Fold-Change-Specific Enrichment Analysis (FSEA): Quantification of Transcriptional Response Magnitude for Functional Gene Groups
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets for FSEA Testing and Application
2.1.1. Real Datasets for FSEA Application
2.1.2. Simulated Datasets for FSEA Testing
- Six groups of DEGs of different sizes (5, 10, 20, 30, 40, and 50 genes) with the fold-change mean equals to 1 (μ = 1) and with the strong correlation within each group (correlation coefficient (ρ) > 0.7). These gene sets simulate pseudo-GO terms;
- One group of DEGs (μ = 1) of 100 genes without any correlation (ρ ~ 0);
- One group of non-DEGs (μ = 0) of 700 genes without any correlation (ρ ~ 0).
2.2. FSEA Method Formal Description
- ;
- , in case of ;
- , in case of ,
2.3. FSEA Implementation
2.4. Comparison of FSEA with GSEA and SEA
3. Results
3.1. FSEA Description
3.2. FSEA Validation
3.3. FSEA Performance on a Cancer-Related Dataset
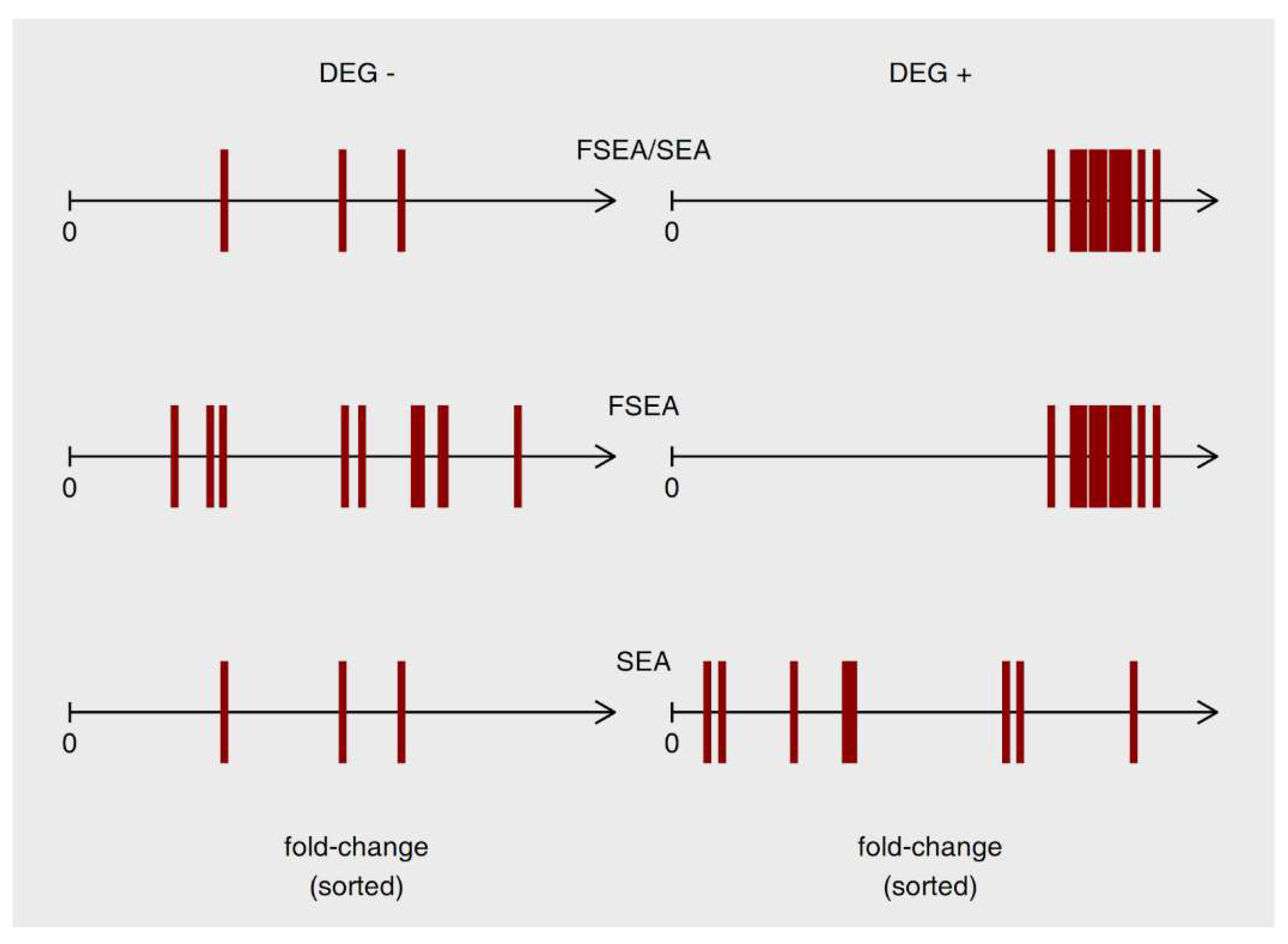
3.3.1. FSEA and SEA: FSEA gives an Additional Dimension to SEA Results
3.3.2. Functional Groups Detected by SEA but Not FSEA: Non-Coordinated Response
3.3.3. Only FSEA: A Quantized Response Invisible for Classical Enrichment Analysis Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Shi Jing, L.; Fathiah Muzaffar Shah, F.; Saberi Mohamad, M.; Moorthy, K.; Deris, S.; Zakaria, Z.; Napis, S. A Review on Bioinformatics Enrichment Analysis Tools Towards Functional Analysis of High Throughput Gene Set Data. Curr. Proteomics 2015, 12, 14–27. [Google Scholar] [CrossRef]
- Omelyanchuk, N.A.; Wiebe, D.S.; Novikova, D.D.; Levitsky, V.G.; Klimova, N.; Gorelova, V.; Weinholdt, C.; Vasiliev, G.V.; Zemlyanskaya, E.V.; Kolchanov, N.A.; et al. Auxin Regulates Functional Gene Groups in a Fold-Change-Specific Manner in Arabidopsis Thaliana Roots. Sci. Rep. 2017, 7, 2489. [Google Scholar] [CrossRef] [PubMed]
- Sharma, E.; Jain, M.; Khurana, J.P. Differential Quantitative Regulation of Specific Gene Groups and Pathways under Drought Stress in Rice. Genomics 2019, 111, 1699–1712. [Google Scholar] [CrossRef]
- Laurent, G.S.; Shtokalo, D.; Tackett, M.R.; Yang, Z.; Vyatkin, Y.; Milos, P.M.; Seilheimer, B.; McCaffrey, T.A.; Kapranov, P. On the Importance of Small Changes in RNA Expression. Methods 2013, 63, 18–24. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. In Statistical Genomics; Mathé, E., Davis, S., Eds.; Springer: New York, NY, USA, 2016; Volume 1418, pp. 93–110. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R Package Rsubread Is Easier, Faster, Cheaper and Better for Alignment and Quantification of RNA Sequencing Reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Repana, D.; Nulsen, J.; Dressler, L.; Bortolomeazzi, M.; Venkata, S.K.; Tourna, A.; Yakovleva, A.; Palmieri, T.; Ciccarelli, F.D. The Network of Cancer Genes (NCG): A Comprehensive Catalogue of Known and Candidate Cancer Genes from Cancer Sequencing Screens. Genome Biol. 2019, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Wiebe, D. FoldGO: Package for Fold-Specific GO Terms Recognition; R Package Version 1.4.0; Bioconductor, 2019. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Sergushichev, A. Fast Gene Set Enrichment Analysis. bioRxiv 2016. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. TopGO: Enrichment Analysis for Gene Ontology; R Package Version 2.38.1; Bioconductor, 2019. [Google Scholar] [CrossRef]
- Carlson, M. Org.Hs.Eg.Db: Genome Wide Annotation for Human; R Package Version 3.8.2; Bioconductor, 2019. [Google Scholar] [CrossRef]
- Cottard, F.; Madi-Berthélémy, P.O.; Erdmann, E.; Schaff-Wendling, F.; Keime, C.; Ye, T.; Kurtz, J.-E.; Céraline, J. Dual Effects of Constitutively Active Androgen Receptor and Full-Length Androgen Receptor for N-Cadherin Regulation in Prostate Cancer. Oncotarget 2017, 8. [Google Scholar] [CrossRef]
- Giannoni, E.; Taddei, M.L.; Morandi, A.; Comito, G.; Calvani, M.; Bianchini, F.; Richichi, B.; Raugei, G.; Wong, N.; Tang, D.; et al. Targeting Stromal-Induced Pyruvate Kinase M2 Nuclear Translocation Impairs Oxphos and Prostate Cancer Metastatic Spread. Oncotarget 2015, 6. [Google Scholar] [CrossRef]
- Yeh, C.-H.; Bellon, M.; Nicot, C. FBXW7: A Critical Tumor Suppressor of Human Cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef]
- Liu, Z.; Ding, Y.; Ye, N.; Wild, C.; Chen, H.; Zhou, J. Direct Activation of Bax Protein for Cancer Therapy: Direct Activation of Bax for Cancer Therapy. Med. Res. Rev. 2016, 36, 313–341. [Google Scholar] [CrossRef]
- Guo, B.; Cao, S.; Tóth, K.; Azrak, R.G.; Rustum, Y.M. Overexpression of Bax Enhances Antitumor Activity of Chemotherapeutic Agents in Human Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2000, 6, 718–724. [Google Scholar]
- Forster, J.; Harriss-Phillips, W.; Douglass, M.; Bezak, E. A Review of the Development of Tumor Vasculature and Its Effects on the Tumor Microenvironment. Hypoxia 2017, 5, 21–32. [Google Scholar] [CrossRef]
- Ren, S.; Shao, Y.; Zhao, X.; Hong, C.S.; Wang, F.; Lu, X.; Li, J.; Ye, G.; Yan, M.; Zhuang, Z.; et al. Integration of Metabolomics and Transcriptomics Reveals Major Metabolic Pathways and Potential Biomarker Involved in Prostate Cancer. Mol. Cell. Proteom. 2016, 15, 154–163. [Google Scholar] [CrossRef]
- Thapa, M.; Dallmann, G. Role of Coenzymes in Cancer Metabolism. Semin. Cell & Dev. Biol. 2020, 98, 44–53. [Google Scholar] [CrossRef]
- Schcolnik-Cabrera, A.; Oldak, B.; Juárez, M.; Cruz-Rivera, M.; Flisser, A.; Mendlovic, F. Calreticulin in Phagocytosis and Cancer: Opposite Roles in Immune Response Outcomes. Apoptosis 2019, 24, 245–255. [Google Scholar] [CrossRef]
- Kissick, H.T.; On, S.T.; Dunn, L.K.; Sanda, M.G.; Asara, J.M.; Pellegrini, K.L.; Noel, J.K.; Arredouani, M.S. The Transcription Factor ERG Increases Expression of Neurotransmitter Receptors on Prostate Cancer Cells. BMC Cancer 2015, 15, 604. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene Expression Patterns of Breast Carcinomas Distinguish Tumor Subclasses with Clinical Implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Sørlie, T. The Impact of Gene Expression Patterns in Breast Cancer. Clin. Chem. 2016, 62, 1150–1151. [Google Scholar] [CrossRef][Green Version]
- Pallai, R.; Bhaskar, A.; Barnett-Bernodat, N.; Gallo-Ebert, C.; Nickels, J.T.; Rice, L.M. Cancerous Inhibitor of Protein Phosphatase 2A Promotes Premature Chromosome Segregation and Aneuploidy in Prostate Cancer Cells through Association with Shugoshin. Tumor Biol. 2015, 36, 6067–6074. [Google Scholar] [CrossRef]
- Cornford, P.; Evans, J.; Dodson, A.; Parsons, K.; Woolfenden, A.; Neoptolemos, J.; Foster, C.S. Protein Kinase C Isoenzyme Patterns Characteristically Modulated in Early Prostate Cancer. Am. J. Pathol. 1999, 154, 137–144. [Google Scholar] [CrossRef]
- Tanaka, Y.; Gavrielides, M.V.; Mitsuuchi, Y.; Fujii, T.; Kazanietz, M.G. Protein Kinase C Promotes Apoptosis in LNCaP Prostate Cancer Cells through Activation of P38 MAPK and Inhibition of the Akt Survival Pathway. J. Biol. Chem. 2003, 278, 33753–33762. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.-J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID Interaction Database: 2019 Update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]




| Qr + | Qr − | Total | |
|---|---|---|---|
| GOi + | A | B | A + B |
| GOi− | C | D | C + D |
| Total | A + C | B + D | N |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiebe, D.S.; Omelyanchuk, N.A.; Mukhin, A.M.; Grosse, I.; Lashin, S.A.; Zemlyanskaya, E.V.; Mironova, V.V. Fold-Change-Specific Enrichment Analysis (FSEA): Quantification of Transcriptional Response Magnitude for Functional Gene Groups. Genes 2020, 11, 434. https://doi.org/10.3390/genes11040434
Wiebe DS, Omelyanchuk NA, Mukhin AM, Grosse I, Lashin SA, Zemlyanskaya EV, Mironova VV. Fold-Change-Specific Enrichment Analysis (FSEA): Quantification of Transcriptional Response Magnitude for Functional Gene Groups. Genes. 2020; 11(4):434. https://doi.org/10.3390/genes11040434
Chicago/Turabian StyleWiebe, Daniil S., Nadezhda A. Omelyanchuk, Aleksei M. Mukhin, Ivo Grosse, Sergey A. Lashin, Elena V. Zemlyanskaya, and Victoria V. Mironova. 2020. "Fold-Change-Specific Enrichment Analysis (FSEA): Quantification of Transcriptional Response Magnitude for Functional Gene Groups" Genes 11, no. 4: 434. https://doi.org/10.3390/genes11040434
APA StyleWiebe, D. S., Omelyanchuk, N. A., Mukhin, A. M., Grosse, I., Lashin, S. A., Zemlyanskaya, E. V., & Mironova, V. V. (2020). Fold-Change-Specific Enrichment Analysis (FSEA): Quantification of Transcriptional Response Magnitude for Functional Gene Groups. Genes, 11(4), 434. https://doi.org/10.3390/genes11040434







