Mutational and Kinetic Analysis of Lesion Recognition by Escherichia coli Endonuclease VIII
Abstract
:1. Introduction
2. Materials and Methods
2.1. Oligodeoxyribonucleotides (ODNs) and Enzymes
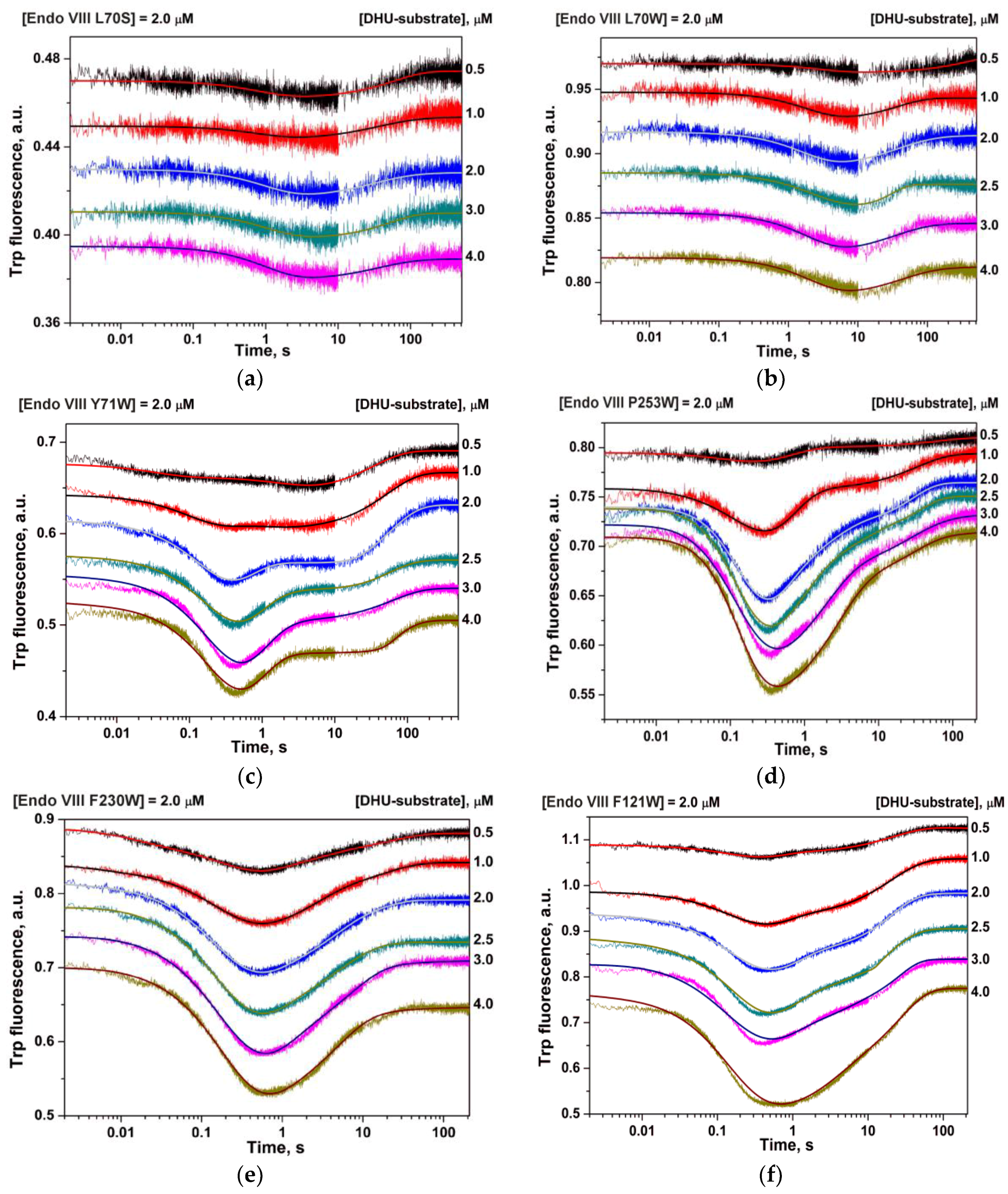
2.2. Stopped-Flow Fluorescence Measurements
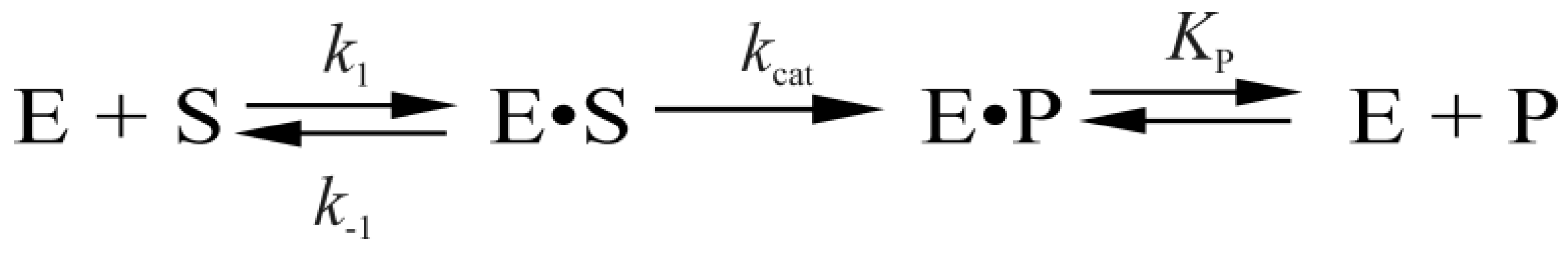
2.3. Data Analysis
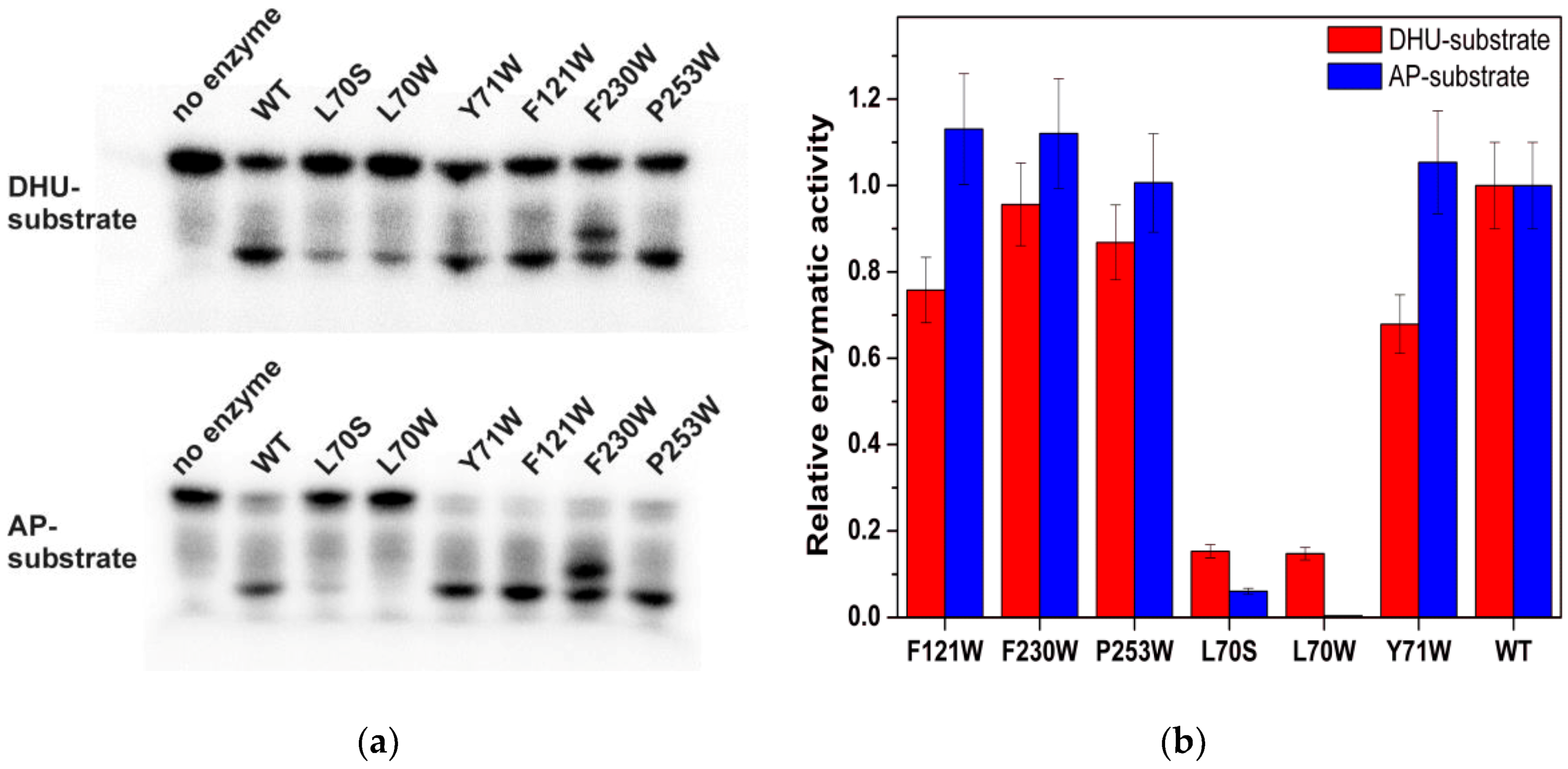
2.4. An Assay of Relative Enzymatic Activity
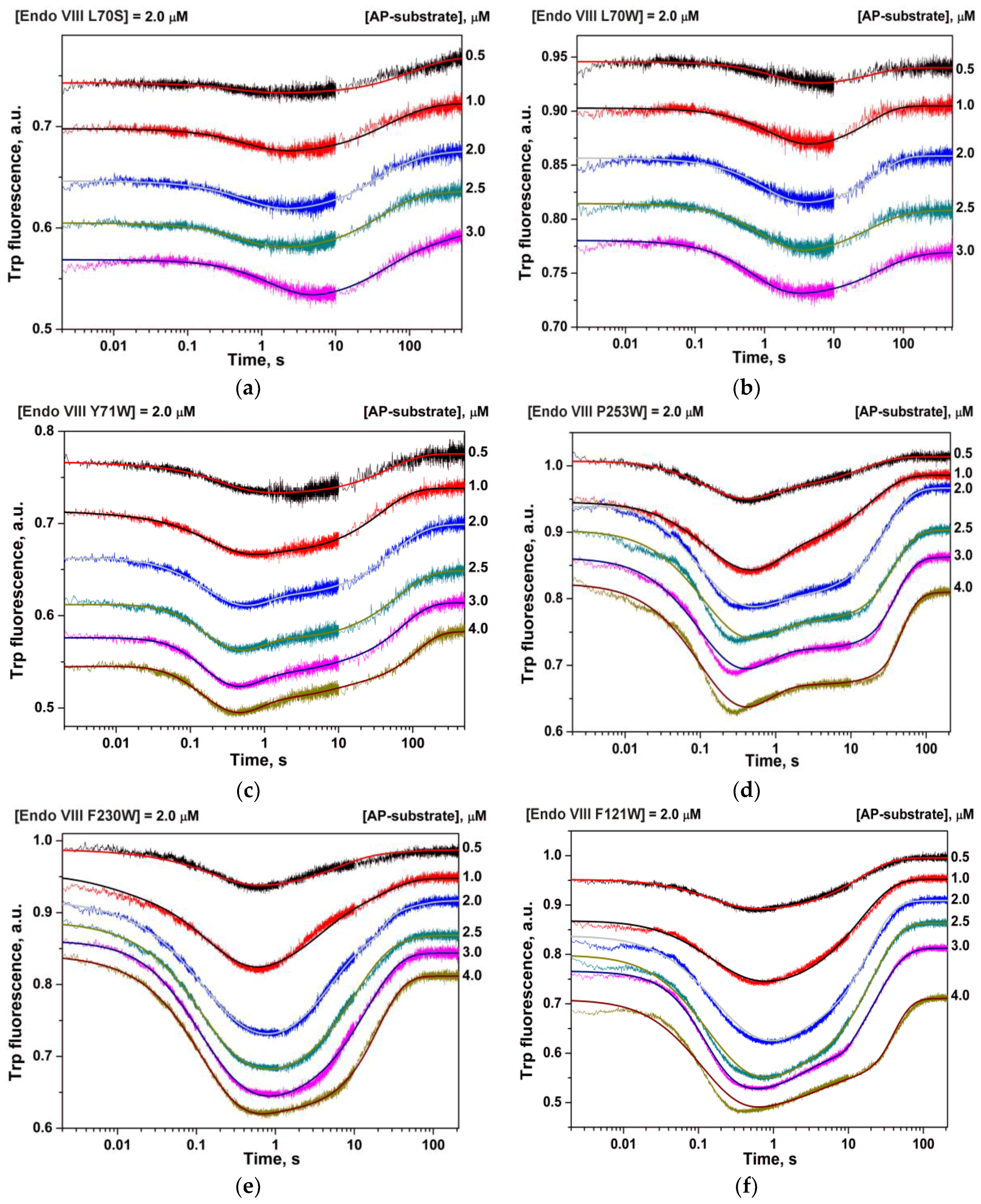
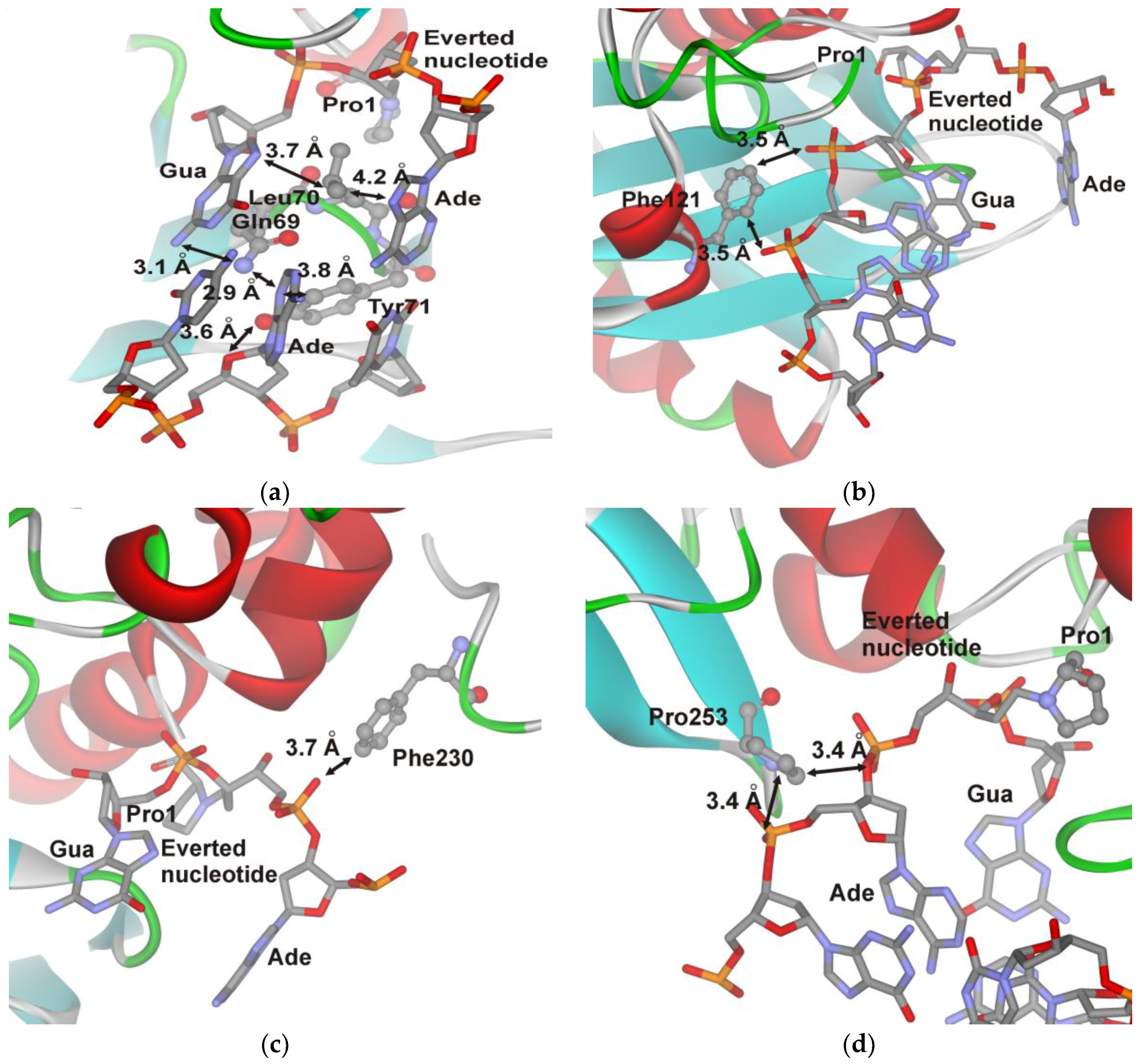
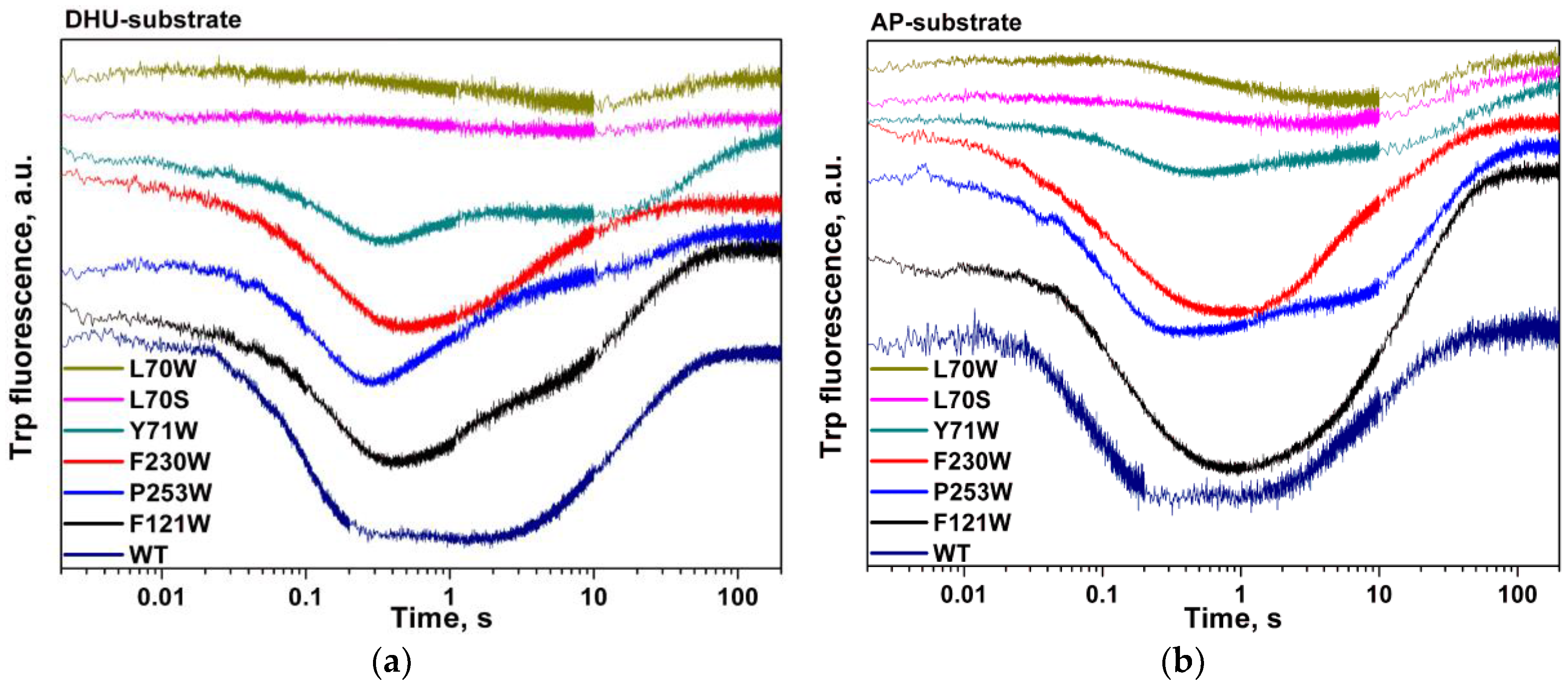
3. Results
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Melamede, R.J.; Hatahet, Z.; Kow, Y.W.; Ide, H.; Wallace, S.S. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry 1994, 33, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Hatahet, Z.; Melamede, R.J.; Kow, Y.W.; Wallace, S.S. Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 1997, 272, 32230–32239. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Jaruga, P.; Dodson, M.L.; Dizdaroglu, M.; Lloyd, R.S. Determination of active site residues in Escherichia coli Endonuclease VIII. J. Biol. Chem. 2002, 277, 2938–2944. [Google Scholar] [CrossRef] [PubMed]
- Kropachev, K.Y.; Zharkov, D.O.; Grollman, A.P. Catalytic mechanism of Escherichia coli Endonuclease VIII: Roles of the intercalation loop and the zinc finger. Biochemistry 2006, 45, 12039–12049. [Google Scholar] [CrossRef] [PubMed]
- Golan, G.; Zharkov, D.O.; Feinberg, H.; Fernandes, A.S.; Zaika, E.I.; Kycia, J.H.; Grollman, A.P.; Shoham, G. Structure of the uncomplexed DNA repair enzyme Endonuclease VIII indicates significant interdomain flexibility. Nucleic Acids Res. 2005, 33, 5006–5016. [Google Scholar] [CrossRef] [PubMed]
- Zharkov, D.O.; Golan, G.; Gilboa, R.; Fernandes, A.S.; Gerchman, S.E.; Kycia, J.H.; Rieger, R.A.; Grollman, A.P.; Shoham, G. Structural analysis of an Escherichia coli Endonuclease VIII covalent reaction intermediate. EMBO J. 2002, 21, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Koval, V.V.; Zharkov, D.O.; Fedorova, O.S. Conformational dynamics of the interaction of Escherichia coli endonuclease VIII with DNA substrates. DNA Repair 2012, 11, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, A.A.; Kuznetsov, N.A.; Vorobjev, Y.N.; Barthes, N.P.; Michel, B.Y.; Burger, A.; Fedorova, O.S. New Environment-Sensitive Multichannel DNA Fluorescent Label for Investigation of the Protein-DNA Interactions. PLoS ONE 2014, 9, e100007. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Vorobjev, Y.N.; Krasnoperov, L.N.; Fedorova, O.S. Thermodynamics of the multi-stage DNA lesion recognition and repair by formamidopyrimidine-DNA glycosylase using pyrrolocytosine fluorescence--stopped-flow pre-steady-state kinetics. Nucleic Acids Res. 2012, 40, 7384–7392. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Bergonzo, C.; Campbell, A.J.; Li, H.; Mechetin, G.V.; de los Santos, C.; Grollman, A.P.; Fedorova, O.S.; Zharkov, D.O.; Simmerling, C. Active destabilization of base pairs by a DNA glycosylase wedge initiates damage recognition. Nucleic Acids Res. 2015, 43, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Dunn, A.R.; Kad, N.M.; Nelson, S.R.; Warshaw, D.M.; Wallace, S.S. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011, 39, 7487–7498. [Google Scholar] [CrossRef] [PubMed]
- Prakash, A.; Doublie, S.; Wallace, S.S. The Fpg/Nei family of DNA glycosylases: Substrates, structures, and search for damage. Prog. Mol. Biol. Transl. Sci. 2012, 110, 71–91. [Google Scholar] [PubMed]
- Lee, A.J.; Warshaw, D.M.; Wallace, S.S. Insights into the glycosylase search for damage from single-molecule fluorescence microscopy. DNA Repair 2014, 20, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.R.; Dunn, A.R.; Kathe, S.D.; Warshaw, D.M.; Wallace, S.S. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Natl. Acad. Sci. USA 2014, 111, E2091–2099. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Wallace, S.S. Visualizing the Search for Radiation-damaged DNA Bases in Real Time. Radiat. Phys. Chem. 2016, 128, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Wallace, S.S. Hide and seek: How do DNA glycosylases locate oxidatively damaged DNA bases amidst a sea of undamaged bases? Free Radic. Biol. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Kladova, O.A.; Kuznetsova, A.A.; Ishchenko, A.A.; Saparbaev, M.K.; Zharkov, D.O.; Fedorova, O.S. Conformational Dynamics of DNA Repair by Escherichia coli Endonuclease III. J. Biol. Chem. 2015, 290, 14338–14349. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, A.A.; Kuznetsov, N.A.; Ishchenko, A.A.; Saparbaev, M.K.; Fedorova, O.S. Step-by-Step Mechanism of DNA Damage Recognition by Human 8-Oxoguanine DNA Glycosylase. Biochim. Biophys. Acta 2014, 1840, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Kuznetsova, A.A.; Vorobjev, Y.N.; Krasnoperov, L.N.; Fedorova, O.S. Thermodynamics of the DNA damage repair steps of human 8-oxoguanine DNA glycosylase. PLoS ONE 2014, 9, e98495. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, R.; Zharkov, D.O.; Golan, G.; Fernandes, A.S.; Gerchman, S.E.; Matz, E.; Kycia, J.H.; Grollman, A.P.; Shoham, G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 2002, 277, 19811–19816. [Google Scholar] [CrossRef] [PubMed]
- Doublie, S.; Bandaru, V.; Bond, J.P.; Wallace, S.S. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc. Natl. Acad. Sci. USA 2004, 101, 10284–10289. [Google Scholar] [CrossRef] [PubMed]
- Koval, V.V.; Kuznetsov, N.A.; Ishchenko, A.A.; Saparbaev, M.K.; Fedorova, O.S. Real-time studies of conformational dynamics of the repair enzyme E. coli formamidopyrimidine-DNA glycosylase and its DNA complexes during catalytic cycle. Mutat. Res. 2010, 685, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Imamura, K.; Averill, A.M.; Wallace, S.S.; Doublie, S. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure 2013, 21, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Prakash, A.; Eckenroth, B.E.; Averill, A.M.; Imamura, K.; Wallace, S.S.; Doublie, S. Structural investigation of a viral ortholog of human NEIL2/3 DNA glycosylases. DNA Repair 2013, 12, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Hoehn, S.T.; Turner, C.J.; Stubbe, J. Solution structure of an oligonucleotide containing an abasic site: Evidence for an unusual deoxyribose conformation. Nucleic Acids Res. 2001, 29, 3413–3423. [Google Scholar] [CrossRef] [PubMed]
- Koval, V.V.; Kuznetsov, N.A.; Zharkov, D.O.; Ishchenko, A.A.; Douglas, K.T.; Nevinsky, G.A.; Fedorova, O.S. Pre-steady-state kinetics shows differences in processing of various DNA lesions by Escherichia coli formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 2004, 32, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Kiryutin, A.S.; Kuznetsova, A.A.; Panov, M.S.; Barsukova, M.O.; Yurkovskaya, A.V.; Fedorova, O.S. The formation of catalytically competent enzyme-substrate complex is not a bottleneck in lesion excision by human alkyladenine DNA glycosylase. J. Biomol. Struct. Dyn. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kuzmic, P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal. Biochem. 1996, 237, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Miroshnikova, A.D.; Kuznetsova, A.A.; Vorobjev, Y.N.; Kuznetsov, N.A.; Fedorova, O.S. Effects of mono- and divalent metal ions on DNA binding and catalysis of human apurinic/apyrimidinic endonuclease 1. Mol. BioSyst. 2016, 12, 1527–1539. [Google Scholar] [CrossRef] [PubMed]
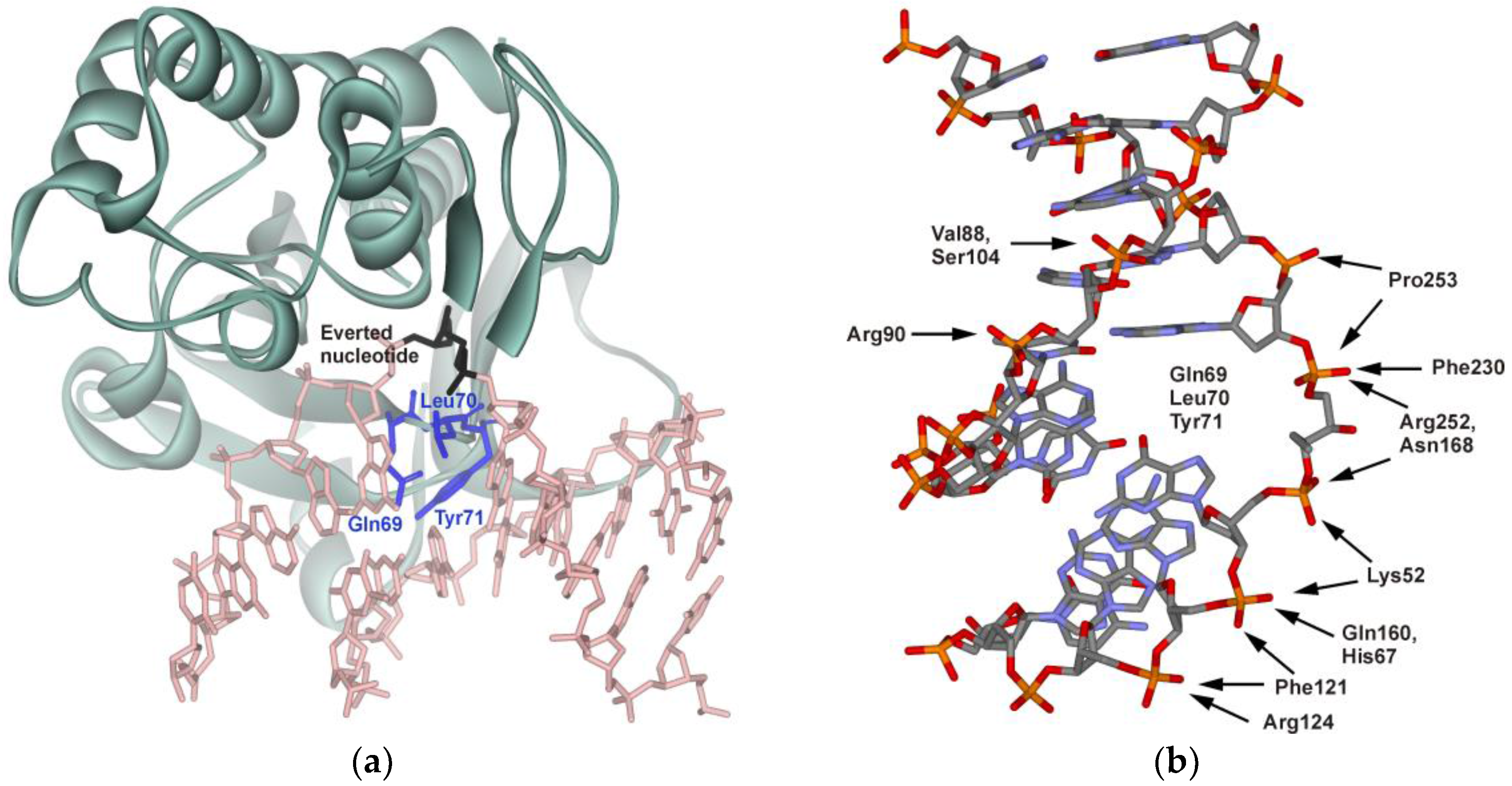

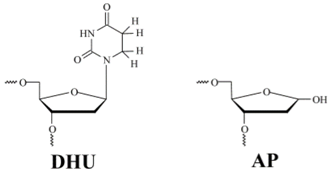
| Shorthand | Sequence |
|---|---|
| DHU-substrate | TCTCTC(DHU)CCTTCC AGAGAG G GGAAGG |
| AP-substrate | TCTCTC(AP)CCTTCC AGAGAG G GGAAGG |
 | |
| Constants | L70S | L70W | Y71W | F121W | F230W | P253W | WT [7] |
|---|---|---|---|---|---|---|---|
| k1, M−1s−1 | (0.09 ± 0.02) × 106 | (0.06 ± 0.01) × 106 | (21 ± 3) × 106 | (39 ± 11) × 106 | (30 ± 2) × 106 | (27 ± 2) × 106 | (36 ± 7) × 106 |
| k−1, s−1 | 1 ± 0.1 | 0.4 ± 0.1 | 330 ± 30 | 120 ± 31 | 310 ± 45 | 310 ± 26 | 410 ± 20 |
| K1, M−1 | 0.9 × 105 | 1.5 × 105 | 0.63 × 105 | 3 × 105 | 0.97 × 105 | 0.87 × 105 | 0.88 × 105 |
| k2, s−1 | 16 ± 3 | 17 ± 4 | 21 ± 3 | 23 ± 3 | 27 ± 2 | ||
| k−2, s−1 | 2.5 ± 0.1 | 0.55 ± 0.25 | 1.2 ± 0.1 | 0.8 ± 0.1 | 1.8 ± 0.3 | ||
| K2 | 6.4 | 31 | 17.5 | 28.7 | 15 | ||
| k3, s−1 | 0.4 ± 0.1 | 0.58 ± 0.15 | 0.82 ± 0.04 | 1.0 ± 0.03 | 1.6 ± 0.1 | ||
| k−3, s−1 | 1.1 ± 0.1 | 0.89 ± 0.14 | 0.66 ± 0.06 | 1.2 ± 0.1 | 1.5 ± 0.2 | ||
| K3 | 0.36 | 0.65 | 1.2 | 0.83 | 1.1 | ||
| Kass, M−1 | 1.45 × 105 | 6 × 106 | 2 × 106 | 2 × 106 | 1.45 × 106 | ||
| kcat, s−1 | 0.09 ± 0.01 | 0.08 ± 0.03 | 0.14 ± 0.03 | 0.26 ± 0.09 | 0.38 ± 0.01 | 0.4 ± 0.03 | 0.35 ± 0.02 |
| Kp, M | (1.0 ± 0.2) × 10−6 | (0.7 ± 0.5) × 10−6 | (0.4 ± 0.1) × 10−6 | (0.6 ± 0.1) × 10−6 | (0.6 ± 0.2) × 10−6 | (0.63 ± 0.08) × 10−6 | (0.7 ± 0.1) × 10−6 |
| Constants | L70S | L70W | Y71W | F121W | F230W | P253W | WT [7] |
|---|---|---|---|---|---|---|---|
| k1, M−1s−1 | (0.09 ± 0.01) × 106 | (0.11 ± 0.02) × 106 | (14 ± 2) × 106 | (16 ± 1) × 106 | (18 ± 8) × 106 | (18 ± 3) × 106 | (12 ± 1) × 106 |
| k−1, s−1 | 0.9 ± 0.03 | 0.6 ± 0.05 | 170 ± 17 | 132 ± 12 | 81 ± 32 | 130 ± 28 | 260 ± 10 |
| K1, M−1 | 1 × 105 | 1.8 × 105 | 0.8 × 105 | 1.2 × 105 | 2.2 × 105 | 1.4 × 105 | 0.5 × 105 |
| k2, s−1 | 21 ± 2 | 35 ± 3 | 25 ± 6 | 37 ± 4 | 30 ± 1 | ||
| k−2, s−1 | 1.4 ± 0.1 | 0.9 ± 0.3 | 1.0 ± 0.3 | 1.9 ± 0.5 | 4.4 ± 0.2 | ||
| K2 | 15 | 39 | 25 | 19 | 6.8 | ||
| k3, s−1 | 0.13 ± 0.01 | 0.8 ± 0.1 | 0.9 ± 0.1 | 0.35 ± 0.02 | 2.1 ± 0.1 | ||
| k−3, s−1 | 1.0 ± 0.1 | 3.0 ± 0.6 | 4.7 ± 1.1 | 1.9 ± 0.2 | 1.1 ± 0.1 | ||
| K3 | 0.13 | 0.27 | 0.19 | 0.18 | 1.9 | ||
| K1K2K3, M−1 | 1 × 105 | 1 × 105 | 1.6 × 105 | 12.6 × 105 | 10.5 × 105 | 4.9 × 105 | 6.5 × 105 |
| kcat, s−1 | 0.09 ± 0.01 | 0.08 ± 0.007 | 0.29 ± 0.03 | 0.41 ± 0.05 | 1.0 ± 0.1 | 0.45 ± 0.08 | 0.79 ± 0.03 |
| Kp, M | (0.18 ± 0.02) × 10−6 | (0.2 ± 0.1) × 10−6 | (0.11 ± 0.01) × 10−6 | (1.5 ± 0.4) × 10−6 | (1.1 ± 0.6) × 10−6 | (1.4 ± 0.2) × 10−6 | (1.8 ± 0.3) × 10−6 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kladova, O.A.; Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Mutational and Kinetic Analysis of Lesion Recognition by Escherichia coli Endonuclease VIII. Genes 2017, 8, 140. https://doi.org/10.3390/genes8050140
Kladova OA, Kuznetsova AA, Fedorova OS, Kuznetsov NA. Mutational and Kinetic Analysis of Lesion Recognition by Escherichia coli Endonuclease VIII. Genes. 2017; 8(5):140. https://doi.org/10.3390/genes8050140
Chicago/Turabian StyleKladova, Olga A., Alexandra A. Kuznetsova, Olga S. Fedorova, and Nikita A. Kuznetsov. 2017. "Mutational and Kinetic Analysis of Lesion Recognition by Escherichia coli Endonuclease VIII" Genes 8, no. 5: 140. https://doi.org/10.3390/genes8050140







