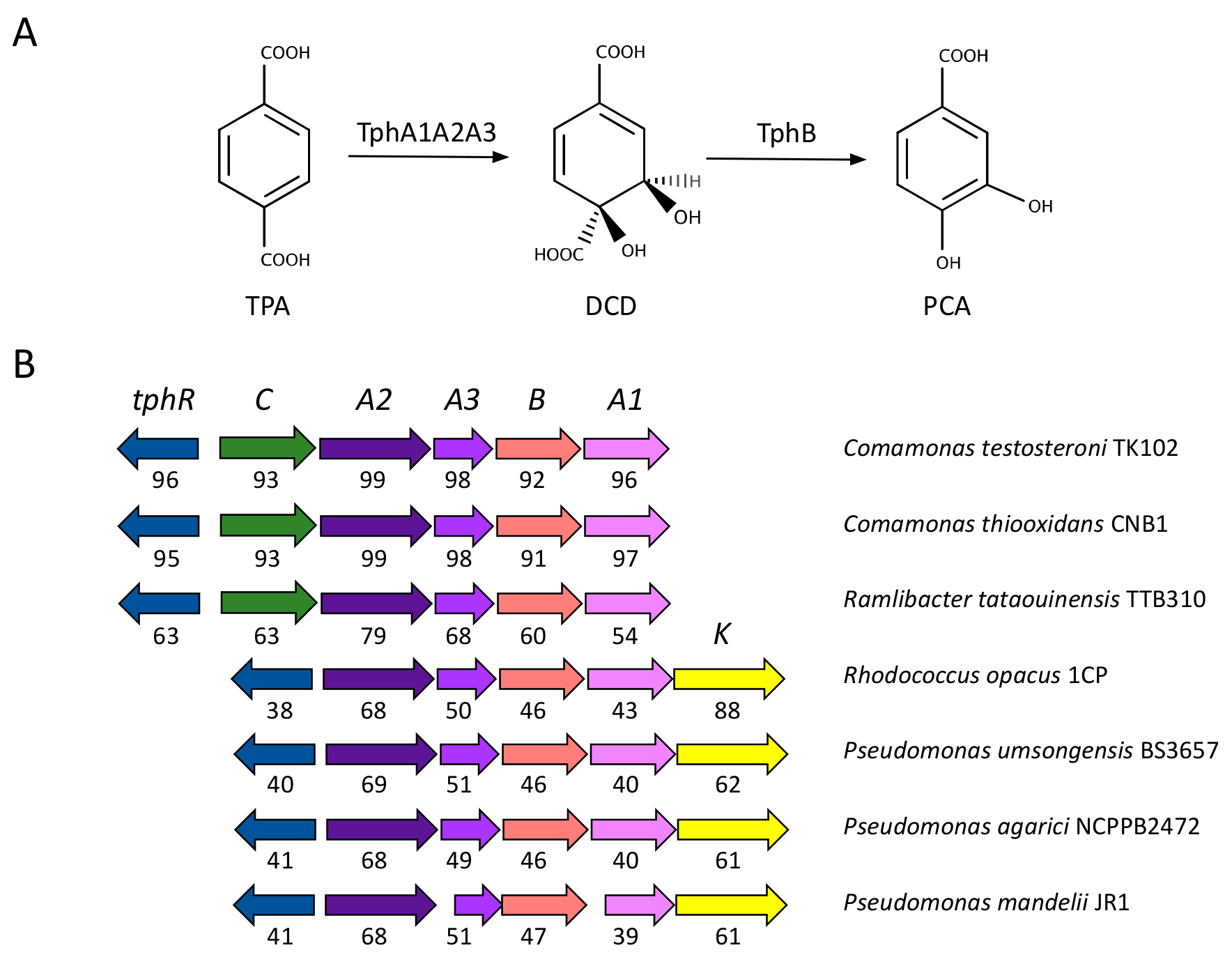
2.2. Metabolism of TPA
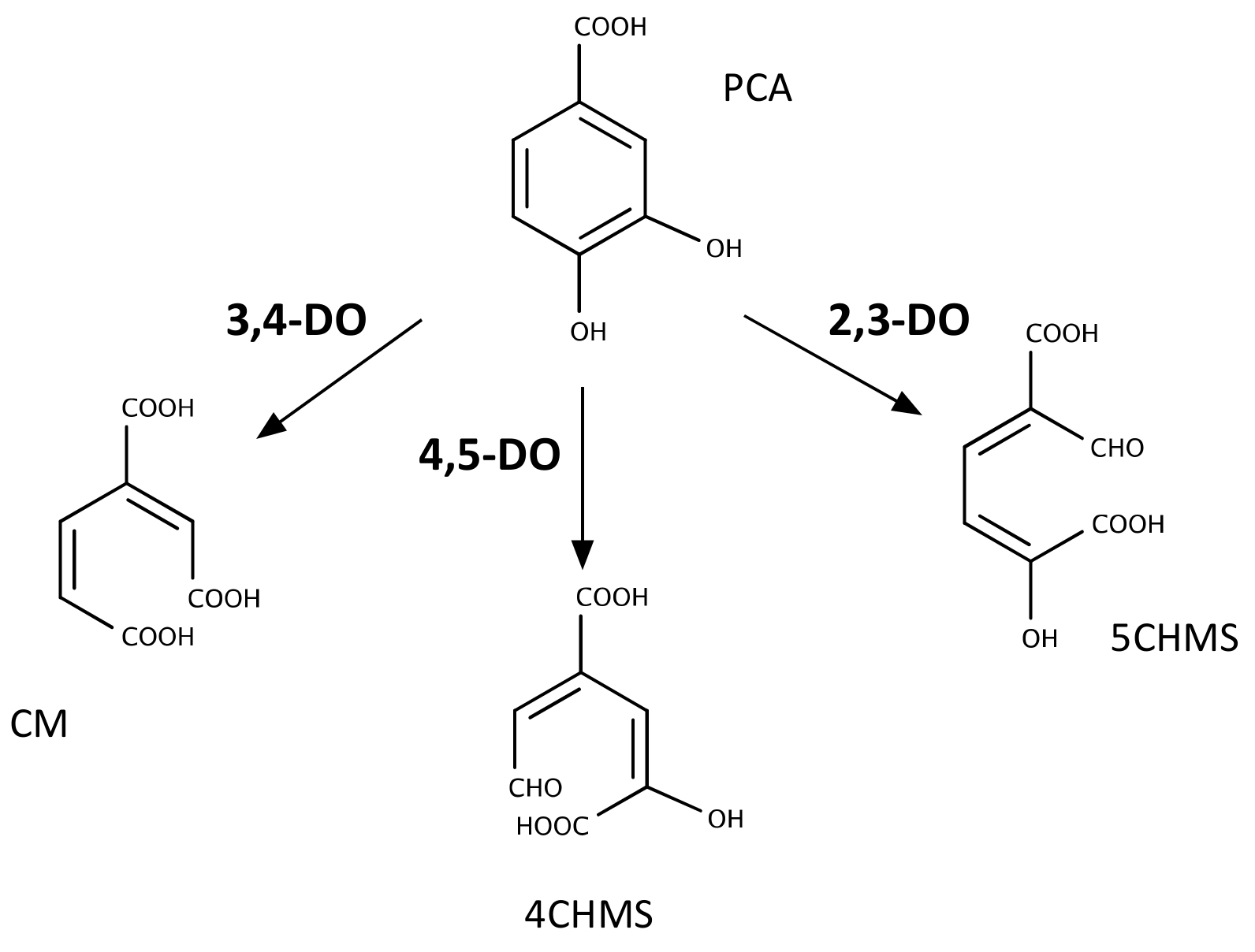
2.3. Metabolism of PCA
2.4. Metabolism of EG
3. Anabolism of Monomers Used for Bio-PET Synthesis
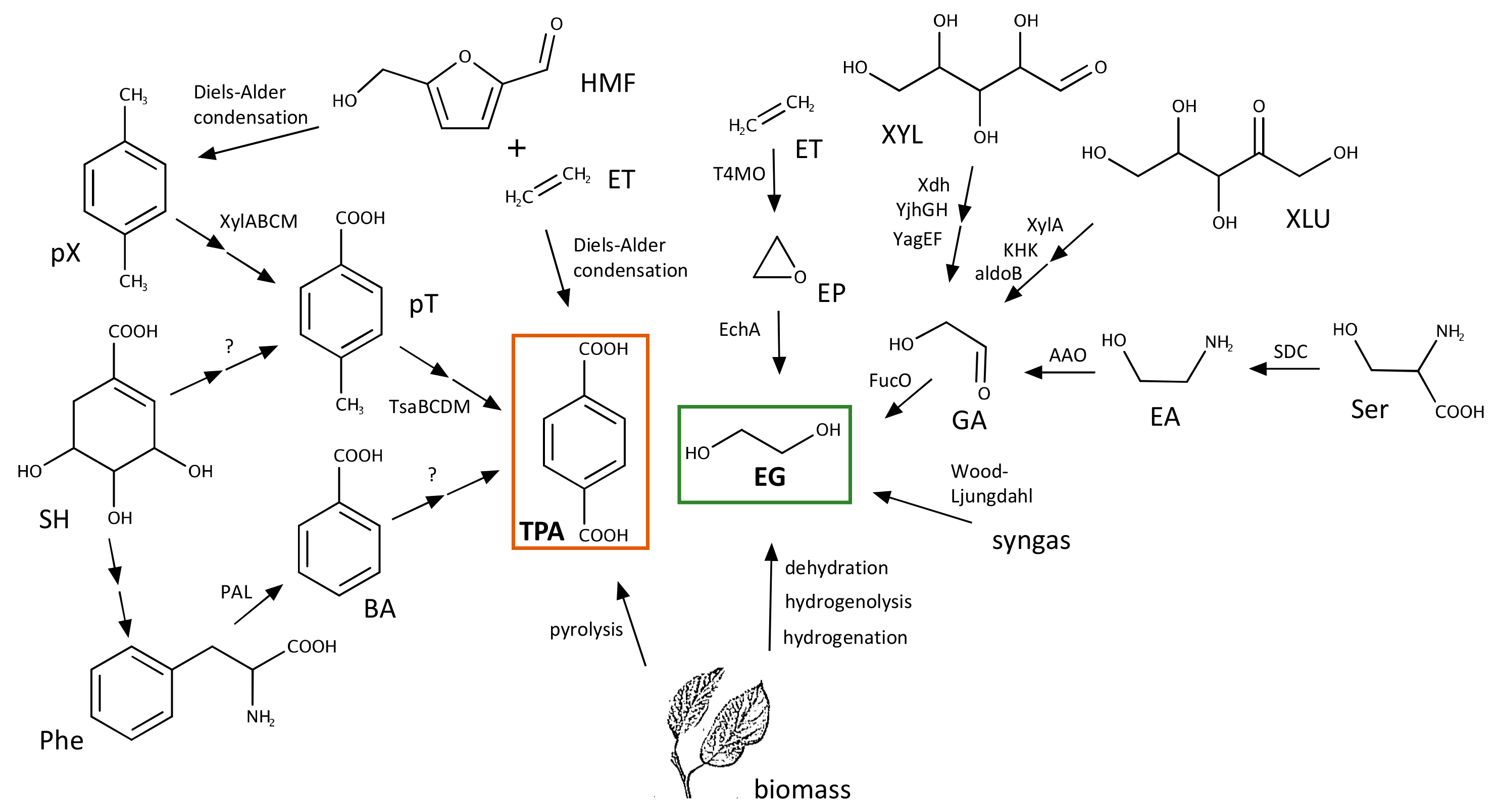
3.1. Biosynthesis of TPA
3.2. Biosynthesis of EG
4. Future Prospects and Concluding Remarks
Funding
Conflicts of Interest
References
- Andrady, A.L.; Neal, M.A. Applications and societal benefits of plastics. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 1977–1984. [Google Scholar] [CrossRef] [PubMed]
- Scalenghe, R. Resource or waste? A perspective of plastics degradation in soil with a focus on end-of-life options. Heliyon 2018, 4, e00941. [Google Scholar] [CrossRef]
- PlasticsEurope: Plastics—The Facts 2018. Available online: https://www.plasticseurope.org/en/resources/publications/619-plastics-facts-2018 (accessed on 16 May 2019).
- Upasen, S.; Wattanachai, P. Packaging to prolong shelf life of preservative-free white bread. Heliyon 2018, 4, e00802. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.G.; Moore, C.J.; van Franeker, J.A.; Moloney, C.L. Monitoring the abundance of plastic debris in the marine environment. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Barnes, D.K.A.; Galgani, F.; Thompson, R.C.; Barlaz, M. Accumulation and fragmentation of plastic debris in global environments. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Derraik, J.G. The pollution of the marine environment by plastic debris: A review. Mar. Pollut. Bull. 2002, 44, 842–852. [Google Scholar] [CrossRef]
- Kubowicz, S.; Booth, A.M. Biodegradability of plastics: Challenges and misconceptions. Environ. Sci. Technol. 2017, 51, 12058–12060. [Google Scholar] [CrossRef]
- Alimba, C.G.; Faggio, C. Microplastics in the marine environment: Current trends in environmental pollution and mechanisms of toxicological profile. Environ. Toxicol. Pharmacol. 2019, 68, 61–74. [Google Scholar] [CrossRef]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef]
- Wierckx, N.; Prieto, M.A.; Pomposiello, P.; de Lorenzo, V.; O’Connor, K.; Blank, L.M. Plastic waste as a novel substrate for industrial biotechnology. Microb. Biotechnol. 2015, 8, 900–903. [Google Scholar] [CrossRef] [PubMed]
- Wierckx, N.; Narancic, T.; Eberlein, C.; Wei, R.; Drzyzga, O.; Magnin, A.; Ballerstedt, H.; Kenny, S.T.; Pollet, E.; Avérous, L.; et al. Plastic biodegradation: Challenges and opportunities. In Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids: Biodegradation and Bioremediation; Springer: Cham, Switzerland, 2018; pp. 1–29. [Google Scholar]
- Narancic, T.; O’Connor, K.E. Microbial biotechnology addressing the plastic waste disaster. Microb. Biotechnol. 2017, 10, 1232–1235. [Google Scholar] [CrossRef] [PubMed]
- Koshti, R.; Mehta, L.; Samarth, N. Biological recycling of polyethylene terephthalate: A mini-review. J. Polym. Environ. 2018, 26, 3520–3529. [Google Scholar] [CrossRef]
- Arroyo, M. Thermoplastic polyesters. In Handbook of Thermoplastics; Olabisi, O., Ed.; Marcel Dekker: New York, NY, USA, 1997; pp. 417–448. [Google Scholar]
- Ji, L.N. Study on preparation process and properties of polyethylene terephthalate (PET). Appl. Mech. Mater. 2013, 312, 406–410. [Google Scholar] [CrossRef]
- NAPCOR. Report on Postconsumer PET Container Recycling Activity in 2017. Available online: https://napcor.com/wp-content/uploads/2018/11/NAPCOR_2017RateReport_FINAL.pdf (accessed on 16 May 2019).
- Nikles, D.E.; Farahat, M.S. New motivation for the depolymerization products derived from poly(ethylene terephthalate) (PET) waste: A review. Macromol. Mater. Eng. 2005, 290, 13–30. [Google Scholar] [CrossRef]
- Al-Sabagh, A.M.; Yehia, F.Z.; Eshaq, G.; Rabie, A.M.; ElMetwally, A.E. Greener routes for recycling of polyethylene terephthalate. Egypt. J. Pet. 2016, 25, 53–64. [Google Scholar] [CrossRef]
- Sinha, V.; Patel, M.; Patel, J. Pet Waste Management by Chemical Recycling: A Review. J. Polym. Environ. 2010, 18, 8–25. [Google Scholar] [CrossRef]
- Furtwengler, P.; Avérous, L. Renewable polyols for advanced polyurethane foams from diverse biomass resources. Polym. Chem. 2018, 9, 4258–4287. [Google Scholar] [CrossRef]
- Webb, H.K.; Arnott, J.; Crawford, R.J.; Ivanova, E.P. Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers 2013, 5, 1–18. [Google Scholar] [CrossRef]
- Roth, C.; Wei, R.; Oeser, T.; Then, J.; Föllner, C.; Zimmermann, W.; Sträter, N. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 2014, 98, 7815–7823. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Oeser, T.; Zimmermann, W. Synthetic polyester-hydrolyzing enzymes from thermophilic actinomycetes. Adv. Appl. Microbiol. 2014, 89, 267–305. [Google Scholar]
- Wei, R.; Zimmermann, W. Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb. Biotechnol. 2017, 10, 1302–1307. [Google Scholar] [CrossRef] [PubMed]
- Kasai, D.; Fujinami, T.; Abe, T.; Mase, K.; Katayama, Y.; Fukuda, M.; Masai, E. Uncovering the protocatechuate 2,3-cleavage pathway genes. J. Bacteriol. 2009, 191, 6758–6768. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Shibayama, T.; Ichikawa, A.; Sakou, Y.; Yamada, S.; Sugisaki, H. Cloning and characterization of the genes encoding enzymes for the protocatechuate meta-degradation pathway of Pseudomonas ochraceae NGJ1. Biosci. Biotechnol. Biochem. 2004, 68, 1434–1441. [Google Scholar] [CrossRef]
- Frazee, R.W.; Livingston, D.M.; LaPorte, D.C.; Lipscomb, J.D. Cloning, sequencing, and expression of the Pseudomonas putida protocatechuate 3,4-dioxygenase genes. J. Bacteriol. 1993, 175, 6194–6202. [Google Scholar] [CrossRef]
- Harwood, C.S.; Parales, R.E. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996, 50, 553–590. [Google Scholar] [CrossRef]
- Trifunović, D.; Schuchmann, K.; Müller, V. Ethylene glycol metabolism in the acetogen Acetobacterium woodii. J. Bacteriol. 2016, 198, 1058–1065. [Google Scholar] [CrossRef]
- Mückschel, B.; Simon, O.; Klebensberger, J.; Graf, N.; Rosche, B.; Altenbuchner, J.; Pfannstiel, J.; Huber, A.; Hauer, B. Ethylene glycol metabolism by Pseudomonas putida. Appl. Environ. Microbiol. 2012, 78, 8531–8539. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, W.; Billig, S. Enzymes for the biofunctionalization of poly(ethylene terephthalate). In Biofunctionalization of Polymers and Their Applications; Nyanhongo, G.S., Steiner, W., Gübitz, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 97–120. [Google Scholar]
- Herrero Acero, E.; Ribitsch, D.; Steinkellner, G.; Gruber, K.; Greimel, K.; Eiteljoerg, I.; Trotscha, E.; Wei, R.; Zimmermann, W.; Zinn, M.; et al. Enzymatic surface hydrolysis of PET: Effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules 2011, 44, 4632–4640. [Google Scholar] [CrossRef]
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.; Hazen, T.; Streit, W.R. New insights into the function and global distribution of polyethylene terephthalate (PET)-Degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ma, Z.; Li, L. Flow-induced crystallization of polymers: Molecular and thermodynamic considerations. Macromolecules 2016, 49, 1505–1517. [Google Scholar] [CrossRef]
- Ronkvist, Å.M.; Xie, W.; Lu, W.; Gross, R.A. Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules 2009, 42, 5128–5138. [Google Scholar] [CrossRef]
- Marten, E.; Müller, R.-J.; Deckwer, W.-D. Studies on the enzymatic hydrolysis of polyesters. II. Aliphatic–aromatic copolyesters. Polym. Degrad. Stab. 2005, 88, 371–381. [Google Scholar] [CrossRef]
- Shirke, A.N.; White, C.; Englaender, J.A.; Zwarycz, A.; Butterfoss, G.L.; Linhardt, R.J.; Gross, R.A. Stabilizing Leaf and branch Compost Cutinase (LCC) with glycosylation: Mechanism and effect on PET hydrolysis. Biochemistry 2018, 57, 1190–1200. [Google Scholar] [CrossRef]
- Guyot, S.; Pottier, L.; Hartmann, A.; Ragon, M.; Hauck Tiburski, J.; Molin, P.; Ferret, E.; Gervais, P. Extremely rapid acclimation of Escherichia coli to high temperature over a few generations of a fed-batch culture during slow warming. Microbiologyopen 2014, 3, 52–63. [Google Scholar] [CrossRef]
- Rudolph, B.; Gebendorfer, K.M.; Buchner, J.; Winter, J. Evolution of Escherichia coli for growth at high temperatures. J. Biol. Chem. 2010, 285, 19029–19034. [Google Scholar] [CrossRef]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Dominick, G.; Duman, R.; El Omari, K.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef]
- Wei, R.; Zimmermann, W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 2017, 10, 1308–1322. [Google Scholar] [CrossRef] [PubMed]
- Barth, M.; Oeser, T.; Wei, R.; Then, J.; Schmidt, J.; Zimmermann, W. Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthalate nanoparticles by a polyester hydrolase from Thermobifida fusca. Biochem. Eng. J. 2015, 93, 222–228. [Google Scholar] [CrossRef]
- Barth, M.; Wei, R.; Oeser, T.; Then, J.; Schmidt, J.; Wohlgemuth, F.; Zimmermann, W. Enzymatic hydrolysis of polyethylene terephthalate films in an ultrafiltration membrane reactor. J. Membr. Sci. 2015, 494, 182–187. [Google Scholar] [CrossRef]
- Carniel, A.; Valoni, É.; Nicomedes, J.; Gomes, A.d.C.; Castro, A.M.d. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Process Biochem. 2017, 59, 84–90. [Google Scholar] [CrossRef]
- Wei, R.; Oeser, T.; Schmidt, J.; Meier, R.; Barth, M.; Then, J.; Zimmermann, W. Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnol. Bioeng. 2016, 113, 1658–1665. [Google Scholar] [CrossRef]
- Choi, K.Y.; Sul, W.J.; Kim, Y.M.; Kim, E.; Kim, D.; Zylstra, G.J.; Chae, J.-C. Molecular and biochemical analysis of phthalate and terephthalate degradation by Rhodococcus sp. strain DK17. FEMS Microbiol. Lett. 2005, 252, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Z.; Zhou, Y.; Zylstra, G.J. Molecular analysis of isophthalate and terephthalate degradation by Comamonas testosteroni YZW-D. Environ. Health Perspect. 1995, 103, 9–12. [Google Scholar] [PubMed]
- Sasoh, M.; Masai, E.; Ishibashi, S.; Hara, H.; Kamimura, N.; Miyauchi, K.; Fukuda, M. Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl. Environ. Microbiol. 2006, 72, 1825–1832. [Google Scholar] [CrossRef] [PubMed]
- Kasai, D.; Kitajima, M.; Fukuda, M.; Masai, E. Transcriptional regulation of the terephthalate catabolism operon in Comamonas sp. strain E6. Appl. Environ. Microbiol. 2010, 76, 6047–6055. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, M.; Kamimura, N.; Toribami, S.; Mori, K.; Kasai, D.; Fukuda, M.; Masai, E. Novel tripartite Aromatic Acid Transporter Essential for Terephthalate Uptake in Comamonas sp. Strain E6. Appl. Environ. Microbiol. 2013, 79, 6148–6155. [Google Scholar] [CrossRef] [PubMed]
- Nichols, N.N.; Harwood, C.S. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 1997, 179, 5056–5061. [Google Scholar] [CrossRef] [PubMed]
- Noda, Y.; Nishikawa, S.; Shiozuka, K.; Kadokura, H.; Nakajima, H.; Yoda, K.; Katayama, Y.; Morohoshi, N.; Haraguchi, T.; Yamasaki, M. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 1990, 172, 2704–2709. [Google Scholar] [CrossRef]
- Oberto, J. SyntTax: A web server linking synteny to prokaryotic taxonomy. BMC Bioinf. 2013, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Wells Jr, T.; Ragauskas, A.J. Biotechnological opportunities with the b-ketoadipate pathway. Trends Biotechnol. 2012, 30, 627–637. [Google Scholar] [CrossRef]
- Johnson, C.W.; Salvachúa, D.; Khanna, P.; Smith, H.; Peterson, D.J.; Beckham, G.T. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016, 3, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Vardon, D.R.; Franden, M.A.; Johnson, C.W.; Karp, E.M.; Guarnieri, M.T.; Linger, J.G.; Salm, M.J.; Strathmann, T.J.; Beckham, G.T. Adipic acid production from lignin. Energy Environ. Sci. 2015, 8, 617–628. [Google Scholar] [CrossRef]
- Child, J.; Willetts, A. Microbial metabolism of aliphatic glycols bacterial metabolism of ethylene glycol. Biochim. Biophys. Acta Gen. Subj. 1978, 538, 316–327. [Google Scholar] [CrossRef]
- Kataoka, M.; Sasaki, M.; Hidalgo, A.-R.G.D.; Nakano, M.; Shimizu, S. Glycolic acid production using ethylene glycol-oxidizing microorganisms. Biosci. Biotechnol. Biochem. 2001, 65, 2265–2270. [Google Scholar] [CrossRef] [PubMed]
- Boronat, A.; Aguilar, J. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: Purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 1979, 140, 320–326. [Google Scholar]
- Wehrmann, M.; Billard, P.; Martin-Meriadec, A.; Zegeye, A.; Klebensberger, J. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. MBio 2017, 8, e00570-17. [Google Scholar] [CrossRef]
- Boronat, A.; Caballero, E.; Aguilar, J. Experimental evolution of a metabolic pathway for ethylene glycol utilization by Escherichia coli. J. Bacteriol. 1983, 153, 134–139. [Google Scholar]
- Franden, M.A.; Jayakody, L.N.; Li, W.-J.; Wagner, N.J.; Cleveland, N.S.; Michener, W.E.; Hauer, B.; Blank, L.M.; Wierckx, N.; Klebensberger, J.; et al. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 2018, 48, 197–207. [Google Scholar] [CrossRef]
- Cusa, E.; Obradors, N.; Baldomà, L.; Badía, J.; Aguilar, J. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 1999, 181, 7479–7484. [Google Scholar] [PubMed]
- Grostern, A.; Sales, C.M.; Zhuang, W.-Q.; Erbilgin, O.; Alvarez-Cohen, L. Glyoxylate metabolism Is a key feature of the metabolic degradation of 1,4-dioxane by Pseudonocardia dioxanivorans strain CB1190. Appl. Environ. Microbiol. 2012, 78, 3298–3308. [Google Scholar] [CrossRef] [PubMed]
- Wanner, B.L.; Wishart, D.; Blattner, F.R.; Thomas, G.H.; Plunkett Guy, I.I.I.; Mori, H.; Keseler, I.M.; Glasner, J.D.; Rudd, K.E.; Serres, M.H.; et al. Escherichia coli K-12: A cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006, 34, 1–9. [Google Scholar]
- Huccetogullari, D.; Luo, Z.W.; Lee, S.Y. Metabolic engineering of microorganisms for production of aromatic compounds. Microb. Cell Fact. 2019, 18, 41. [Google Scholar] [CrossRef]
- Osterhout, R.E.; Burgard, A.P.; Pharkya, P.; Burk, P. Microorganisms and Methods for the Biosynthesis of Aromatics, 2,4-Pentadienoate and 1,3-Butadiene. U.S. Patent 8715957 B2, 26 July 2011. [Google Scholar]
- Delépine, B.; Duigou, T.; Carbonell, P.; Faulon, J.-L. RetroPath2.0: A retrosynthesis workflow for metabolic engineers. Metab. Eng. 2018, 45, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.S.; Hertweck, C.; Hopke, J.N.; Izumikawa, M.; Kalaitzis, J.A.; Nilsen, G.; O’Hare, T.; Piel, J.; Shipley, P.R.; Xiang, L.; et al. Plant-like biosynthetic pathways in bacteria: From benzoic acid to chalcone. J. Nat. Prod. 2002, 65, 1956–1962. [Google Scholar] [CrossRef]
- Lind, W.; Campbell, R. Preparation of Potassium Terephthalate. U.S. Patent 3761515 A, 21 October 1971. [Google Scholar]
- Bernhard, R. Production of Terephthalic Acid. U.S. Patent 2823229 A, 20 June 1956. [Google Scholar]
- Graglia, M.; Kanna, N.; Esposito, D. Lignin refinery: Towards the preparation of renewable aromatic building blocks. ChemBioEng Rev. 2015, 2, 377–392. [Google Scholar] [CrossRef]
- Luo, Z.W.; Lee, S.Y. Biotransformation of p-xylene into terephthalic acid by engineered Escherichia coli. Nat. Commun. 2017, 8, 15689. [Google Scholar] [CrossRef] [PubMed]
- Franklin, F.C.; Bagdasarian, M.; Bagdasarian, M.M.; Timmis, K.N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 1981, 78, 7458–7462. [Google Scholar] [CrossRef] [PubMed]
- Harayama, S.; Rekik, M.; Wubbolts, M.; Rose, K.; Leppik, R.A.; Timmis, K.N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J. Bacteriol. 1989, 171, 5048–5055. [Google Scholar] [CrossRef] [PubMed]
- Junker, F.; Kiewitz, R.; Cook, A.M. Characterization of the p-toluenesulfonate operon tsaMBCD and tsaR in Comamonas testosteroni T-2. J. Bacteriol. 1997, 179, 919–927. [Google Scholar] [CrossRef]
- Dedov, A.G.; Loktev, A.S.; Karavaev, A.A.; Moiseev, I.I. A novel direct catalytic production of p-xylene from isobutanol. Mendeleev Commun. 2018, 28, 352–353. [Google Scholar] [CrossRef]
- Peters, M.; Taylor, J.; Jenni, M.; Manzer, L.; Henton, D. Integrated process to selectively convert renewable isobutanol to p-xylene. U.S. Patent US 20110087000 A1, 6 October 2010. [Google Scholar]
- Chang, R.; Zhu, L.; Jin, F.; Fan, M.; Liu, J.; Jia, Q.; Tang, C.; Li, Q. Production of bio-based p-xylene via catalytic pyrolysis of biomass over metal oxide-modified HZSM-5 zeolites. J. Chem. Technol. Biotechnol. 2018, 93, 3292–3301. [Google Scholar] [CrossRef]
- Pacheco, J.J.; Davis, M.E. Synthesis of terephthalic acid via Diels-Alder reactions with ethylene and oxidized variants of 5-hydroxymethylfurfural. Proc. Natl. Acad. Sci. USA 2014, 111, 8363–8367. [Google Scholar] [CrossRef] [PubMed]
- Shiramizu, M.; Toste, F.D. On the Diels–Alder approach to solely biomass-derived polyethylene terephthalate (PET): Conversion of 2,5-dimethylfuran and acrolein into p-xylene. Chem. A Eur. J. 2011, 17, 12452–12457. [Google Scholar] [CrossRef] [PubMed]
- Maneffa, A.; Priecel, P.; Lopez-Sanchez, J.A. Biomass-derived renewable aromatics: Selective routes and outlook for p-xylene commercialisation. ChemSusChem 2016, 9, 2736–2748. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.L.; Chang, C.-C.; Do, P.; Nikbin, N.; Caratzoulas, S.; Vlachos, D.G.; Lobo, R.F.; Fan, W.; Dauenhauer, P.J. Cycloaddition of biomass-derived furans for catalytic production of renewable p-xylene. ACS Catal. 2012, 2, 935–939. [Google Scholar] [CrossRef]
- Van de Poel, B.; Cooper, E.D.; Delwiche, C.F.; Chang, C. An evolutionary perspective on the plant hormone ethylene. In Ethylene in Plants; Springer: Dordrecht, The Netherlands; pp. 109–134.
- Digiacomo, F.; Girelli, G.; Aor, B.; Marchioretti, C.; Pedrotti, M.; Perli, T.; Tonon, E.; Valentini, V.; Avi, D.; Ferrentino, G.; et al. Ethylene-producing bacteria that ripen fruit. ACS Synth. Biol. 2014, 3, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Steeman, A. PET is PET—Petro-PET or Bio-PET. Available online: https://bestinpackaging.com/2011/07/13/pet-is-pet-petro-pet-or-bio-pet/ (accessed on 3 March 2019).
- Salusjärvi, L.; Havukainen, S.; Koivistoinen, O.; Toivari, M. Biotechnological production of glycolic acid and ethylene glycol: Current state and perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ramos, K.R.M.; Valdehuesa, K.N.G.; Nisola, G.M.; Lee, W.-K.; Chung, W.-J. Biosynthesis of ethylene glycol in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 3409–3417. [Google Scholar] [CrossRef] [PubMed]
- Alkim, C.; Cam, Y.; Trichez, D.; Auriol, C.; Spina, L.; Vax, A.; Bartolo, F.; Besse, P.; François, J.M.; Walther, T. Optimization of ethylene glycol production from (d)-xylose via a synthetic pathway implemented in Escherichia coli. Microb. Cell Fact. 2015, 14, 127. [Google Scholar] [CrossRef]
- Cam, Y.; Alkim, C.; Trichez, D.; Trebosc, V.; Vax, A.; Bartolo, F.; Besse, P.; François, J.M.; Walther, T. Engineering of a synthetic metabolic pathway for the assimilation of (d)-xylose into value-added chemicals. ACS Synth. Biol. 2016, 5, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Cabulong, R.B.; Valdehuesa, K.N.G.; Ramos, K.R.M.; Nisola, G.M.; Lee, W.-K.; Lee, C.R.; Chung, W.-J. Enhanced yield of ethylene glycol production from d-xylose by pathway optimization in Escherichia coli. Enzyme Microb. Technol. 2017, 97, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Uranukul, B.; Woolston, B.M.; Fink, G.R.; Stephanopoulos, G. Biosynthesis of monoethylene glycol in Saccharomyces cerevisiae utilizing native glycolytic enzymes. Metab. Eng. 2019, 51, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Zhang, H.; De Mey, M.; Lim, C.G.; Li, Z.-J.; Stephanopoulos, G. Engineering a novel biosynthetic pathway in Escherichia coli for production of renewable ethylene glycol. Biotechnol. Bioeng. 2016, 113, 376–383. [Google Scholar] [CrossRef]
- Islam, M.A.; Hadadi, N.; Ataman, M.; Hatzimanikatis, V.; Stephanopoulos, G. Exploring biochemical pathways for mono-ethylene glycol (MEG) synthesis from synthesis gas. Metab. Eng. 2017, 41, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.H.; Koryakina, I.; Case, A.E.; Toney, M.D.; Atsumi, S. Biological conversion of gaseous alkenes to liquid chemicals. Metab. Eng. 2016, 38, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Ji, N.; Zhang, T.; Zheng, M.; Wang, A.; Wang, H.; Wang, X.; Shu, Y.; Stottlemyer, A.L.; Chen, J.G. Catalytic conversion of cellulose into ethylene glycol over supported carbide catalysts. Catal. Today 2009, 147, 77–85. [Google Scholar] [CrossRef]
- Sun, J.; Liu, H. Selective hydrogenolysis of biomass-derived xylitol to ethylene glycol and propylene glycol on supported Ru catalysts. Green Chem. 2011, 13, 135–142. [Google Scholar] [CrossRef]
- Pang, J.; Zheng, M.; Wang, A.; Zhang, T. Catalytic hydrogenation of corn stalk to ethylene glycol and 1,2-propylene glycol. Ind. Eng. Chem. Res. 2011, 50, 6601–6608. [Google Scholar] [CrossRef]
- Song, T.; Xu, Y.; Ye, Y.; Chen, Y.; Shen, S. Electricity generation from terephthalic acid using a microbial fuel cell. J. Chem. Technol. Biotechnol. 2009, 84, 356–360. [Google Scholar] [CrossRef]
- The Coca Cola Company Coca-Cola Produces World’s First PET Bottle Made Entirely From Plants. Available online: https://www.coca-colacompany.com/press-center/press-releases/coca-cola-produces-worlds-first-pet-bottle-made-entirely-from-plants (accessed on 5 March 2019).


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvador, M.; Abdulmutalib, U.; Gonzalez, J.; Kim, J.; Smith, A.A.; Faulon, J.-L.; Wei, R.; Zimmermann, W.; Jimenez, J.I. Microbial Genes for a Circular and Sustainable Bio-PET Economy. Genes 2019, 10, 373. https://doi.org/10.3390/genes10050373
Salvador M, Abdulmutalib U, Gonzalez J, Kim J, Smith AA, Faulon J-L, Wei R, Zimmermann W, Jimenez JI. Microbial Genes for a Circular and Sustainable Bio-PET Economy. Genes. 2019; 10(5):373. https://doi.org/10.3390/genes10050373
Chicago/Turabian StyleSalvador, Manuel, Umar Abdulmutalib, Jaime Gonzalez, Juhyun Kim, Alex A. Smith, Jean-Loup Faulon, Ren Wei, Wolfgang Zimmermann, and Jose I. Jimenez. 2019. "Microbial Genes for a Circular and Sustainable Bio-PET Economy" Genes 10, no. 5: 373. https://doi.org/10.3390/genes10050373
APA StyleSalvador, M., Abdulmutalib, U., Gonzalez, J., Kim, J., Smith, A. A., Faulon, J.-L., Wei, R., Zimmermann, W., & Jimenez, J. I. (2019). Microbial Genes for a Circular and Sustainable Bio-PET Economy. Genes, 10(5), 373. https://doi.org/10.3390/genes10050373





