RNA-Seq Study of Microbially Induced Hemocyte Transcripts from Larval Heliothis virescens (Lepidoptera: Noctuidae)
Abstract
:1. Introduction
2. Results and Discussion
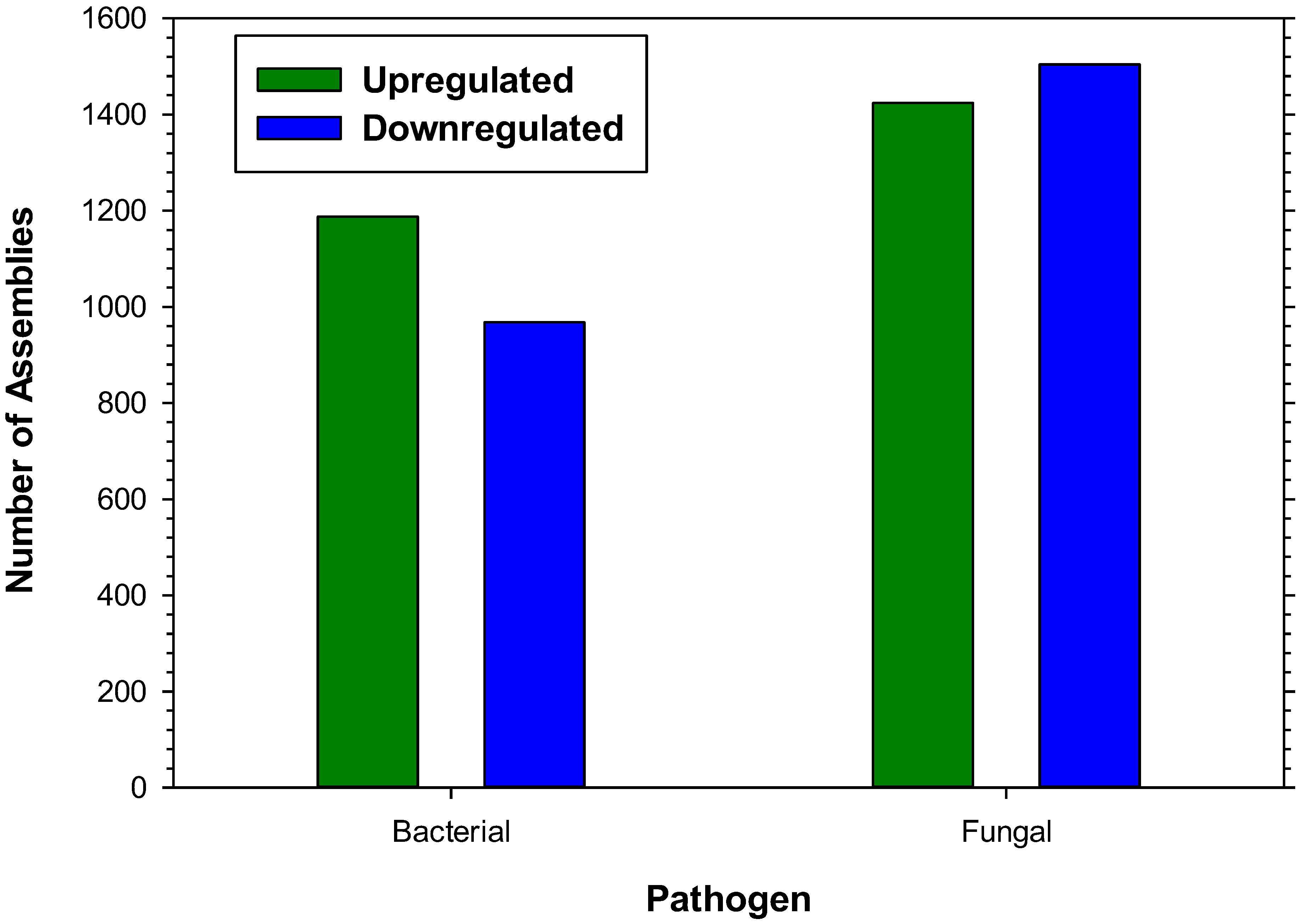
2.1. Transcriptome Assembly
| Gene ID | Contig ID(HvUMC.) | Contr | Bac | Bac Fold Change | Fung | Fung Fold Change | Contig Length | % Cov | SpeciesTop Blast | e-Value | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Up-Regulated Genes | |||||||||||
| β-1,3-Glucan Recognition Protein 2b | 2.6025.Contig1 | 47 | 138 | 2.9 | 146 | 3.1 | 239 | 77 | Helicoverpa armigera | 4E-27 | |
| C-type Lectin 10 | 2.28786.Contig1 | 32 | 112 | 3.5 | 92 | 2.9 | 487 | 52 | Bombyx mori | 2E-38 | |
| C-type Lectin | 1.single.v21-20824 | 1,723 | 4,150 | 2.4 | 7,281 | 4.2 | 3,126 | 46 | Aedes aegypti | E-122 | |
| Cyclophilin | 1.single.v31-289707 | 4 | 19 | 4.8 | 13 | 3.3 | 591 | 53 | Aedes aegypti | 1E-50 | |
| Dscam, Isoform AY | 1.single.v21-784756 | 6 | 12 | 2.0 | 18 | 3.0 | 159 | 84 | Drosophila melanogaster | 4E-17 | |
| Dscam | 1.single.v21-781454 | 6 | 14 | 2.3 | 26 | 4.3 | 192 | 92 | Pediculus h. corporus | 5E-27 | |
| F-box/LRR-repeat protein 16 | 1.single.v21-728991 | 2 | 8 | 4.0 | 12 | 6.0 | 187 | 88 | Camponotus floridanus | 4E-24 | |
| Galactose Binding Lectin, Soluble 9 | 1.single.Rem21-3740 | 142 | 508 | 3.6 | 194 | 1.4 | 696 | 51 | Mus musculus | 1E-19 | |
| Hemocytin | 1.single.Rem27-385 | 1,152 | 3,816 | 3.3 | 5,276 | 4.6 | 484 | 39 | Harpegnathos saltator | 8E-31 | |
| Immulectin-2 | 6.26705.Contig1 | 66 | 58 | 0.9 | 844 | 12.8 | 1,005 | 36 | Manduca sexta | 1E-49 | |
| Immulectin-3 | 1.single.v23-434605 | 17 | 51 | 3.0 | 44 | 2.6 | 235 | 60 | Manduca sexta | 1E-6 | |
| Integrin alpha 3 | 3.22533.Contig1 | 1,133 | 2,519 | 2.2 | 4,186 | 3.7 | 3,312 | 60 | Pseudoplusia includens | 0 | |
| Integrin Beta 1 Subunit | 1.single.Rem23-17883 | 31 | 126 | 4.1 | 66 | 2.1 | 130 | 100 | Spodoptera exigua | 3E-12 | |
| Integrin-linked Kinase | 4.1761.Contig1 | 31 | 96 | 3.1 | 102 | 3.3 | 633 | 78 | Glossina morsitans | 1E-63 | |
| Lectin 3 | 1.single.v21-686389 | 11 | 43 | 3.9 | 27 | 2.5 | 245 | 36 | Lonomia obliqua | 2E-8 | |
| Leucine Rich Protein | 1.single.v37-139619 | 8 | 15 | 1.9 | 32 | 4.0 | 227 | 36 | Aedes aegypti | 2E-8 | |
| Lipopolysaccharide Binding Protein | 3.9650.Contig1 | 2,442 | 7,490 | 3.1 | 8,921 | 3.7 | 158 | 78 | Helicoverpa armigera | 1E-11 | |
| NF-Kappa B Essential Modulator | 1.single.Rem21-38499 | 12 | 39 | 3.3 | 24 | 2.0 | 176 | 48 | Aedes aegypti | 4E-6 | |
| Paralytic Peptide Binding Protein 2 | 64.547.Contig4 | 3 | 197 | 65.7 | 1 | 0.33 | 247 | 42 | Bombyx mori | 7E-5 | |
| Peroxinectin | 2.12813.Contig1 | 6 | 25 | 4.2 | 32 | 5.3 | 190 | 45 | Ixodes scapularis | 2E-7 | |
| PGRP | 6.1191.Contig1 | 302 | 1,969 | 6.5 | 6,633 | 22.0 | 1,366 | 87 | Helicoverpa armigera | 3E-88 | |
| PGRP-SA | 1.single.v37-159335 | 71 | 171 | 2.4 | 249 | 3.5 | 232 | 78 | Tribolium castaneum | 3E-13 | |
| Scavenger Receptor | 1.single.v29-351108 | 3 | 9 | 3.0 | 14 | 4.7 | 172 | 71 | Culex quinquefasciatus | 2E-18 | |
| Toll Precursor | 15.16134.Contig1 | 178 | 550 | 3.1 | 708 | 4.0 | 2,095 | 28 | Pediculus h. corporis | 4E-58 | |
| Toll receptor 18-Wheeler | 1.single.v37-162137 | 32 | 41 | 1.3 | 98 | 3.1 | 144 | 52 | Spodoptera frugiperda | 7E-5 | |
| Down-Regulated Genes | |||||||||||
| Galectin-12 | 9.8877.v39-13988 | 139 | 358 | 2.6 | 11 | 0.08 | 218 | 48 | Harpegnathos saltator | 3E-12 | |
| Lectin 5 | 17.2309.Contig1 | 266 | 36 | 0.14 | 361 | 1.4 | 848 | 45 | Lonomia obliqua | 2E-36 | |
| NADP-Leukotriene B4 12- hydroxydehydrogenase | 1.single.v25-664428 | 12 | 5 | 0.42 | 2 | 0.17 | 424 | 63 | Culex quinquefasciatus | 2E-45 | |
| Gene ID | Contig ID(HvUMC.) | Contr | Bac | Bac Fold Change | Fung | Fung Fold Change | Contig Length | % Cov | SpeciesTop BLAST | e-Value | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Up-Regulated Genes | |||||||||||
| Brasiliensin Thrombin Inhibitor | 1.single.v23-492316 | 114 | 195 | 1.7 | 453 | 4.0 | 792 | 41 | Triatoma basiliensis | 6E-32 | |
| DOPA Decarboxylase | 1.single.v21-586620 | 76 | 120 | 1.6 | 621 | 8.2 | 1,394 | 94 | Mamestra brassicae | 0 | |
| Hemocyte Protein-Glutamine Gamma- Glutamyltransferase | 1.single.v23-275987 | 232 | 374 | 1.6 | 1,449 | 6.2 | 2,292 | 34 | Camponotus floridanus | 0 | |
| Hemolymph Proteinase 18 | 3.3815.Contig1 | 490 | 710 | 1.4 | 4,941 | 10.1 | 1,350 | 41 | Manduca sexta | 8E-74 | |
| Hemolymph Proteinase 19 | 1.single.v21-731703 | 14 | 54 | 3.9 | 22 | 1.6 | 369 | 69 | Manduca sexta | 2E-49 | |
| Immulectin-2 | 6.26705.Contig1 | 66 | 58 | 0.9 | 844 | 12.8 | 1,005 | 36 | Manduca sexta | 1E-49 | |
| Immulectin-3 | 1.single.v23-434605 | 17 | 51 | 3.0 | 44 | 2.6 | 235 | 60 | Manduca sexta | 1E-6 | |
| Laccase-4-like | 1.single.Rem21-26133 | 6 | 31 | 5.2 | 13 | 2.2 | 292 | 56 | Bombus terrestris | 1E-22 | |
| Laccase-7-like | 2.7276.Contig1 | 9 | 33 | 3.7 | 14 | 1.6 | 269 | 55 | Acyrthosiphon pisum | 8E-23 | |
| Prophenoloxidase Activating Enzyme | 3.18472.Contig1 | 40 | 166 | 4.2 | 168 | 4.2 | 135 | 95 | Helicoverpa armigera | 4E-16 | |
| Reeler | 1.single.v21-527354 | 7 | 189 | 27.0 | 549 | 78.4 | 502 | 77 | Bombyx mori | 1E-61 | |
| Serpin-like Protein | 1.single.v21-800532 | 2 | 4 | 2.0 | 20 | 10.0 | 167 | 83 | Antheraea mylitta | 7E-19 | |
| Transglutaminase | 1.single.Rem21-50794 | 2 | 6 | 3.0 | 13 | 6.5 | 185 | 76 | Apis mellifera | 2E-18 | |
| Yellow 2 | 1.single.Rem21-90085 | 2 | 1 | 0.5 | 11 | 5.5 | 124 | 57 | Bombyx mori | 6E-6 | |
| Yellow-b | 1.single.v23-9110 | 832 | 2,954 | 3.6 | 2,802 | 3.4 | 1,833 | 78 | Heliconius melpomene | 0 | |
| Yellow-f | 1.single.v39-31578 | 49 | 147 | 3.0 | 154 | 3.1 | 559 | 85 | Bombyx mori | 6E-92 | |
| Down-Regulated Genes | |||||||||||
| Hemolymph Proteinase 17 | 1.single.v21-676209 | 395 | 19 | 0.05 | 72 | 0.18 | 1,979 | 51 | Manduca sexta | 1E-168 | |
| Hemolymph Proteinase 20 | 2.20519.Contig1 | 15 | 3 | 0.20 | 4 | 0.27 | 253 | 51 | Manduca sexta | 3E-17 | |
| Phenylalanine Hydroxylase | 1.single.v25-669762 | 14 | 3 | 0.21 | 2 | 0.14 | 250 | 95 | Papilio xuthus | 3E-41 | |
| Gene ID | Contig ID (HvUMC.) | Contr | Bac | Bac Fold Change | Fung | Fung Fold Change | Contig Length | % Cov | Species Top BLAST | e-Value | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Up-Regulated Genes | |||||||||||
| Antibacterial Protein 3Tox | 35.7580.Contig4 | 63 | 1,115 | 17.7 | 2,810 | 44.6 | 270 | 77 | Heliothis virescens | 1E-24 | |
| Antimicrobial Protein 5Tox | 24.13350.Contig4 | 16 | 451 | 28.2 | 538 | 33.6 | 314 | 64 | Bombyx mori | 2E-27 | |
| Antimicrobial Protein 6Tox | 24.13350.Contig5 | 16 | 273 | 17.1 | 766 | 47.9 | 267 | 55 | Bombyx mori | 2E-19 | |
| Attacin A Precursor | 3.25069.Contig1 | 4 | 156 | 39.0 | 587 | 146.8 | 735 | 100 | Heliothis virescens | E-68 | |
| Carboxylesterase | 3.2554.Contig1 | 9 | 49 | 5.4 | 92 | 10.2 | 445 | 87 | Helicoverpa armigera | 3E-70 | |
| Cecropin 3 | 34.474.Contig1 | 123 | 1,336 | 10.9 | 1,333 | 10.8 | 366 | 86 | Helicoverpa armigera | 3E-31 | |
| Cecropin A2 | 34.474.Contig3 | 275 | 917 | 3.3 | 598 | 2.2 | 301 | 66 | Helicoverpa armigera | 3E-14 | |
| Cecropin D | 11.9800.Contig1 | 25 | 246 | 9.8 | 411 | 16.4 | 345 | 74 | Helicoverpa armigera | 2E-17 | |
| Chymotrypsin Inhibitor CI-8A | 1.single.v37-179791 | 285 | 450 | 1.6 | 1,453 | 5.1 | 493 | 71 | Bombyx mori | 3E-65 | |
| Cobatoxin B Long Form | 1.single.v29-23061 | 2,365 | 25,592 | 10.8 | 21,264 | 9 | 555 | 51 | Spodoptera frugiperda | 9E-16 | |
| Gallerimycin | 6.18346.Contig1 | 21 | 372 | 17.7 | 846 | 40.3 | 362 | 78 | Helicoverpa armigera | 9E-27 | |
| Gloverin-like Antibacterial Protein | 5.6087.Contig1 | 13 | 420 | 32.3 | 645 | 49.6 | 426 | 82 | Heliothis virescens | 5E-68 | |
| Hemolin | 3.4762.Contig1 | 313 | 3,201 | 10.2 | 6,907 | 22.1 | 1,239 | 96 | Heliothis virescens | 0 | |
| Immune Inducible Protein | 3.5943.Contig1 | 38 | 759 | 20.0 | 1,773 | 46.7 | 294 | 79 | Helicoverpa armigera | 2E-22 | |
| Inducible Metalloproteinase Inhibitor | 1.single.v21-505063 | 133 | 518 | 3.9 | 178 | 1.3 | 693 | 53 | Galleria mellonela | 1E-17 | |
| I-type Lysozyme | 3.8206.Contig1 | 290 | 509 | 1.8 | 1,038 | 3.6 | 795 | 57 | Sitophilus zeamais | 6E-42 | |
| Heliocin Precursor | 12.13910.Contig1 | 26 | 361 | 13.9 | 813 | 31.3 | 583 | 86 | Heliothis virescens | 3E-49 | |
| Kazal-type Inhibitor | 2.5500.Contig1 | 4 | 21 | 5.3 | 5 | 1.3 | 351 | 48 | Panstrongylus megistus | 6E-20 | |
| Lysozyme | 3.27236.Contig1 | 6,536 | 16,912 | 2.6 | 23,565 | 3.6 | 355 | 99 | Heliothis virescens | 3E-67 | |
| Metalloproteinase Inhibitor 3 | 1.single.v21-774328 | 56 | 102 | 1.8 | 211 | 3.8 | 1,054 | 42 | Tribolium castaneum | 2E-39 | |
| Nimrod-like Protein | 4.7692.Contig1 | 315 | 1,043 | 3.3 | 2,122 | 6.7 | 843 | 47 | Tribolium castaneum | 2E-47 | |
| Viresin | 2.22031.Contig1 | 28 | 35 | 1.2 | 194 | 6.9 | 448 | 90 | Heliothis virescens | 9E-61 | |
| Virescein Precursor | 2.19097.Contig1 | 0 | 19 | 19.0 | 65 | 65.0 | 193 | 100 | Heliothis virescens | 3E-6 | |
| Down-Regulated Genes | |||||||||||
| Adamts-7 Metallopeptidase | 1.single.v37-182137 | 33 | 6 | 0.18 | 3 | 0.09 | 167 | 69 | Aedes aegypti | 7E-16 | |
| Chemosensory Protein | 1.single.Rem23-43567 | 22 | 3 | 0.14 | 19 | 0.87 | 304 | 100 | Heliothis virescens | 4E-33 | |
| Immune-related Hdd13 | 3.8673.Contig1 | 106 | 28 | 0.26 | 18 | 0.17 | 340 | 56 | Hyphantria cunea | 1E-31 | |
| Odorant Binding Protein | 1.single.v29-513279 | 26 | 2 | 0.08 | 30 | 1.15 | 179 | 98 | Heliothis virescens | 3E-27 | |
| Gene ID | Contig ID(HvUMC.) | Contr | Bac | Bac Fold Change | Fung | Fung Fold Change | Contig Length | % Cov | SpeciesTop BLAST | e-Value | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Up-Regulated Genes | |||||||||||
| Antennal Cytochrome P450 CYP4 | 1.single.v37-177294 | 5 | 49 | 9.8 | 1 | 0.20 | 271 | 78 | Mamestra brassicae | 2E-28 | |
| Antennal Cytochrome P450 CYP9 | 1.single.v27-352481 | 27 | 32 | 1.2 | 280 | 10.4 | 701 | 78 | Mamestra brassicae | 1E-103 | |
| Autophagy-like Protein Atg12 | 2.26567.Contig1 | 4 | 9 | 2.3 | 17 | 4.3 | 265 | 69 | Biston betularia | 1E-10 | |
| Autophagy-related Protein B2-like | 1.single.v29-386662 | 2 | 6 | 3.0 | 8 | 4.0 | 714 | 50 | Acromyrmex echinatior | 6E-51 | |
| Autophagy-related Protein 9A | 2.9906.Contig1 | 3 | 8 | 2.7 | 15 | 5.0 | 170 | 60 | Danio rerio | 3E-13 | |
| Calcium & Integrin-binding Protein 1 | 1.single.v35-260454 | 11 | 33 | 3.0 | 19 | 1.7 | 231 | 61 | Harpegnathos saltator | 1E-13 | |
| Calnexin 99A, Isoform C | 1.single.v39-34103 | 29 | 96 | 3.3 | 81 | 2.8 | 241 | 73 | Drosophila melanogaster | 1E-30 | |
| Calpain Protein | 6.18445.Rem27-3414 | 42 | 126 | 3.0 | 90 | 2.1 | 205 | 85 | Bombyx mori | 1E-5 | |
| Calponin/Transgelin | 3.3375.Contig1 | 121 | 134 | 1.1 | 466 | 3.9 | 854 | 91 | Aedes aegypti | 9E-84 | |
| CarE | 1.single.v21-683558 | 167 | 271 | 1.6 | 536 | 3.2 | 152 | 80 | Spodoptera exigua | 0 | |
| Carboxyl/choline Esterase | 1.single.v25-663787 | 11 | 48 | 4.4 | 58 | 5.3 | 242 | 75 | Helicoverpa armigera | 6E-30 | |
| Catalase | 1.single.Rem23-39568 | 39 | 58 | 1.5 | 384 | 9.8 | 152 | 92 | Spodoptera litura | 3E-15 | |
| Fasciclin-1 | 1.single.v29-874 | 3489 | 29527 | 8.5 | 15189 | 4.4 | 1459 | 56 | Danaus plexippus | 5E-94 | |
| Ferritin Heavy Chain | 1.single.v23-133164 | 10 | 60 | 6.0 | 0 | 0 | 159 | 92 | Trichoplusia ni | 8E-20 | |
| Glutathione Synthetase | 3.11637.Contig1 | 5 | 36 | 7.2 | 16 | 3.2 | 275 | 60 | Harpegnathos saltator | 1E-6 | |
| Glutathione S-transferase | 1.single.v21-548409 | 16 | 72 | 4.5 | 238 | 14.9 | 639 | 73 | Amyelois transitella | 2E-86 | |
| Gossypol-induced Cytochrome P450 | 1.single.v35-24142 | 3111 | 13869 | 4.5 | 5967 | 1.9 | 1908 | 79 | Helicoverpa armigera | 0 | |
| Hemicentin-like Protein 2 | 1.single.v33-251 | 235 | 1084 | 4.6 | 651 | 2.8 | 363 | 50 | Spodoptera frugiperda | 2E-20 | |
| Innexin 1 | 1.single.Rem21-52588 | 76 | 243 | 3.2 | 359 | 4.7 | 200 | 82 | Pediculus h. corporis | 1E-25 | |
| Innexin 3 | 3.4996.Rem23-8776 | 35 | 168 | 4.8 | 245 | 7.0 | 263 | 68 | Harpegnathos saltator | 5E-25 | |
| Laminin A Chain | 2.31786.Contig1 | 100 | 317 | 3.2 | 142 | 1.4 | 844 | 69 | Aedes aegypti | 2E-64 | |
| Laminin-like Protein Epi-1 | 1.single.v25-592818 | 11 | 34 | 3.1 | 40 | 3.6 | 236 | 58 | Camponotus floridanus | 2E-16 | |
| NADPH Oxidase 5-like | 1.single.v21-32103 | 31 | 143 | 4.6 | 187 | 6.0 | 382 | 77 | Acyrthosiphon pisum | 2E-49 | |
| Papilin | 3.16799.Contig1 | 120 | 742 | 6.2 | 556 | 4.6 | 1366 | 40 | Harpegnathos saltator | 5E-63 | |
| Peroxidasin | 1.single.v21-283343 | 806 | 1896 | 2.4 | 2986 | 3.7 | 4327 | 50 | Tribolium castaneum | 0 | |
| Selenium-binding protein | 2.31409.Contig1 | 154 | 388 | 2.5 | 785 | 5.1 | 355 | 59 | Pediculus h. corporus | 8E-30 | |
| Talin-2-like | 2.27492.Contig1 | 26 | 63 | 2.4 | 89 | 3.4 | 1280 | 75 | Bombus terrestris | 1E-178 | |
| Teneurin-3 Isoform 1 | 1.single.v21-374519 | 217 | 652 | 3.0 | 703 | 3.2 | 597 | 88 | Apis mellifera | 4E-98 | |
| Transferrin | 4.13853.Contig1 | 28 | 101 | 3.6 | 187 | 6.7 | 1071 | 33 | Aedes aegypti | 2E-44 | |
| Down-Regulated Genes | |||||||||||
| Apoptosis Inducing Factor, Putative | 1.single.v21-402220 | 272 | 286 | 1.1 | 54 | 0.20 | 391 | 62 | Ixodes scapularis | 7E-16 | |
| Calnexin, Putative | 1.single.Rem21-38232 | 26 | 18 | 0.69 | 5 | 0.19 | 159 | 68 | Ixodes scapularis | 1E-14 | |
| Ferrochelatase precursor | 1705.v27-585704 | 16 | 3 | 0.188 | 3 | 0.188 | 149 | 66 | Chironomus sp. | 7E-11 | |
| Flavin-dependent Monooxygenase | 1.single.v21-786790 | 79 | 24 | 0.30 | 36 | 0.46 | 501 | 97 | Helicoverpa armigera | 2E-87 | |
2.2. Pattern Recognition Receptors
2.3. Signal Transduction
2.4. Melanization, Nodulation and Encapsulation
2.5. Antimicrobials and Other Effectors
2.7. Cellular Response Factors
3. Experimental Section
3.1. Insects, Infection, and RNA Isolation
3.2. Sequence Generation
3.3. De Novo Exome Assembly
3.4. Identification of Differentially Regulated Genes
4. Conclusions
Acknowledgments
References
- Cho, S.; Mitchell, A.; Mitter, C.; Regier, J.; Matthews, M.; Robertson, R. Molecular phylogenetics of heliothine moths (Lepidoptera: Noctuidae: Heliothinae), with comments on the evolution of host range and pest status. Syst. Entomol. 2008, 33, 581–594. [Google Scholar] [CrossRef]
- Albernaz, K.C.; Silva-Brandao, K.L.; Fresia, P.; Consoli, F.L.; Omoto, C. Genetic variability and demographic history of Heliothis virescens (Lepidoptera: Noctuidae) populations from Brazil inferred by mtDNA sequences. Bull. Entomol. Res. 2012, 103, 333–343. [Google Scholar]
- Fitt, G.P. The ecology of Heliothis species in relation to agroecosystems. Annu. Rev. Entomol. 1989, 34, 17–52. [Google Scholar] [CrossRef]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef]
- Hunter, W.B.; VanEngelsdorp, D.; Hayes, J.; Westervelt, D.; Glick, E.; Williams, M.; Sela, I.; Maori, E.; Pettis, J.; Cox-Foster, D.; et al. Large-Scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLoS Pathog. 2010, 6, e1001160. [Google Scholar] [CrossRef]
- Govind, G.; Mittapalli, O.; Griebel, T.; Allmann, S.; Bocker, S.; Baldwin, I.T. Unbiased transcriptional comparisons of generalist and specialist herbivores feeding on progressively defenseless Nicotiana attenuata plants. PLoS One 2010, 5, e8735. [Google Scholar]
- Shelby, K.S.; Popham, H.J.R. Analysis of ESTs generated from immune-stimulated hemocytes of larval Heliothis virescens. J. Invertebr. Pathol. 2009, 101, 86–95. [Google Scholar] [CrossRef]
- Vogel, H.; Heidel, A.; Heckel, D.G.H; Groot, A.T. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genomics 2010, 11. [Google Scholar] [CrossRef]
- Kafatos, F.; Waterhouse, R.; Zdobnov, E.; Christophides, G. Comparative Genomics of Insect Immunity. In Insect Infection and Immunity: Evolution, Ecology, and Mechanisms; Rolff, J., Reynolds, S.E., Eds.; Oxford University Press: New York, NY, USA, 2009; pp. 86–105. [Google Scholar]
- Strand, M.R. Insect Hemocytes and Their Role in Immunity. In Insect Immunology; Beckage, N.E., Ed.; Elsevier: New York, NY, USA, 2008; pp. 49–68. [Google Scholar]
- Jiang, H.; Vilcinskas, A.; Kanost, M.R. Immunity in Lepidopteran Insects. In Invertebrate Immunity; Soderhall, K., Ed.; Springer: New York, NY, USA, 2011; Volume 708, pp. 163–180, Advances in Experimental Biology and Medicine. [Google Scholar]
- Blander, J.M.; Sander, L.E. Beyond pattern recognition: Five immune checkpoints for scaling the microbial threat. Nat. Rev. Immunol. 2012, 12, 215–225. [Google Scholar] [CrossRef]
- Karlsson, J.; Oldenvi, S.; Fahlander, C.; Daenthanasanmak, A.; Steiner, H. Growing bacteria shed elicitors of Drosophila humoral immunity. J. Innate Immun. 2012, 4, 111–116. [Google Scholar] [CrossRef]
- Altincicek, B.; Berisha, A.; Mukherjee, K.; Spengler, B.; Rommp, A.; Vilcinskas, A. Identification of collagen IV derived danger/alarm signals in insect immunity by nanoLC-FTICR MS. Biol. Chem. 2009, 390, 1303–1311. [Google Scholar] [CrossRef]
- Broderick, N.A.; Welchman, D.P.; Lemaitre, B. Recognition and Response to Microbial Infection in Drosophila. In Insect Infection and Immunity: Evolution, Ecology, and Mechanisms; Rolff, J., Reynolds, S.E., Eds.; Oxford University Press: New York, NY, USA, 2009; pp. 13–33. [Google Scholar]
- Kanost, M.R.; Nardi, J.B. Innate Immune Responses of Manduca Sexta. In Molecular Biology and Genetics of the Lepidoptera; Goldsmith, M.R., Marec, F., Eds.; CRC Press: Boca Raton, FL, USA, 2009; pp. 271–292. [Google Scholar]
- Huang, J.; Wu, S.-F.; Li, X.-H.; Adamo, S.A.; Ye, G.-Y. The characterization of a concentration-sensitive–adrenergic-like octopamine receptor found on insect immune cells and its possible role in mediating stress hormone effects on immune function. Brain Behav. Immun. 2012, 26, 942–950. [Google Scholar] [CrossRef]
- Garcia, E.S.; Castro, D.P.; Figueiredo, M.B.; Genta, F.A.; Azambuja, P. Trypanosoma rangeli: A new perspective for styding the modulation of immune reactions of Rhodnius prolixus. Parasit. Vectors 2009, 2, 33. [Google Scholar] [CrossRef]
- Hrsyl, P.; Dobes, P.; Wang, Z.; Hauling, T.; Wilhelmsson, C.; Theopold, U. Clotting factors and eicosanoids protect against nematode infections. J. Innate Immunol. 2011, 3, 65–70. [Google Scholar] [CrossRef]
- Terenius, O.; Popham, H.J.R.; Shelby, K.S. Bacterial, but not baculoviral infections stimulate hemolin expression in noctuid moths. Dev. Comp. Immunol. 2009, 33, 1176–1185. [Google Scholar] [CrossRef]
- Shelby, K.S.; Popham, H.J.R. Cloning and characterization of the secreted hemocytic prophenoloxidases of Heliothis virescens. Arch. Insect Biochem. Physiol. 2008, 69, 127–142. [Google Scholar]
- Breitenbach, J.E.; Shelby, K.S.; Popham, H.J.R. Baculovirus induced transcripts in hemocytes from the noctuid moth, Heliothis virescens. Viruses 2011, 3, 2047–2064. [Google Scholar] [CrossRef]
- Beldade, P.; McMillan, W.O.; Papanicolaou, A. Butterfly genomics eclosing. Heredity 2008, 100, 150–157. [Google Scholar] [CrossRef]
- Zhu, Y.; Ragan, E.J.; Kanost, M.R. Leureptin: A soluble, extracellular leucine-rich repeat protein from Manduca sexta that binds lipopolysaccharide. Insect Biochem. Mol. Biol. 2010, 40, 713–722. [Google Scholar] [CrossRef]
- Mohamed, A.A.M.; Kim, Y. Target-Specific feeding toxicity of β1 integrin dsRNA against diamondback moth, Plutella xylostella. Arch. Insect Biochem. Physiol. 2011, 78, 216–230. [Google Scholar] [CrossRef]
- Ghosh, J.; Lun, C.M.; Majeske, A.J.; Sacchi, S.; Schrankel, C.S.; Smith, L.C. Invertebrate immune diversity. Dev. Comp. Immunol. 2011, 35, 959–9774. [Google Scholar] [CrossRef]
- Zdybicka-Barabas, A.; Staczek, S.; Mak, P.; Piersiak, T.; Skrzyrpiek, T.; Cytrynska, M. The effect of Galleria mellonella apolipophorin III on yeasts and filamentous fungi. J. Insect Physiol. 2012, 58, 164–177. [Google Scholar] [CrossRef]
- Hanada, Y.; Sekimizu, K.; Kaito, C. Silkworm apolipophorin protein inhibits Staphylococcus aureus virulence. J. Biol. Chem. 2011, 286, 39360–39369. [Google Scholar]
- Gupta, L.; Noh, J.Y.; Jo, Y.H.; Oh, S.H.; Kumar, S.; Noh, M.Y.; Lee, Y.S.; Cha, S.J.; Seo, S.J.; Kim, I.; et al. Apolipophorin-III mediates antiplasmodial epithelial responses in Anopheles gambiae (G3) mosquitoes. PLoS One 2010, 5, e15410. [Google Scholar]
- Tsuzuki, S.; Ochiai, M.; Matsumoto, H.; Kurata, S.; Ohnishi, A.; Hayakawa, Y. Drosophila growth-blocking peptide-like factor mediates acute immune reactions during infections and non-infectious stress. Nat. Sci. Rep. 2012, 2. [Google Scholar] [CrossRef]
- Park, J.-A.; Kim, Y. Eicosanoid biosynthesis is activated via Toll, but not Imd signal pathway in response to fungal infection. J. Invertbr. Pathol. 2012, 110, 382–388. [Google Scholar] [CrossRef]
- Falabella, P.; Riviello, L.; Pascale, M.; Di Lelio, I.; Tettamanti, G.; Grimaldi, A.; Iannone, C.; Monti, M.; Pucci, P.; Tamburro, A.M.; et al. Functional amyloids in insect immune response. Insect Biochem. Mol. Biol. 2012, 42, 203–211. [Google Scholar] [CrossRef]
- Bao, Y.-Y.; Xue, J.; Wu, W.-J.; Wang, Y.; Lv, Z.-Y.; Zhang, C.-X. An immune-induced Reeler protein is involved in the Bombyx mori melanization cascade. Insect Biochem. Mol. Biol. 2011, 441, 696–706. [Google Scholar]
- Luo, K.; Turnbull, M.W. Characterization of nonjunctional hemichannels in caterpillar cells. J. Insect Sci. 2011, 11, 6. [Google Scholar]
- Das, S.B.N.A.; Dong, Y.; Garver, L.; Dimoupoulos, G. Specificity of the Innate Immune System: A Closer look at the Mosquito Pattern-Recognition Receptor Repertoire. In Insect Infection and Immunity: Evolution, Ecology, and Mechanisms; Rolff, J., Reynolds, S.E., Eds.; Oxford University Press: New York, NY, USA, 2009; pp. 69–85. [Google Scholar]
- Gennaro, R.; Zanetti, M.; Benincasa, M.; Podda, E.; Miani, M. Pro-Rich antimicrobial peptides from animals: Structure, biological functions and mechanism of action. Curr. Pharm. Des. 2002, 8, 763–778. [Google Scholar] [CrossRef]
- Lamberty, M.; Ades, S.; Uttenweiler-Joseph, S.; Brookhart, G.; Bushey, D.; Hoffmann, J.A.; Bulet, P. Insect immunity. Isolation from the lepidopteran Heliothis virescens of a novel insect defensin with potent antifungal activity. J. Biol. Chem. 1999, 274, 9320–9326. [Google Scholar]
- Chung, K.T.; Ourth, D.D. Viresin: A novel antibacterial protein from immune hemolymph of Heliothis virescens pupae. Eur. J. Biochem. 2000, 267, 677–683. [Google Scholar] [CrossRef]
- Destoumieux-Garzon, D.; Brehelin, M.; Bulet, P.; Boublik, Y.; Girad, P.-A.; Baghdiguian, S.; Zumbihl, R.; Escoubas, J.-M. Spodoptera frugiperda X-tox protein, an immune related defensin rosary, has lost the function of ancestral defensins. PLoS One 2009, 4, e6795. [Google Scholar]
- Girard, P.-A.; Boublik, Y.; Wheat, C.W.; Volkoff, A.-N.; Cousserans, F.; Brehelin, M.; Escoubas, J.-M. X-Tox: An atypical defensin derived family of immune-related proteins specific to Lepidoptera. Dev. Comp. Immunol. 2008, 32, 575–585. [Google Scholar] [CrossRef]
- Shandala, T.; Woodcock, J.M.P.Y.; Biggs, L.; Skoulakis, E.M.C.; Brooks, D.A.; Lopez, A.F. Drosophila 14-3-3ε has a crucial role in anti-microbial peptide secretion and innate immunity. J. Cell Sci. 2012, 123, 2165–2174. [Google Scholar]
- Vogel, H.; Altincicek, B.; Glockner, G.; Vilcinskas, A. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics 2011, 12. [Google Scholar] [CrossRef]
- Sabin, L.R.; Hanna, S.L.; Cherry, S. Innate antiviral immunity in Drosophila. Curr. Opin. Immunol. 2010, 22, 4–9. [Google Scholar]
- Flenniken, M.L.; Kunitomi, M.; Tassetto, M.; Andino, R. The Antiviral Role of RNA Interference. In Insect Viruses; Asgari, S., Johnson, K.N., Eds.; Caister Academic Press: Norfolk, UK, 2010; pp. 367–388. [Google Scholar]
- Aronstein, K.; Oppert, B.; Lorenzen, M.D. RNAi in Agriculturaly-Important Arthropods. In RNA Processing; Grabowski, P., Ed.; InTech: Rijeka, Croatia, 2011; pp. 157–180. [Google Scholar]
- Zha, W.; Peng, X.; Chen, R.; Du, B.; Zhu, L.; He, G. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS One 2011, 6, e20504. [Google Scholar]
- Renwick, J.; Reeves, E.P.; Wientjes, F.B.; Kavanagh, K. Translocation of proteins homologous to human neutrophil p47phox and p67phox to the cell membrane in activated hemocytes of Galleria mellonella. Dev. Comp. Immunol. 2007, 31, 347–359. [Google Scholar] [CrossRef]
- Popham, H.J.R.; Sun, R.; Shelby, K.S.; Robertson, J.D. Changes in trace metals in hemolymph of baculovirus-infected noctuid larvae. Biol. Trace Elem. Res. 2012, 146, 325–334. [Google Scholar] [CrossRef]
- Popham, H.J.R.; Sun, R.; Shelby, K.S.; Robertson, J.D. Iron levels change in larval Heliothis virescens Tissues following baculovirus infection. Biol. Trace Elem. Res. 2012, 148, 356–362. [Google Scholar] [CrossRef]
- Gao, K.; Deng, X.; Qian, H.; Wu, P.; Qin, G.; Guo, X. Cloning, characterization, and expression analysis of a novel BmGDAP1 gene from silkworm, Bombyx mori, involved in cytoplastic polyhedrosis virus infect. Gene 2012, 497, 208–213. [Google Scholar] [CrossRef]
- Kang, L.; Shi, H.; Liu, X.; Zhang, C.; Yao, Q.; Wang, Y.; Chang, C.; Shi, J.; Cao, J.; Kong, J.; et al. Arginine kinase is highly expressed in a resistant strain of silkworm (Bombyx mori, Lepidoptera): Implication of its role in resistance to Bombyx mori nucleopolyhedrovirus. Comp. Biochem. Physiol. 2011, 158B, 230–234. [Google Scholar]
- Navarro-Cerrillo, G.; Ferré, J.; de Maagd, R.A.; Herrero, S. Functional interactions between members of the REPAT family of insect pathogen-induced proteins. Insect Mol. Biol. 2012, 21, 335–342. [Google Scholar] [CrossRef]
- Mao, Y.-B.; Tao, X.-Y.; Xue, X.-Y.; Wang, L.-J.; Chen, X.-Y. Cotton plants expressing CYP6AE14 dsRNAi show enhanced resistance to bollworms. Transgenic Res. 2011, 20, 655–673. [Google Scholar] [CrossRef]
- Ratzka, A.; Vogel, H.; Kliebenstein, D.J.; Mitchell-Olds, T.; Kroymann, J. Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. USA 2002, 99, 11223–11228. [Google Scholar]
- Schramm, K.; Vassao, D.G.; Reichelt, M.; Gershenzon, J.; Wittstock, U. Metabolism of glucosinolate-derived isothiocyanates to glutathione conjugates in generalist lepidopteran herbivores. Insect Biochem. Mol. Biol. 2012, 42, 174–182. [Google Scholar] [CrossRef]
- Isom, S.C.; Spollen, W.G.; Blake, S.M.; Bauer, B.K.; Springer, G.K.; Prather, R.S. Transcriptional profiling of day 12 porcine embryonic disc and trophectoderm samples using ultra-deep sequencing technologies. Mol. Reprod. Dev. 2010, 77, 812–819. [Google Scholar] [CrossRef]
- Popham, H.J.R.; Shelby, K.S.; Popham, T.W. Effect of dietary Se supplementation on resistance to baculovirus infection. Biol. Control 2005, 32, 419–426. [Google Scholar] [CrossRef]
- Shelby, K.S.; Popham, H.J.R. Plasma phenoloxidase of larval Heliothis virescens is virucidal. J. Insect Sci. 2006, 6. [Google Scholar] [CrossRef]
- Walker, W.B.; Allen, M.L. RNAi-Mediated knockdown of IAP in Lygus lineolaris induces mortality in adult and pre-adult life stages. Entomol. Exp. Appl. 2011, 138, 83–92. [Google Scholar] [CrossRef]
- Hamilton, C.; Bulmer, M.S. Molecular antifungal defenses in subterranean termites: RNAi reveals in vivo roles of termicins and GNBPs against a naturally encountered pathogen. Dev. Comp. Immunol. 2012, 36, 371–377. [Google Scholar]
- Zhao, P.; Dong, Z.; Duan, J.; Wang, G.; Wang, L.; Li, Y.; Xiang, Z.; Xia, Q. Genome-Wide identification and immune response analysis of serine protease inhibitor genes in the silkworm, Bombyx mori. PLoS One 2012, 7, e31168. [Google Scholar]
- Bulmer, M.S.; Bachelet, I.; Raman, R.; Rosengaus, R.B.; Sasisekharan, R. Targeting an antimicrobial effector function in insect immunity as a pest control strategy. Proc. Natl. Acad. Sci USA 2009, 106, 12652–12657. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shelby, K.S.; Popham, H.J.R. RNA-Seq Study of Microbially Induced Hemocyte Transcripts from Larval Heliothis virescens (Lepidoptera: Noctuidae). Insects 2012, 3, 743-762. https://doi.org/10.3390/insects3030743
Shelby KS, Popham HJR. RNA-Seq Study of Microbially Induced Hemocyte Transcripts from Larval Heliothis virescens (Lepidoptera: Noctuidae). Insects. 2012; 3(3):743-762. https://doi.org/10.3390/insects3030743
Chicago/Turabian StyleShelby, Kent S., and Holly J. R. Popham. 2012. "RNA-Seq Study of Microbially Induced Hemocyte Transcripts from Larval Heliothis virescens (Lepidoptera: Noctuidae)" Insects 3, no. 3: 743-762. https://doi.org/10.3390/insects3030743
APA StyleShelby, K. S., & Popham, H. J. R. (2012). RNA-Seq Study of Microbially Induced Hemocyte Transcripts from Larval Heliothis virescens (Lepidoptera: Noctuidae). Insects, 3(3), 743-762. https://doi.org/10.3390/insects3030743




