Rabbits as a Reservoir of Multidrug-Resistant Escherichia coli: Clonal Lineages and Public Health Impact
Abstract
:1. Introduction
2. Results
2.1. Bacteria Isolation
2.2. Antibiotic Resistance Phenotypes
2.3. Molecular Characterization and Multilocus Sequence Typing (MLST)
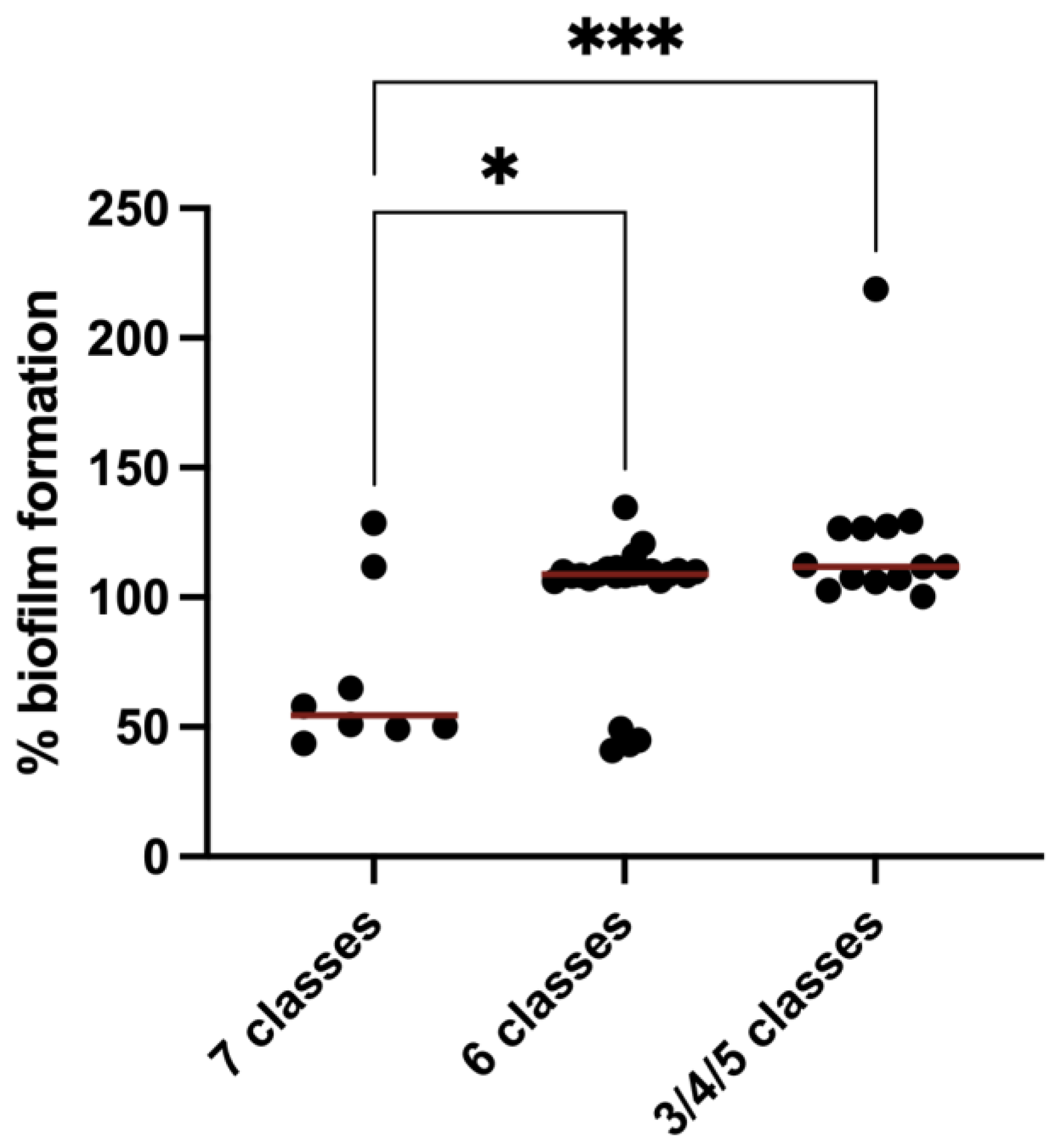
2.4. Quantification of Biofilm Formation
3. Discussion
3.1. Antibiotic Resistance in Rabbit Farm Environments
3.2. Genetic Diversity of CTX-Resistant E. coli in Rabbit Farms
3.3. Biofilm Formation in MDR E. coli Isolated from Rabbit Farms
4. Materials and Methods
4.1. Sample Collection, Isolation, and Identification of Escherichia coli Isolates
4.2. Antimicrobial Susceptibility Testing
4.3. Characterization of Antimicrobial Resistance Genes and Virulence Genotyping
4.4. Phylogenetic Diversity and Clonal Relationship
4.5. Biofilm Formation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clemente, L.; Leão, C.; Moura, L.; Albuquerque, T.; Amaro, A. Prevalence and Characterization of ESBL/AmpC Producing Escherichia coli from Fresh Meat in Portugal. Antibiotics 2021, 10, 1333. [Google Scholar] [CrossRef]
- Homeier-Bachmann, T.; Kleist, J.F.; Schütz, A.K.; Bachmann, L. Distribution of ESBL/AmpC- Escherichia coli on a Dairy Farm. Antibiotics 2022, 11, 940. [Google Scholar] [CrossRef]
- Tseng, C.-H.; Liu, C.-W.; Liu, P.-Y. Extended-Spectrum β-Lactamases (ESBL) Producing Bacteria in Animals. Antibiotics 2023, 12, 661. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Silva, V.; de Lurdes Enes Dapkevicius, M.; Caniça, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as Commensal and Pathogenic Bacteria among Food-Producing Animals: Health Implications of Extended Spectrum β-Lactamase (ESBL) Production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef] [PubMed]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in Prevalence and Characteristics of ESBL/PAmpC Producing E. Coli in Food in Germany. Vet. Microbiol. 2019, 233, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Widodo, A.; Effendi, M.H.; Khairullah, A.R. Extended-Spectrum Beta -Lactamase (ESBL)-Producing Escherichia coli from Livestock. Syst. Rev. Pharm. 2020, 11, 382–392. [Google Scholar]
- Silva, A.; Silva, V.; Pereira, J.E.; Maltez, L.; Igrejas, G.; Valentão, P.; Falco, V.; Poeta, P. Antimicrobial Resistance and Clonal Lineages of Escherichia coli from Food-Producing Animals. Antibiotics 2023, 12, 1061. [Google Scholar] [CrossRef]
- Suay-García, B.; Galán, F.; Rodríguez-Iglesias, M.A.; Pérez-Gracia, M.T. Detection and Characterization of Extended-Spectrum Beta-Lactamases-Producing Escherichia coli in Animals. Vector-Borne Zoonotic Dis. 2019, 19, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Crovato, S.; Menegon, F.; Mascarello, G.; Pinto, A.; Nadin, A.; Piovan, G.; Ricaldi, G.; Di Martino, G.; Pozza, G. Development of a Training Strategy Aimed at Increasing Veterinarians’ Awareness of the Proper Use of Antibiotics on Rabbit Farms. Animals 2023, 13, 2411. [Google Scholar] [CrossRef]
- Saxmose Nielsen, S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Depner, K.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Gortázar Schmidt, C.; Michel, V.; et al. Health and Welfare of Rabbits Farmed in Different Production Systems. EFSA J. 2020, 18, e05944. [Google Scholar] [CrossRef]
- Nielsen, S.S.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Gortazar Schmidt, C.; Herskin, M.; Michel, V.; et al. Assessment of Animal Diseases Caused by Bacteria Resistant to Antimicrobials: Rabbits. EFSA J. 2021, 19, e06999. [Google Scholar] [CrossRef]
- Ben Rhouma, R.; Jouini, A.; Klibi, A.; Hamrouni, S.; Boubaker, A.; Kmiha, S.; Maaroufi, A. Molecular Characterisation of Antimicrobial Resistance and Virulence Genes in Escherichia coli Strains Isolated from Diarrhoeic and Healthy Rabbits in Tunisia. World Rabbit Sci. 2020, 28, 81–91. [Google Scholar] [CrossRef]
- Massella, E.; Giacometti, F.; Bonilauri, P.; Reid, C.J.; Djordjevic, S.P.; Merialdi, G.; Bacci, C.; Fiorentini, L.; Massi, P.; Bardasi, L.; et al. Antimicrobial Resistance Profile and Expec Virulence Potential in Commensal Escherichia coli of Multiple Sources. Antibiotics 2021, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, J.; Ju, Z.; Chang, W.; Sun, S. Molecular Characterization of Antimicrobial Resistance in Escherichia coli from Rabbit Farms in Tai’an, China. BioMed Res. Int. 2018, 2018, 8607647. [Google Scholar] [CrossRef] [PubMed]
- Attili, A.R.; Bellato, A.; Robino, P.; Galosi, L.; Papeschi, C.; Rossi, G.; Fileni, E.; Linardi, M.; Cuteri, V.; Chiesa, F.; et al. Analysis of the Antibiotic Resistance Profiles in Methicillin-Sensitive S. aureus Pathotypes Isolated on a Commercial Rabbit Farm in Italy. Antibiotics 2020, 9, 673. [Google Scholar] [CrossRef] [PubMed]
- Agnoletti, F.; Brunetta, R.; Bano, L.; Drigo, I.; Mazzolini, E. Longitudinal Study on Antimicrobial Consumption and Resistance in Rabbit Farming. Int. J. Antimicrob. Agents 2018, 51, 197–205. [Google Scholar] [CrossRef]
- Di Martino, G.; Crovato, S.; Pinto, A.; Dorotea, T.; Mascarello, G.; Brunetta, R.; Agnoletti, F.; Bonfanti, L. Farmers’ Attitudes towards Antimicrobial Use and Awareness of Antimicrobial Resistance: A Comparative Study among Turkey and Rabbit Farmers. Ital. J. Anim. Sci. 2019, 18, 194–201. [Google Scholar] [CrossRef]
- Food, E.; Authority, S. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2017/2018. EFSA J. 2020, 18, e07867. [Google Scholar] [CrossRef]
- Sousa, M.; Silva, V.; Silva, A.; Silva, N.; Ribeiro, J.; Tejedor-Junco, M.T.; Capita, R.; Chenouf, N.S.; Alonso-Calleja, C.; Rodrigues, T.M.; et al. Staphylococci among Wild European Rabbits from the Azores: A Potential Zoonotic Issue? J. Food Prot. 2020, 83, 1110–1114. [Google Scholar] [CrossRef]
- Jenckel, M.; Hall, R.N.; Strive, T. Pathogen Profiling of Australian Rabbits by Metatranscriptomic Sequencing. Transbound. Emerg. Dis. 2022, 69, e2629–e2640. [Google Scholar] [CrossRef]
- Kylie, J.; Brash, M.; Whiteman, A.; Tapscott, B.; Slavic, D.; Weese, J.S.; Turner, P.V. Biosecurity Practices and Causes of Enteritis on Ontario Meat Rabbit Farms. Can. Vet. J. 2017, 58, 571–578. [Google Scholar] [PubMed]
- Vaz-Moreira, I.; Ferreira, C.; Nunes, O.C.; Manaia, C.M. Part III: Socio-Economical Perspectives and Impact of AR. In Sources of Antibiotic Resistance: Zoonotic, Human, Environment; Wiley: Hoboken, NJ, USA, 2020; pp. 211–238. [Google Scholar]
- Qian, J.; Wu, Z.; Zhu, Y.; Liu, C. One Health: A Holistic Approach for Food Safety in Livestock. Sci. One Health 2022, 1, 100015. [Google Scholar] [CrossRef]
- Scott, H.M.; Acuff, G.; Bergeron, G.; Bourassa, M.W.; Gill, J.; Graham, D.W.; Kahn, L.H.; Morley, P.S.; Salois, M.J.; Simjee, S.; et al. Critically Important Antibiotics: Criteria and Approaches for Measuring and Reducing Their Use in Food Animal Agriculture. Ann. N. Y. Acad. Sci. 2019, 1441, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Bessalah, S.; Fairbrother, J.M.; Salhi, I.; Vanier, G.; Khorchani, T.; Seddik, M.M.; Hammadi, M. Characterization and Antimicrobial Susceptibility of Escherichia coli Isolated from Healthy Farm Animals in Tunisia. Anim. Biotechnol. 2021, 32, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.; Igrejas, G.; Figueiredo, N.; Gonçalves, A.; Radhouani, H.; Rodrigues, J.; Poeta, P. Molecular Characterization of Antimicrobial Resistance in Enterococci and Escherichia coli Isolates from European Wild Rabbit (Oryctolagus cuniculus). Sci. Total Environ. 2010, 408, 4871–4876. [Google Scholar] [CrossRef] [PubMed]
- Marinho, C.; Igrejas, G.; Gonçalves, A.; Silva, N.; Santos, T.; Monteiro, R.; Gonçalves, D.; Rodrigues, T.; Poeta, P. Azorean Wild Rabbits as Reservoirs of Antimicrobial Resistant Escherichia coli. Anaerobe 2014, 30, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Cummins, E.A.; Snaith, A.E.; McNally, A.; Hall, R.J. The Role of Potentiating Mutations in the Evolution of Pandemic Escherichia coli Clones. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Freitas-Silva, J.; Inácio, Â.S.; Mourão, J.; Antunes, P.; Mendes, Â.; de Carvalho, A.P.; Vasconcelos, V.; Peixe, L.; da Costa, P.M. Occurrence of Mcr-1 in Escherichia coli from Rabbits of Intensive Farming. Vet. Microbiol. 2018, 227, 78–81. [Google Scholar] [CrossRef]
- Fuga, B.; Sellera, F.P.; Cerdeira, L.; Esposito, F.; Cardoso, B.; Fontana, H.; Moura, Q.; Cardenas-Arias, A.; Sano, E.; Ribas, R.M.; et al. WHO Critical Priority Escherichia coli as One Health Challenge for a Post-Pandemic Scenario: Genomic Surveillance and Analysis of Current Trends in Brazil. Microbiol. Spectr. 2022, 10, e0125621. [Google Scholar] [CrossRef]
- Nobili, G.; Franconieri, I.; Basanisi, M.G.; La Bella, G.; Tozzoli, R.; Caprioli, A.; La Salandra, G. Short Communication: Isolation of Shiga Toxin-Producing Escherichia coli in Raw Milk and Mozzarella Cheese in Southern Italy. J. Dairy Sci. 2016, 99, 7877–7880. [Google Scholar] [CrossRef]
- Zając, M.; Sztromwasser, P.; Bortolaia, V.; Leekitcharoenphon, P.; Cavaco, L.M.; Ziȩtek-Barszcz, A.; Hendriksen, R.S.; Wasyl, D. Occurrence and Characterization of Mcr-1-Positive Escherichia coli Isolated From Food-Producing Animals in Poland, 2011–2016. Front. Microbiol. 2019, 10, 1753. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Hu, B.; Xu, Y.; Sun, H.; Zhao, A.; Ba, P.; Fu, S.; Fan, R.; Jin, Y.; Wang, H.; et al. Molecular and Phylogenetic Characterization of Non-O157 Shiga Toxin-Producing Escherichia coli Strains in China. Front. Cell. Infect. Microbiol. 2016, 6, 143. [Google Scholar] [CrossRef]
- Vignaroli, C.; Luna, G.M.; Rinaldi, C.; Di Cesare, A.; Danovaro, R.; Biavasco, F. New Sequence Types and Multidrug Resistance among Pathogenic Escherichia coli Isolates from Coastal Marine Sediments. Appl. Environ. Microbiol. 2012, 78, 3916–3922. [Google Scholar] [CrossRef]
- Nesporova, K.; Wyrsch, E.R.; Valcek, A.; Bitar, I.; Chaw, K.; Harris, P.; Hrabak, J.; Literak, I.; Djordjevic, S.P.; Dolejskaa, M. Escherichia coli Sequence Type 457 Is an Emerging Extended-Spectrum-Lactam-Resistant Lineage with Reservoirs in Wildlife and Food-Producing Animals. Antimicrob. Agents Chemother. 2021, 65, e01118-20. [Google Scholar] [CrossRef] [PubMed]
- Sano, E.; Esposito, F.; Fontana, H.; Fuga, B.; Cardenas-Arias, A.; Moura, Q.; Cardoso, B.; Costa, G.C.V.; Bosqueiro, T.C.M.; Sinhorini, J.A.; et al. One Health Clones of Multidrug-Resistant Escherichia coli Carried by Synanthropic Animals in Brazil. One Health 2023, 16, 100476. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, I.; Tejedor-Junco, M.T.; González-Martín, M.; Corbera, J.A.; Suárez-Pérez, A.; Silva, V.; Igrejas, G.; Torres, C.; Poeta, P. Molecular Diversity of Extended-Spectrum β-Lactamase-Producing Escherichia coli from Vultures in Canary Islands. Environ. Microbiol. Rep. 2020, 12, 540–547. [Google Scholar] [CrossRef]
- Yu, Y.; Cui, C.Y.; Kuang, X.; Chen, C.; Wang, M.G.; Liao, X.P.; Sun, J.; Liu, Y.H. Prevalence of Tet(X4) in Escherichia coli From Duck Farms in Southeast China. Front. Microbiol. 2021, 12, 716393. [Google Scholar] [CrossRef]
- Sun, L.; Meng, N.; Wang, Z.; Hong, J.; Jiao, X.; Dai, Y.; Wang, Z.; Wang, J. Genomic Characterization of ESBL/AmpC-Producing Escherichia coli in Stray Dogs Sheltered in Yangzhou, China. Infect. Drug Resist. 2022, 15, 7741–7750. [Google Scholar] [CrossRef] [PubMed]
- Juraschek, K.; Deneke, C.; Schmoger, S.; Grobbel, M.; Malorny, B.; Käsbohrer, A.; Schwarz, S.; Meemken, D.; Hammerl, J.A. Phenotypic and Genotypic Properties of Fluoroquinolone-Resistant, Qnr-Carrying Escherichia coli Isolated from the German Food Chain in 2017. Microorganisms 2021, 9, 1308. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, D.; Hu, J.; Zhang, Y.; Tan, B.K.; Lin, S. Control Measurements of Escherichia coli Biofilm: A Review. Foods 2022, 11, 2469. [Google Scholar] [CrossRef]
- Barilli, E.; Vismarra, A.; Frascolla, V.; Rega, M.; Bacci, C. Escherichia coli Strains Isolated from Retail Meat Products: Evaluation of Biofilm Formation Ability, Antibiotic Resistance, and Phylogenetic Group Analysis. J. Food Prot. 2020, 83, 233–240. [Google Scholar] [CrossRef]
- Sivaranjani, M.; McCarthy, M.C.; Sniatynski, M.K.; Wu, L.; Dillon, J.A.R.; Rubin, J.E.; White, A.P. Biofilm Formation and Antimicrobial Susceptibility of Escherichia coli Associated with Colibacillosis Outbreaks in Broiler Chickens From Saskatchewan. Front. Microbiol. 2022, 13, 841516. [Google Scholar] [CrossRef]
- Kaleva, M.D.; Ilieva, Y.; Zaharieva, M.M.; Dimitrova, L.; Kim, T.C.; Tsvetkova, I.; Georgiev, Y.; Orozova, P.; Nedev, K.; Najdenski, H. Antimicrobial Resistance and Biofilm Formation of Escherichia coli Isolated from Pig Farms and Surroundings in Bulgaria. Microorganisms 2023, 11, 1909. [Google Scholar] [CrossRef]
- Ray, R.; Singh, P. Prevalence and Implications of Shiga Toxin-Producing Escherichia coli in Farm and Wild Ruminants. Pathogens 2022, 11, 1332. [Google Scholar] [CrossRef]
- EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters 2022. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 10.0. 2020. Available online: http://www.eucast.org (accessed on 3 February 2024).
- Carvalho, I.; Cunha, R.; Martins, C.; Martínez-Álvarez, S.; Chenouf, N.S.; Pimenta, P.; Pereira, A.R.; Ramos, S.; Sadi, M.; Martins, Â.; et al. Antimicrobial Resistance Genes and Diversity of Clones among Faecal ESBL-Producing Escherichia coli Isolated from Healthy and Sick Dogs Living in Portugal. Antibiotics 2021, 10, 1013. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Gautom, R.K. Rapid Pulsed-Field Gel Electrophoresis Protocol for Typing of Escherichia coli O157:H7 and Other Gram-Negative Organisms in 1 Day. J. Clin. Microbiol. 1997, 35, 2977–2980. [Google Scholar] [CrossRef] [PubMed]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ—A Tool for Analyzing DNA Fingerprint Gel Images. BMC Bioinform. 2015, 16, 270. [Google Scholar] [CrossRef] [PubMed]
- Guiral, E.; Pons, M.J.; Vubil, D.; Marí-Almirall, M.; Sigaúque, B.; Soto, S.M.; Alonso, P.L.; Ruiz, J.; Vila, J.; Mandomando, I. Epidemiology and Molecular Characterization of Multidrug-Resistant Escherichia coli Isolates Harboring BlaCTX-M Group 1 Extended-Spectrum β-Lactamases Causing Bacteremia and Urinary Tract Infection in Manhiça, Mozambique. Infect. Drug Resist. 2018, 11, 927–936. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and Virulence in Escherichia coli: An Evolutionary Perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of Multiple Methods for Quantification of Microbial Biofilms Grown in Microtiter Plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Correia, E.; Pereira, J.E.; González-Machado, C.; Capita, R.; Alonso-Calleja, C.; Igrejas, G.; Poeta, P. Biofilm Formation of Staphylococcus aureus from Pets, Live-Stock, and Wild Animals: Relationship with Clonal Lineages and Antimicrobial Resistance. Antibiotics 2022, 11, 772. [Google Scholar] [CrossRef] [PubMed]

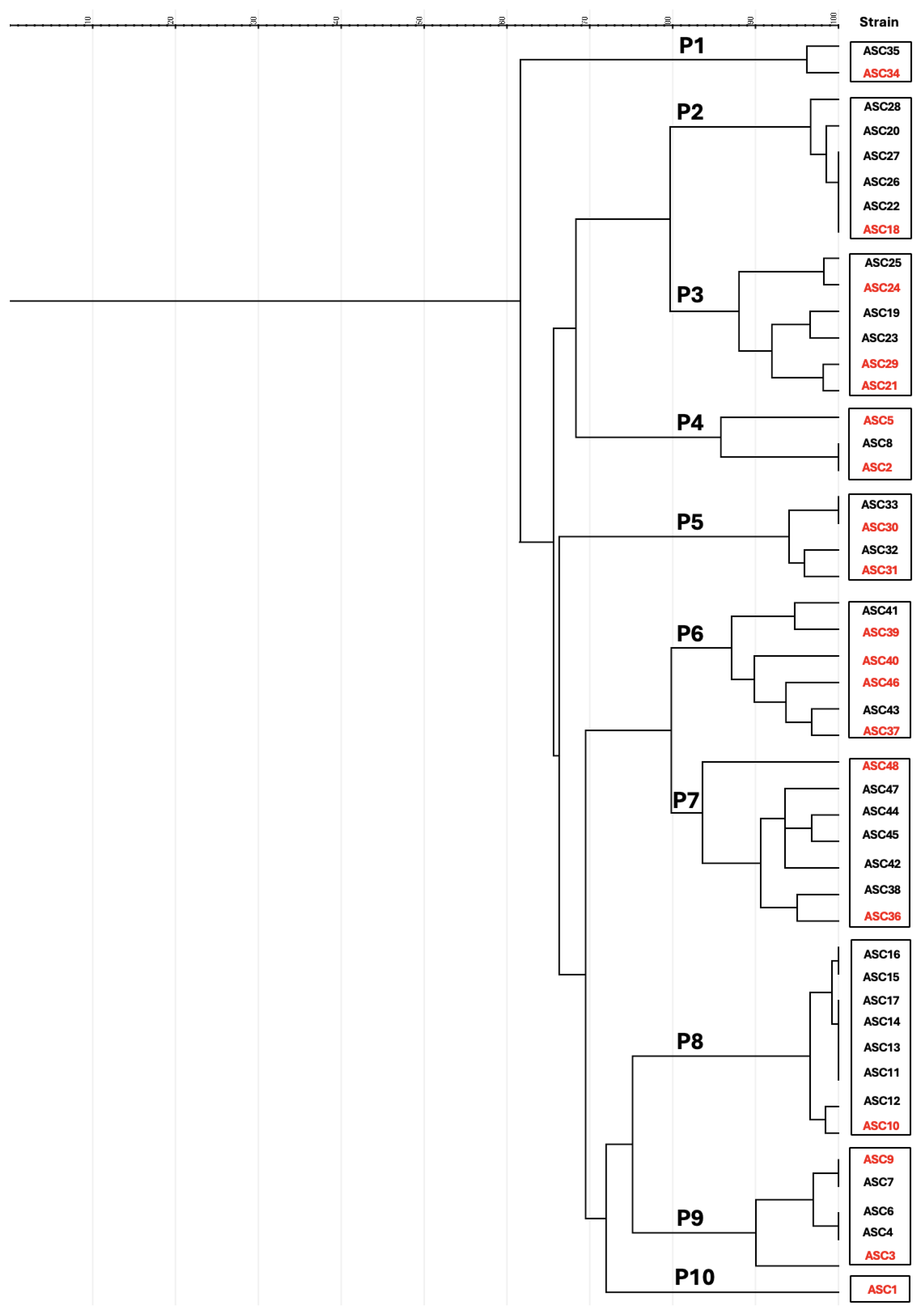

| Isolate | Farm | Resistance Phenotype | Resistance Genotype | Phylogenetic Group | Integrase Gene | Virulence Genes | PFGE Pattern |
|---|---|---|---|---|---|---|---|
| ASC4 | 3 | AMP-S-TOB-CTX-NA-CIP-SXT-C-TE | sul1-sul3-qnrS- -strA-strB- blaCTX-3G-tetB-aac(6)-Ib | A | int1 | fimA-bfp | P9 |
| ASC6 | 3 | ATM-AMP-S-TOB-CTX-NA-CIP-SXT-C-TE | sul1-sul3-tetA-qnrS-qnrA-—strA-strB- blaCTX-3G-tetB-aac(6)-Ib-blaTEM | A | int1 | papG-III-fimA | P9 |
| ASC7 | 3 | ATM-AMP-AK-CN-S-TOB-CTX-NA-CIP-SXT-C-TE | sul1-sul3-tetA-tetB-qnrS-qnrA- -aac(3)-IV-aac(3)-II- strA- blaCTX-3G- aac(6)-Ib-blaTEM | A | int1 | papG-III-fimA-bfp | P9 |
| ASC8 | 3 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul1-sul3-tetA -strA- strB-blaCTX-3G | B1 | int1 | fimA | P4 |
| ASC11 | 4 | AUG-ATM-AMP-S-CTX-CAZ-SXT-C-TE | sul2-sul3-tetA- strA-strB- blaCTX-3G-blaTEM | A | int1 | papG-III-fimA-bfp | P8 |
| ASC12 | 4 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul2-sul3-tetA-strA-strB- blaCTX-3G-blaTEM | A | int1 | papG-III-fimA-bfp | P8 |
| ASC13 | 4 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul2-sul3-tetA- strA-strB- blaCTX-3G-blaTEM | A | int1 | papG-III-fimA-bfp | P8 |
| ASC14 | 4 | AUG-ATM-AMP-S-CTX-SXT-C-TE | sul2-sul3-tetA- strA-strB- blaCTX-3G-blaTEM | A | int1 | papG-III-fimA-bfp | P8 |
| ASC15 | 4 | AUG-ATM-AMP-S-CTX-SXT-C-TE | sul1-sul2-sul3-tetA- tetB- strA-strB- blaCTX-3G- blaTEM | A | int1 | papG-III-fimA-bfp | P8 |
| ASC16 | 4 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul1-sul3-tetA-cmlA -strA-strB- blaCTX-3G-blaCTX-M9-blaTEM | A | int1 | papG-III-fimA-bfp | P8 |
| ASC17 | 4 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul3-tetA-cmlA- strA-strB-blaCTX-3G-blaCTX-M9-aadA5-blaTEM | A | int1 | papG-III-fimA-bfp | P8 |
| ASC19 | 5 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul1-sul3-tetA-cmlA- strA-strB- blaCTX-M9 | B1 | int1 | papG-III-fimA | P3 |
| ASC20 | 5 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul3-cmlA- strA-strB-blaCTX-3G-blaCTX-M9-aadA5-blaTEM | B1 | int1 | papG-III | P2 |
| ASC22 | 5 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul3-cmlA -strA-strB-blaCTX-M9-blaTEM | B1 | int1 | papG-III | P2 |
| ASC23 | 5 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul2-sul3-cmlA-strA- blaCTX-M9-aadA5-blaTEM | B1 | - | papG-III | P3 |
| ASC25 | 5 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul2-sul3-tetA-cmlA-blaCTXM-strA-strB- blaTEM | B1 | int1 | papG-III | P3 |
| ASC26 | 5 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul1-sul2-sul3-cmlA- strA-strB- blaCTX-M9-blaTEM | B1 | int1 | papG-III | P2 |
| ASC27 | 5 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul1-sul2-sul3-tetA-cmlA- strA-strB- blaCTX-M9-blaTEM | B1 | int1 | papG-III | P2 |
| ASC28 | 5 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul1-sul3-tetA-cmlA-strA-strB- blaCTX-M9-blaTEM | B1 | int1 | papG-III | P2 |
| ASC32 | 6 | ATM-AMP-S-TOB-CTX-CAZ | sul3- strA-strB-blaCTX-3G-blaCTX-M9 | B1 | - | papG-III | P5 |
| ASC33 | 6 | ATM-AMP-S-TOB-CTX-SXT-CAZ | sul3-strA- blaCTX-3G-aadA5-blaTEM | B1 | - | - | P5 |
| ASC35 | 13 | ATM-AMP-S-CTX-NA-CIP-SXT-C-TE | qnrS-qnrA-parC-cmlA-tetA- blaCTX-3G-blaCTX-M9-aac(6)-Ib-aadA5-blaSHV | D | int1 | papG-III | P1 |
| ASC38 | 13 | ATM-AMP-S-CTX-C-TE | cmlA-tetA- tetB-blaCTX-M9-blaTEM-blaSHV | A | - | papG-III-bfp | P7 |
| ASC41 | 13 | ATM-AMP-S-CTX-NA-CIP-SXT-TE | qnrS-qnrA-tetA- blaCTX-M9 blaCTX-3G-aac(6)-Ib-blaTEM | A | int1 | papG-III-fimA | P6 |
| ASC42 | 13 | ATM-AMP-S-CTX-NA-CIP-SXT-TE | qnrA -tetA- tetB-blaCTX-M9 blaCTX-3G-aac(6)-Ib-blaTEM | A | int1 | papG-III-fimA-bfp | P7 |
| ASC43 | 13 | ATM-AMP-S-CTX-NA-CIP-SXT-TE | qnrS-qnrA- tetA-tetB blaCTX-3G-aac(6)-Ib-aadA5 | A | int1 | papG-III-fimA | P6 |
| ASC44 | 13 | ATM-AMP-CTX-TE | tetB-blaCTX-M9-blaCTX-3G | A | - | papG-III-fimA-bfp | P7 |
| ASC45 | 13 | ATM-AMP-CTX-TE | tetB-blaCTX-M9-blaCTX-3G-blaTEM | A | - | papG-III-fimA-bfp | P7 |
| ASC47 | 13 | ATM-AMP-S-TOB-CTX-TE | tetB-blaCTX-M9-blaCTX-3G | A | - | papG-III-fimA-bfp | P7 |
| Isolate | Farm | MLST | Resistance Phenotype | Resistance Genotype | Phylogenetic Group | Integrase Gene | Virulence Genes | PFGE Pattern |
|---|---|---|---|---|---|---|---|---|
| ASC1 | 2 | ST10 | AUG-ATM-AMP-S-TOB-CTX-CAZ-NA-CIP-SXT-C-TE | sul2-sul3-tetA-cmlA -strA-strB- blaCTX-3G-tetB-aac(6)-Ib | A | intI1 | fimA-bfp | P10 |
| ASC2 | 3 | ST1611 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul1-sul3-tetA -strA-strB- blaCTX-3G | B1 | intI1 | papG-III-fimA | P4 |
| ASC3 | 3 | ST8470 | AUG-ATM-AMP-S-TOB-CTX-NA-CIP-SXT-C-TE | sul1-sul3-tetA-qnrS-qnrA-strA-strB- blaCTX-3G-aac(6)-Ib-blaTEM | A | int1 | papG-III-fimA-bfp | P9 |
| ASC5 | 3 | ST1611 | AUG-ATM-AMP-CN-S-TOB-CTX-SXT-C-TE | sul1-sul3-tetA-aac(3)-IV-aac(3)-II- strA-strB- blaCTX-3G | B1 | - | papG-I-fimA II | P4 |
| ASC9 | 3 | ST8470 | AUG-ATM-AMP-S-TOB-CTX-NA-CIP-SXT-C-TE | sul1-sul3-tetA-qnrS-qnrA- strA-strB- blaCTX-3G-tetB-blaTEM | A | - | fimA-cnf1 | P9 |
| ASC10 | 4 | ST10 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul1-sul2-sul3-tetA- strA-strB- blaCTX-3G-blaTEM | A | int1 | papG-III-fimA-bfp | P8 |
| ASC18 | 5 | ST2825 | AUG-ATM-AMP-AK-CN-S-TOB-CTX-SXT-C-TE | sul1-sul2-aac(3)-IV-aac(3)-II-cmlA-strA-strB- blaCTX-3G-blaCTX-M9 | B1 | int1 | papG-III-fimA | P2 |
| ASC21 | 5 | ST2825 | AUG-ATM-AMP-S-TOB-CTX-SXT-C-TE | sul2-cmlA-strA-strB- blaCTX-M9-blaTEM | B1 | int1 | papG-III-fimA | P3 |
| ASC24 | 5 | ST2825 | AUG-ATM-AMP-CN-S-TOB-CTX-SXT-C-TE | sul2-sul3-tetA-aac(3)-IV-aac(3)-II-cmlA- strA-strB-blaCTX-3G-blaCTX-M9-aadA5-blaTEM | B1 | int1 | papG-III | P3 |
| ASC29 | 5 | ST2825 | AUG-ATM-AMP-AK-CN-S-TOB-CTX-SXT-C-TE | sul1-sul3-tetA-aac(3)-IV-aac(3)-II-cmlA strA-strB-blaCTX-M9-blaTEM | B1 | int1 | papG-III | P3 |
| ASC30 | 6 | ST8823 | ATM-AMP-S-TOB-CTX-CAZ | sul3-strB-blaCTX-3G-blaCTX-M9 | B1 | - | - | P5 |
| ASC31 | 6 | ST8823 | ATM-AMP-S-TOB-CTX-CAZ | sul3- strA-strB-blaCTX-3G | B1 | - | papG-III | P5 |
| ASC34 | 13 | ST457 | ATM-AMP-S-CTX-NA-CIP-SXT-C-TE | tetA-qnrA -qnrS-cmlA- blaCTX-3G-aac(6)-Ib-aadA5-blaTEM | D | int1 | papG-III-bfp | P1 |
| ASC36 | 13 | ST2325 | ATM-AMP-CTX-TE | tetB-blaCTX-3G- blaCTX-M9-blaTEM | A | - | papG-III-bfp | P7 |
| ASC37 | 13 | ST2325 | ATM-AMP-S-CTX-CAZ-NA-CIP-SXT-TE | tetA-qnrS-qnrA-blaCTX-M9-blaSHV | A | int1 | papG-III | P6 |
| ASC39 | 13 | ST2325 | ATM-AMP-S-CTX-TE | tetB-blaCTX-M9-aadA5 | A | - | papG-III-bfp | P6 |
| ASC40 | 13 | ST2325 | ATM-AMP-S-CTX-C-TE | cmlA-tetA-tetB-blaCTX-M9-blaCTX-3G-aadA5 | A | - | papG-III-bfp | P6 |
| ASC46 | 13 | ST2325 | ATM-AMP-S-TOB-CTX-C-TE | cmlA-tetA- tetB-blaCTX-M9 | A | - | papG-III-fimA-bfp | P6 |
| ASC48 | 13 | ST2325 | ATM-AMP-S-CTX-C-TE | cmlA-tetA- blaCTX-M9-blaCTX-3G-blaTEM- blaSHV | A | - | papG-III-fimA-bfp | P7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, A.; Silva, V.; Tavares, T.; López, M.; Rojo-Bezares, B.; Pereira, J.E.; Falco, V.; Valentão, P.; Igrejas, G.; Sáenz, Y.; et al. Rabbits as a Reservoir of Multidrug-Resistant Escherichia coli: Clonal Lineages and Public Health Impact. Antibiotics 2024, 13, 376. https://doi.org/10.3390/antibiotics13040376
Silva A, Silva V, Tavares T, López M, Rojo-Bezares B, Pereira JE, Falco V, Valentão P, Igrejas G, Sáenz Y, et al. Rabbits as a Reservoir of Multidrug-Resistant Escherichia coli: Clonal Lineages and Public Health Impact. Antibiotics. 2024; 13(4):376. https://doi.org/10.3390/antibiotics13040376
Chicago/Turabian StyleSilva, Adriana, Vanessa Silva, Teresa Tavares, María López, Beatriz Rojo-Bezares, José Eduardo Pereira, Virgílio Falco, Patrícia Valentão, Gilberto Igrejas, Yolanda Sáenz, and et al. 2024. "Rabbits as a Reservoir of Multidrug-Resistant Escherichia coli: Clonal Lineages and Public Health Impact" Antibiotics 13, no. 4: 376. https://doi.org/10.3390/antibiotics13040376








