Metabolomics and Lipidomics Analyses Aid Model Classification of Type 2 Diabetes in Non-Human Primates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Housing and Husbandry
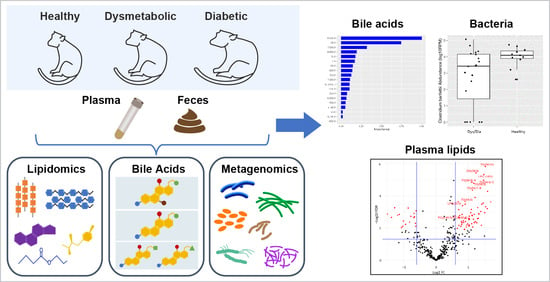
2.2. One-Time Plasma and Feces Collection
2.3. Animal Phenotype Characterization
2.4. Sample Processing and LC-MS Data Acquisition
2.5. Metabolite Panels
2.5.1. Targeted Panels
2.5.2. Untargeted Metabolite Panels
2.6. Untargeted and Targeted Data Processing
- Filtered contaminating peaks in blank samples from biological data files.
- Excluded extreme outlier technical replicates in QC samples.
- Calculated the RSD% of QC samples, and flagged metabolites showing RSD > 30%
- Measurements below detection were imputed with one half of the lowest observed peak intensity.
- Batch/injection order was corrected using QC samples.
- Data were normalized using the sum of the known metabolites or “mTIC”
- Post-normalized peak intensities were scaled and/or log transformed.
2.7. Metabolomics and Lipidomics Statistics Analysis and Modeling
2.8. Metagenomics Analysis
3. Results
3.1. Clinical and Demographic Adjustments for Animal Confounding Factors
3.2. Plasma Lipids and Fecal Metabolites Independently Aid in Animal Disease Phenotype Classification
3.3. Combined Univariate and Multivariate Results Reveal Specific Plasma Lipids That Are Differential between Healthy, Dys, and Dia Animals
3.4. Strong Association between HbA1c and Plasma Long-Chain Polyunsaturated TGs
3.5. Fecal Secondary Bile Acids Can Distinguish Healthy from Dys/Dia Animals and Are Associated with T2D
3.6. Metagenomics Results Reveal Several Bacterial Species to Be Associated with T2D
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Merino, J.; Udler, M.S.; Leong, A.; Meigs, J.B. A Decade of Genetic and Metabolomic Contributions to Type 2 Diabetes Risk Prediction. Curr. Diabetes Rep. 2017, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.A.; Volpi, E.; Fujita, S.; Cadenas, J.G.; Rasmussen, B.B. Dysregulation of muscle fatty acid metabolism in type 2 diabetes is independent of malonyl-CoA. Diabetologia 2006, 49, 2144–2152. [Google Scholar] [CrossRef] [PubMed]
- McGarry, J.D. Banting lecture 2001: Dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes 2002, 51, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.B.; Petersen, K.F.; Shulman, G.I. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol. Rev. 2007, 87, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Noble, D.; Mathur, R.; Dent, T.; Meads, C.; Greenhalgh, T. Risk models and scores for type 2 diabetes: Systematic review. BMJ 2011, 343, d7163. [Google Scholar] [CrossRef]
- Kuzuya, T.; Katano, Y.; Nakano, I.; Hirooka, Y.; Itoh, A.; Ishigami, M.; Hayashi, K.; Honda, T.; Goto, H.; Fujita, Y.; et al. Regulation of branched-chain amino acid catabolism in rat models for spontaneous type 2 diabetes mellitus. Biochem. Biophys. Res. Commun. 2008, 373, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A Branched-Chain Amino Acid-Related Metabolic Signature that Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef]
- She, P.; Van Horn, C.; Reid, T.; Hutson, S.M.; Cooney, R.N.; Lynch, C.J. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am. J. Physiol.-Endocrinol. Metab. 2007, 293, E1552–E1563. [Google Scholar] [CrossRef]
- Wijekoon, E.P.; Skinner, C.; Brosnan, M.E.; Brosnan, J.T. Amino acid metabolism in the Zucker diabetic fatty rat: Effects of insulin resistance and of type 2 diabetes. Can. J. Physiol. Pharmacol. 2004, 82, 506–514. [Google Scholar] [CrossRef]
- Guasch-Ferre, M.; Hruby, A.; Toledo, E.; Clish, C.B.; Martinez-Gonzalez, M.A.; Salas-Salvado, J.; Hu, F.B. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care 2016, 39, 833–846. [Google Scholar] [CrossRef]
- Harwood, H.J., Jr.; Listrani, P.; Wagner, J.D. Nonhuman primates and other animal models in diabetes research. J. Diabetes Sci. Technol. 2012, 6, 503–514. [Google Scholar] [CrossRef]
- Wagner, J.D.; Kavanagh, K.; Ward, G.M.; Auerbach, B.J.; Harwood, H.J.; Kaplan, J.R. Old World Nonhuman Primate Models of Type 2 Diabetes Mellitus. ILAR J. 2006, 47, 259–271. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Wang, D.; Wei, L. Practical and Critical Instruction for Nonhuman Primate Diabetic Models. Transplant. Proc. 2013, 45, 1856–1865. [Google Scholar] [CrossRef] [PubMed]
- Pound, L.D.; Kievit, P.; Grove, K.L. The nonhuman primate as a model for type 2 diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 2014, 21, 89–94. [Google Scholar] [CrossRef]
- Barupal, D.K.; Fiehn, O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017, 7, 14567. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.C.; Bodkin, N.L. Beta-cell hyperresponsiveness: Earliest event in development of diabetes in monkeys. Am. J. Physiol. 1990, 259, R612–R617. [Google Scholar] [CrossRef] [PubMed]
- Tigno, X.T.; Gerzanich, G.; Hansen, B.C. Age-related changes in metabolic parameters of nonhuman primates. J. Gerontol. A Biol. Sci. Med. Sci. 2004, 59, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Marigliano, M.; Casu, A.; Bertera, S.; Trucco, M.; Bottino, R. Hemoglobin A1C Percentage in Nonhuman Primates: A Useful Tool to Monitor Diabetes before and after Porcine Pancreatic Islet Xenotransplantation. J. Transpl. 2011, 2011, 965605. [Google Scholar] [CrossRef]
- Kind, T.; Liu, K.H.; Lee, D.Y.; DeFelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef]
- Skogerson, K.; Wohlgemuth, G.; Barupal, D.K.; Fiehn, O. The volatile compound BinBase mass spectral database. BMC Bioinform. 2011, 12, 321. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Holman, R.T.; Smythe, L.; Johnson, S. Effect of sex and age on fatty acid composition of human serum lipids. Am. J. Clin. Nutr. 1979, 32, 2390–2399. [Google Scholar] [CrossRef] [PubMed]
- Koikkalainen, J.; Pölönen, H.; Mattila, J.; Van Gils, M.; Soininen, H.; Lötjönen, J. Improved Classification of Alzheimer’s Disease Data via Removal of Nuisance Variability. PLoS ONE 2012, 7, e31112. [Google Scholar] [CrossRef] [PubMed]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Prieto, B.; Eriksson, L.; Trygg, J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). J. Chemom. 2014, 28, 623–632. [Google Scholar] [CrossRef]
- Lee, J.; Little, T.D. A practical guide to propensity score analysis for applied clinical research. Behav. Res. Ther. 2017, 98, 76–90. [Google Scholar] [CrossRef]
- Rosenbaum, P.R.; Rubin, D.B. The Central Role of the Propensity Score in Observational Studies for Causal Effects. Biometrika 1983, 70, 41–55. [Google Scholar] [CrossRef]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef]
- Manor, O.; Borenstein, E. MUSiCC: A marker genes based framework for metagenomic normalization and accurate profiling of gene abundances in the microbiome. Genome Biol. 2015, 16, 53. [Google Scholar] [CrossRef]
- Spitler, K.M.; Davies, B.S.J. Aging and plasma triglyceride metabolism. J. Lipid Res. 2020, 61, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Despres, J.P.; Moorjani, S.; Tremblay, A.; Ferland, M.; Lupien, P.J.; Nadeau, A.; Bouchard, C. Relation of high plasma triglyceride levels associated with obesity and regional adipose tissue distribution to plasma lipoprotein-lipid composition in premenopausal women. Clin. Investig. Med. 1989, 12, 374–380. [Google Scholar]
- Suvitaival, T.; Bondia-Pons, I.; Yetukuri, L.; Poho, P.; Nolan, J.J.; Hyotylainen, T.; Kuusisto, J.; Oresic, M. Lipidome as a predictive tool in progression to type 2 diabetes in Finnish men. Metabolism 2018, 78, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sobczak, A.I.S.; Blindauer, C.A.; Stewart, A.J. Changes in Plasma Free Fatty Acids Associated with Type-2 Diabetes. Nutrients 2019, 11, 2022. [Google Scholar] [CrossRef] [PubMed]
- Field, B.C.; Gordillo, R.; Scherer, P.E. The Role of Ceramides in Diabetes and Cardiovascular Disease Regulation of Ceramides by Adipokines. Front. Endocrinol. 2020, 11, 569250. [Google Scholar] [CrossRef] [PubMed]
- Haus, J.M.; Kashyap, S.R.; Kasumov, T.; Zhang, R.; Kelly, K.R.; Defronzo, R.A.; Kirwan, J.P. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes 2009, 58, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Hatch, G.M.; Wang, Y.; Yu, F.; Wang, M. The relationship between phospholipids and insulin resistance: From clinical to experimental studies. J. Cell Mol. Med. 2019, 23, 702–710. [Google Scholar] [CrossRef]
- Floegel, A.; Stefan, N.; Yu, Z.; Muhlenbruch, K.; Drogan, D.; Joost, H.G.; Fritsche, A.; Haring, H.U.; Hrabe de Angelis, M.; Peters, A.; et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 2013, 62, 639–648. [Google Scholar] [CrossRef]
- Tirosh, A.; Shai, I.; Bitzur, R.; Kochba, I.; Tekes-Manova, D.; Israeli, E.; Shochat, T.; Rudich, A. Changes in triglyceride levels over time and risk of type 2 diabetes in young men. Diabetes Care 2008, 31, 2032–2037. [Google Scholar] [CrossRef]
- Barber, M.N.; Risis, S.; Yang, C.; Meikle, P.J.; Staples, M.; Febbraio, M.A.; Bruce, C.R. Plasma lysophosphatidylcholine levels are reduced in obesity and type 2 diabetes. PLoS ONE 2012, 7, e41456. [Google Scholar] [CrossRef]
- Drobnik, W.; Liebisch, G.; Audebert, F.X.; Frohlich, D.; Gluck, T.; Vogel, P.; Rothe, G.; Schmitz, G. Plasma ceramide and lysophosphatidylcholine inversely correlate with mortality in sepsis patients. J. Lipid Res. 2003, 44, 754–761. [Google Scholar] [CrossRef]
- Heimerl, S.; Fischer, M.; Baessler, A.; Liebisch, G.; Sigruener, A.; Wallner, S.; Schmitz, G. Alterations of plasma lysophosphatidylcholine species in obesity and weight loss. PLoS ONE 2014, 9, e111348. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.S.; Shearer, J. Metabolomics and Type 2 Diabetes: Translating Basic Research into Clinical Application. J. Diabetes Res. 2016, 2016, 3898502. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Kwak, S.Y.; Jo, G.; Song, T.J.; Shin, M.J. Serum metabolite profile associated with incident type 2 diabetes in Koreans: Findings from the Korean Genome and Epidemiology Study. Sci. Rep. 2018, 8, 8207. [Google Scholar] [CrossRef] [PubMed]
- Hanamatsu, H.; Ohnishi, S.; Sakai, S.; Yuyama, K.; Mitsutake, S.; Takeda, H.; Hashino, S.; Igarashi, Y. Altered levels of serum sphingomyelin and ceramide containing distinct acyl chains in young obese adults. Nutr. Diabetes 2014, 4, e141. [Google Scholar] [CrossRef]
- Lemaitre, R.N.; Yu, C.; Hoofnagle, A.; Hari, N.; Jensen, P.N.; Fretts, A.M.; Umans, J.G.; Howard, B.V.; Sitlani, C.M.; Siscovick, D.S.; et al. Circulating Sphingolipids, Insulin, HOMA-IR, and HOMA-B: The Strong Heart Family Study. Diabetes 2018, 67, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Jager, K.J.; Zoccali, C.; Macleod, A.; Dekker, F.W. Confounding: What it is and how to deal with it. Kidney Int. 2008, 73, 256–260. [Google Scholar] [CrossRef] [PubMed]
- van Stralen, K.J.; Dekker, F.W.; Zoccali, C.; Jager, K.J. Confounding. Nephron Clin. Pract. 2010, 116, c143–c147. [Google Scholar] [CrossRef]
- Doumatey, A.P.; Adeyemo, A.; Zhou, J.; Lei, L.; Adebamowo, S.N.; Adebamowo, C.; Rotimi, C.N. Gut Microbiome Profiles Are Associated with Type 2 Diabetes in Urban Africans. Front. Cell. Infect. Microbiol. 2020, 10, 63. [Google Scholar] [CrossRef]
- Maskarinec, G.; Raquinio, P.; Kristal, B.S.; Setiawan, V.W.; Wilkens, L.R.; Franke, A.A.; Lim, U.; Le Marchand, L.; Randolph, T.W.; Lampe, J.W.; et al. The gut microbiome and type 2 diabetes status in the Multiethnic Cohort. PLoS ONE 2021, 16, e0250855. [Google Scholar] [CrossRef]
- Sanna, S.; van Zuydam, N.R.; Mahajan, A.; Kurilshikov, A.; Vich Vila, A.; Vosa, U.; Mujagic, Z.; Masclee, A.A.M.; Jonkers, D.; Oosting, M.; et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019, 51, 600–605. [Google Scholar] [CrossRef]
- Connors, J.; Dunn, K.A.; Allott, J.; Bandsma, R.; Rashid, M.; Otley, A.R.; Bielawski, J.P.; Van Limbergen, J. The relationship between fecal bile acids and microbiome community structure in pediatric Crohn’s disease. ISME J. 2020, 14, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zhang, D.; Wang, Z.; Sun, J.; Xu, B.; Chen, Y.; Ding, L.; Huang, X.; Lv, X.; Lu, J.; et al. Insulin Resistance is Associated with Total Bile Acid Level in Type 2 Diabetic and Nondiabetic Population: A Cross-Sectional Study. Medicine 2016, 95, e2778. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef] [PubMed]






| Characteristic | Dia | Dys | Healthy |
|---|---|---|---|
| N = 24 | N = 16 | N = 17 | |
| Demographics | |||
| Female, Male | 8, 16 | 5, 11 | 3, 14 |
| Age | 19.8 (4.6) | 15.5 (3.2) | 15.3 (5.8) |
| BW, kg | 13.5 (4.0) | 15.3 (3.6) | 11.8 (3.5) |
| Clinical assessment | |||
| Glucose, mg/dL | 164.9 (40.1) | 75.7 (31.7) | 62.5 (5.2) |
| HbA1c, % | 10.0 (2.2) | 6.2 (1.6) | 4.4 (0.3) |
| TG, mg/dL | 360.5 (284.5) | 157.1 (85.0) | 62.0 (36.8) |
| Disease State | Class | N | p-Value |
|---|---|---|---|
| Healthy vs. Dys | PC | 127 | 0.0142 |
| TG | 93 | 0.6353 * | |
| Dys vs. Dia | PC | 127 | 0.7640 |
| TG | 93 | <0.001 *** |
| Animal Phenotype | AUROC [95% CI] |
|---|---|
| Dys vs. Dia | 0.59 [0.40, 0.79] |
| Healthy vs. Dys | 0.67 [0.47, 0.87] |
| Healthy vs. Dys/Dia | 0.76 [0.64, 0.89] |
| Species | AUROC [95% CI] | Dys/Dia vs. Healthy |
|---|---|---|
| Roseburia inulinivorans | 0.31 [0.13, 0.50] | Down |
| Clostridium bartlettii | 0.28 [0.10, 0.46] | Down |
| Ruminococcus obeum | 0.28 [0.07, 0.49] | Down |
| Streptococcus pasteurianus | 0.26 [0.09, 0.43] | Down |
| Streptococcus lutetiensis | 0.26 [0.08, 0.44] | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, P.; Conarello, S.; Wyche, T.P.; Zhang, N.R.; Chng, K.; Kang, J.; Sana, T.R. Metabolomics and Lipidomics Analyses Aid Model Classification of Type 2 Diabetes in Non-Human Primates. Metabolites 2024, 14, 159. https://doi.org/10.3390/metabo14030159
Tao P, Conarello S, Wyche TP, Zhang NR, Chng K, Kang J, Sana TR. Metabolomics and Lipidomics Analyses Aid Model Classification of Type 2 Diabetes in Non-Human Primates. Metabolites. 2024; 14(3):159. https://doi.org/10.3390/metabo14030159
Chicago/Turabian StyleTao, Peining, Stacey Conarello, Thomas P. Wyche, Nanyan Rena Zhang, Keefe Chng, John Kang, and Theodore R. Sana. 2024. "Metabolomics and Lipidomics Analyses Aid Model Classification of Type 2 Diabetes in Non-Human Primates" Metabolites 14, no. 3: 159. https://doi.org/10.3390/metabo14030159