Urine and Serum Metabolomics Analyses May Distinguish between Stages of Renal Cell Carcinoma
Abstract
:1. Introduction
2. Results
2.1. Samples
2.2. 1H NMR Spectral Analysis of Serum and Urine Samples
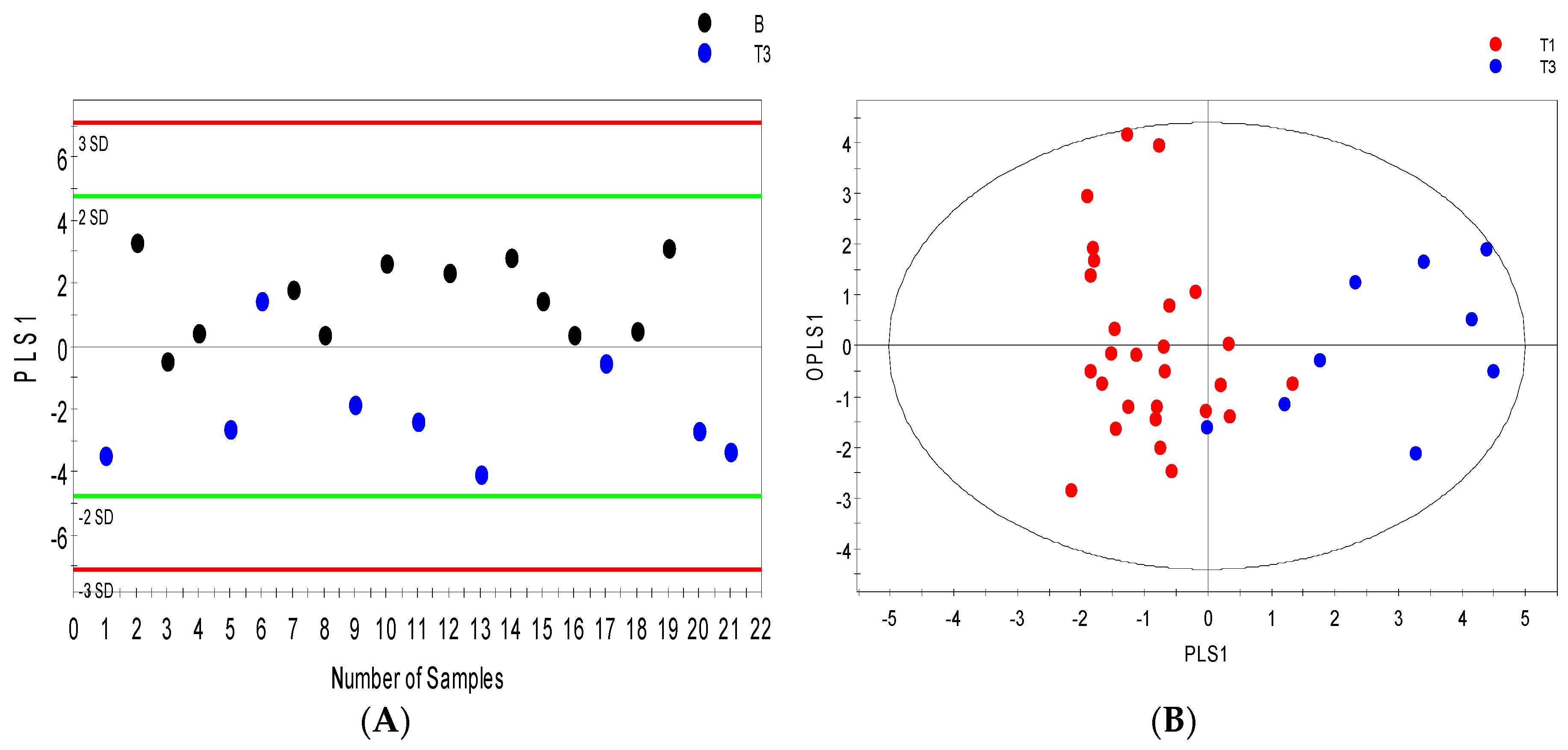
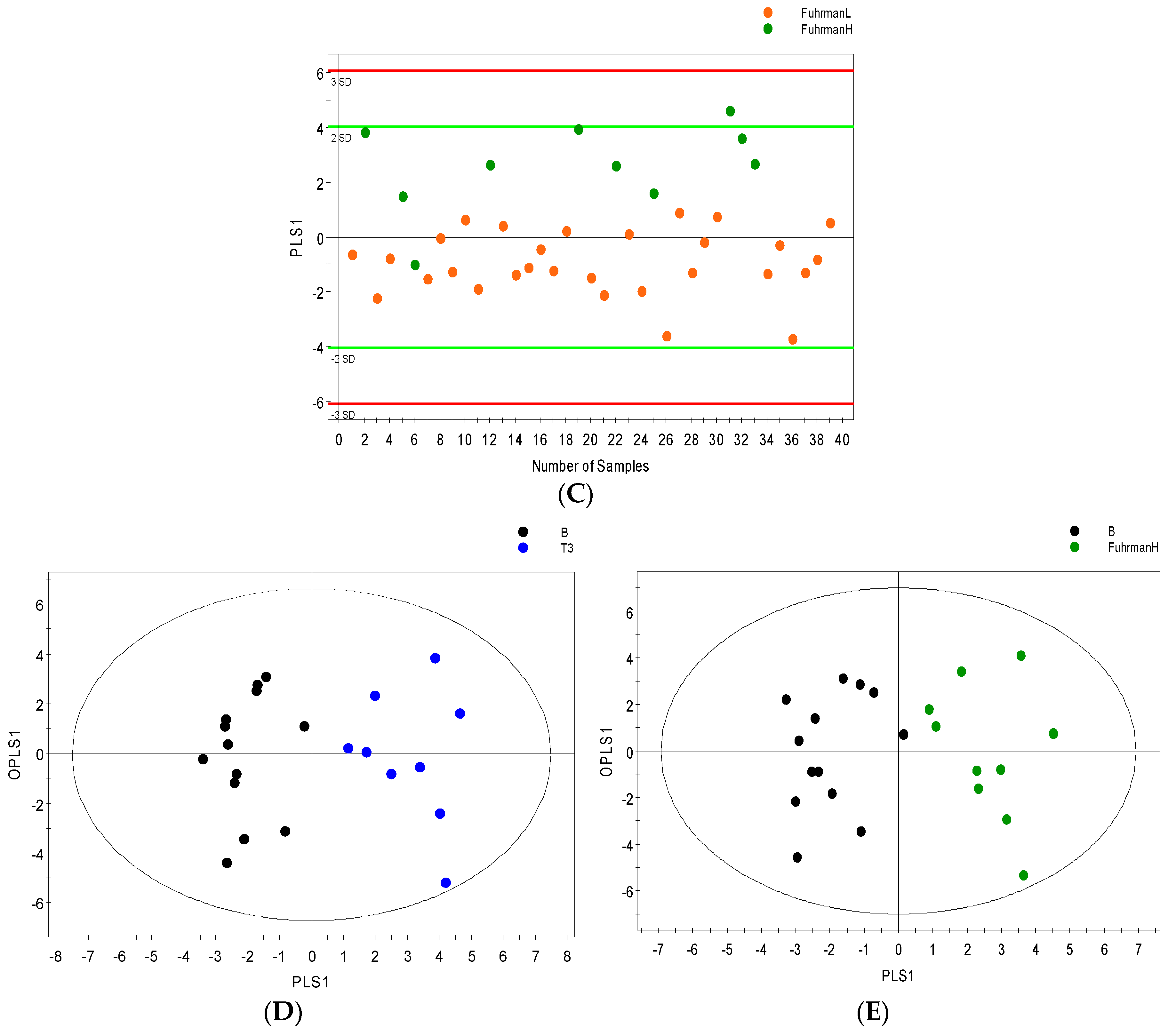
2.3. Distinguishing between Benign and Cancerous Renal Masses in 1H NMR and GCMS Datasets
2.4. Integrative 1H NMR and GCMS Data Analysis
2.5. Differential Metabolites
2.6. Internal Validation and AUC
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. 1H NMR Analysis
4.3. GCMS Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Linehan, W.M.; Srinivasan, R.; Schmidt, L.S. The genetic basis of kidney cancer: A metabolic disease. Nat. Rev. Urol. 2010, 7, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.C.; Vukina, J.; Smith, A.B.; Meyer, A.M.; Wheeler, S.B.; Kuo, T.M.; Tan, H.J.; Woods, M.E.; Raynor, M.C.; Wallen, E.M.; et al. Preoperatively misclassified, surgically removed benign renal masses: A systematic review of surgical series and United States population level burden estimate. J. Urol. 2015, 193, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Ball, M.W.; Bezerra, S.M.; Gorin, M.A.; Cowan, M.; Pavlovich, C.P.; Pierorazio, P.M.; Netto, G.J.; Allaf, M.E. Grade heterogeneity in small renal masses: Potential implications for renal mass biopsy. J. Urol. 2015, 193, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Linehan, W.M.; Ricketts, C.J. The metabolic basis of kidney cancer. Semin. Cancer Biol. 2013, 23, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Yang, O.C.; Maxwell, P.H.; Pollard, P.J. Renal cell carcinoma: Translational aspects of metabolism and therapeutic consequences. Kidney Int. 2013, 84, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.; Monteiro, M.; Jeronimo, C.; Henrique, R.; Belo, L.; Bastos, M.L.; Guedes de Pinho, P.; Carvalho, M. Renal cell carcinoma: A critical analysis of metabolomic biomarkers emerging from current model systems. Transl. Res. 2016, 180, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Catchpole, G.; Platzer, A.; Weikert, C.; Kempkensteffen, C.; Johannsen, M.; Krause, H.; Jung, K.; Miller, K.; Willmitzer, L.; Selbig, J.; et al. Metabolic profiling reveals key metabolic features of renal cell carcinoma. J. Cell. Mol. Med. 2011, 15, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Dong, B.; Jia, J.; Zhu, H.; Diao, C.; Yan, Z.; Huang, Y.; Li, X. Application of ex vivo 1H NMR metabonomics to the characterization and possible detection of renal cell carcinoma metastases. J. Cancer Res. Clin. Oncol. 2012, 138, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Wettersten, H.I.; Hakimi, A.A.; Morin, D.; Bianchi, C.; Johnstone, M.E.; Donohoe, D.R.; Trott, J.F.; Aboud, O.A.; Stirdivant, S.; Neri, B.; et al. Grade-Dependent Metabolic Reprogramming in Kidney Cancer Revealed by Combined Proteomics and Metabolomics Analysis. Cancer Res. 2015, 75, 2541–2552. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Ji, J.; Zhao, L.; Chen, M.; Shi, A.; Pan, L.; Huang, Y.; Zhang, H.; Dong, B.; Gao, H. Prediction and diagnosis of renal cell carcinoma using nuclear magnetic resonance-based serum metabolomics and self-organizing maps. Oncotarget 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Huang, Z.; Gao, Y.; Yan, X.; Xing, J.; Hang, W. LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J. Proteome Res 2011, 10, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Zira, A.N.; Theocharis, S.E.; Mitropoulos, D.; Migdalis, V.; Mikros, E. 1H NMR metabonomic analysis in renal cell carcinoma: A possible diagnostic tool. J. Proteome Res. 2010, 9, 4038–4044. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Aronov, P.; Zakharkin, S.O.; Anderson, D.; Perroud, B.; Thompson, I.M.; Weiss, R.H. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol. Cell. Proteom. 2009, 8, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Ganti, S.; Weiss, R.H. Urine metabolomics for kidney cancer detection and biomarker discovery. Urol. Oncol. 2011, 29, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, M.S.; Barros, A.S.; Pinto, J.; Carvalho, M.; Pires-Luis, A.S.; Henrique, R.; Jeronimo, C.; Bastos, M.L.; Gil, A.M.; Guedes de Pinho, P. Nuclear Magnetic Resonance metabolomics reveals an excretory metabolic signature of renal cell carcinoma. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Taylor, S.L.; Ganti, S.; Guo, L.; Osier, M.V.; Weiss, R.H. Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. OMICS 2011, 15, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Palmnas, M.S.; Vogel, H.J. The future of NMR metabolomics in cancer therapy: Towards personalizing treatment and developing targeted drugs? Metabolites 2013, 3, 373–396. [Google Scholar] [CrossRef] [PubMed]
- Chatham, J.C.; Blackband, S.J. Nuclear magnetic resonance spectroscopy and imaging in animal research. ILAR J. 2001, 42, 189–208. [Google Scholar] [CrossRef] [PubMed]
- Hornak, J.P. Basics of NMR. Available online: http://www.cis.rit.edu/htbooks/nmr/index.html (accessed on 19 January 2016).
- Smolinska, A.; Blanchet, L.; Buydens, L.M.; Wijmenga, S.S. NMR and pattern recognition methods in metabolomics: From data acquisition to biomarker discovery: A review. Anal. Chim. Acta 2012, 750, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Settle, F.A.; Majors, R.E.; Moody, J.R.; Hites, A.R. Gas chromatography mass spectrometry. In Handbook of Instrumental Techniques for Analytical Chemistry; Settle, F.A., Ed.; Prentice Hall: Upper Saddle River, NJ, USA, 1997; Chapter 31; pp. 609–626. [Google Scholar]
- Trock, B.J. Application of metabolomics to prostate cancer. Urol. Oncol. 2011, 29, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Marshall, D.D.; Lei, S.; Worley, B.; Huang, Y.; Garcia-Garcia, A.; Franco, R.; Dodds, E.D.; Powers, R. Combining DI-ESI-MS and NMR datasets for metabolic profiling. Metabolomics 2015, 11, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, M.S.; Carvalho, M.; de Lourdes Bastos, M.; de Pinho, P.G. Biomarkers in renal cell carcinoma: A metabolomics approach. Metabolomics 2014, 10. [Google Scholar] [CrossRef]
- Monteiro, M.; Carvalho, M.; Henrique, R.; Jeronimo, C.; Moreira, N.; de Lourdes Bastos, M.; de Pinho, P.G. Analysis of volatile human urinary metabolome by solid-phase microextraction in combination with gas chromatography-mass spectrometry for biomarker discovery: Application in a pilot study to discriminate patients with renal cell carcinoma. Eur. J. Cancer 2014, 50, 1993–2002. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Park, B.; Jeong, B.C.; Seo, S.I.; Jeon, S.S.; Choi, H.Y.; Adami, H.O.; Lee, J.E.; Lee, H.M. Body mass index and survival in patients with renal cell carcinoma: A clinical-based cohort and meta-analysis. Int. J. Cancer 2013, 132, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Vavallo, A.; Simone, S.; Lucarelli, G.; Rutigliano, M.; Galleggiante, V.; Grandaliano, G.; Gesualdo, L.; Campagna, M.; Cariello, M.; Ranieri, E.; et al. Pre-existing type 2 diabetes mellitus is an independent risk factor for mortality and progression in patients with renal cell carcinoma. Medicine 2014, 93. [Google Scholar] [CrossRef] [PubMed]
- Ragone, R.; Sallustio, F.; Piccinonna, S.; Rutigliano, M.; Vanessa, G.; Palazzo, S.; Lucarelli, G.; Ditonno, P.; Battaglia, M.; Fanizzi, P.F.; et al. Renal Cell Carcinoma: A Study through NMR-Based Metabolomics Combined with Transcriptomics. Diseases 2016, 4. [Google Scholar] [CrossRef]
- Slupsky, C.M.; Steed, H.; Wells, T.H.; Dabbs, K.; Schepansky, A.; Capstick, V.; Faught, W.; Sawyer, M.B. Urine Metabolite Analysis Offers Potential Early Diagnosis of Ovarian and Breast Cancers. Clin. Cancer Res. 2010, 16, 5835–5841. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Cai, G.; Su, M.; Chen, T.; Liu, Y.; Xu, Y.; Ni, Y.; Zhao, A.; Cai, S.; Xu, L.X.; et al. Urinary metabonomic study on colorectal cancer. J. Proteome Res. 2010, 9, 1627–1634. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lane, A.N.; Ricketts, C.J.; Sourbier, C.; Wei, M.-H.; Shuch, B.; Pike, L.; Wu, M.; Rouault, T.A.; Boros, L.G.; Fan, T.W.M.; Linehan, W.M. Metabolic Reprogramming for Producing Energy and Reducing Power in Fumarate Hydratase Null Cells from Hereditary Leiomyomatosis Renal Cell Carcinoma. PLoS ONE 2013, 8, e72179. [Google Scholar] [CrossRef] [PubMed]
- Hinder, L.M.; Vivekanandan-Giri, A.; McLean, L.L.; Pennathur, S.; Feldman, E.L. Decreased glycolytic and tricarboxylic acid cycle intermediates coincide with peripheral nervous system oxidative stress in a murine model of type 2 diabetes. J. Endocrinol. 2013, 216, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pause, A.; Lee, S.; Worrell, R.A.; Chen, D.Y.; Burgess, W.H.; Linehan, W.M.; Klausner, R.D. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 2156–2161. [Google Scholar] [CrossRef] [PubMed]
- Pinthus, J.H.; Whelan, K.F.; Gallino, D.; Lu, J.P.; Rothschild, N. Metabolic features of clear-cell renal cell carcinoma: Mechanisms and clinical implications. Can. Urol. Assoc. J. 2011, 5, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr. The von Hippel-Lindau protein, HIF hydroxylation, and oxygen sensing. Biochem. Biophys. Res. Commun. 2005, 338, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Sudarshan, S.; Karam, J.A.; Brugarolas, J.; Thompson, R.H.; Uzzo, R.; Rini, B.; Margulis, V.; Patard, J.J.; Escudier, B.; Linehan, W.M. Metabolism of kidney cancer: From the lab to clinical practice. Eur. Urol. 2013, 63, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Lucarelli, G.; Galleggiante, V.; Rutigliano, M.; Sanguedolce, F.; Cagiano, S.; Bufo, P.; Lastilla, G.; Maiorano, E.; Ribatti, D.; Giglio, A.; et al. Metabolomic profile of glycolysis and the pentose phosphate pathway identifies the central role of glucose-6-phosphate dehydrogenase in clear cell-renal cell carcinoma. Oncotarget 2015, 6, 13371–13386. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar]
- Suburu, J.; Chen, Y.Q. Lipids and prostate cancer. Prostaglandins Other Lipid Mediat. 2012, 98, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Swierczynski, J.; Hebanowska, A.; Sledzinski, T. Role of abnormal lipid metabolism in development, progression, diagnosis and therapy of pancreatic cancer. World J. Gastroenterol. 2014, 20, 2279–2303. [Google Scholar] [CrossRef] [PubMed]
- Koochekpour, S.; Majumdar, S.; Azabdaftari, G.; Attwood, K.; Scioneaux, R.; Subramani, D.; Manhardt, C.; Lorusso, G.D.; Willard, S.S.; Thompson, H.; et al. Serum glutamate levels correlate with Gleason score and glutamate blockade decreases proliferation, migration, and invasion and induces apoptosis in prostate cancer cells. Clin. Cancer Res. 2012, 18, 5888–5901. [Google Scholar] [CrossRef] [PubMed]
- Gameiro, P.A.; Yang, J.; Metelo, A.M.; Perez-Carro, R.; Baker, R.; Wang, Z.; Arreola, A.; Rathmell, W.K.; Olumi, A.; Lopez-Larrubia, P.; et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Meta. 2013, 17, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Pelicano, H.; Huang, P. Cancer metabolism: Is glutamine sweeter than glucose? Cancer Cell 2010, 18, 199–200. [Google Scholar] [CrossRef] [PubMed]
- Schicho, R.; Shaykhutdinov, R.; Ngo, J.; Nazyrova, A.; Schneider, C.; Panaccione, R.; Kaplan, G.G.; Vogel, H.J.; Storr, M. Quantitative Metabolomic Profiling of Serum, Plasma, and Urine by 1H NMR Spectroscopy Discriminates between Patients with Inflammatory Bowel Disease and Healthy Individuals. J. Proteome Res. 2012, 11, 3344–3357. [Google Scholar] [CrossRef] [PubMed]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, E.; Slupsky, C.M. Targeted profiling: Quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 2006, 78, 4430–4442. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application in 1H NMR Metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Hummel, J.; Selbig, J.; Walther, D.; Kopka, J. The Golm metabolome database: A database for GC-MS based metabolite profiling. In Metabolomics; Nielsen, J., Jewett, M., Eds.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2007; Volume 18, pp. 75–95. [Google Scholar]
- Egdington, E.S. Randomization Tests; Marcel Dekker Inc.: New York, NY, USA, 1987. [Google Scholar]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Umetrics AB. User Guide to SIMCA; Version 14; Umetrics AB: Malmo, Sweden, 2015. [Google Scholar]
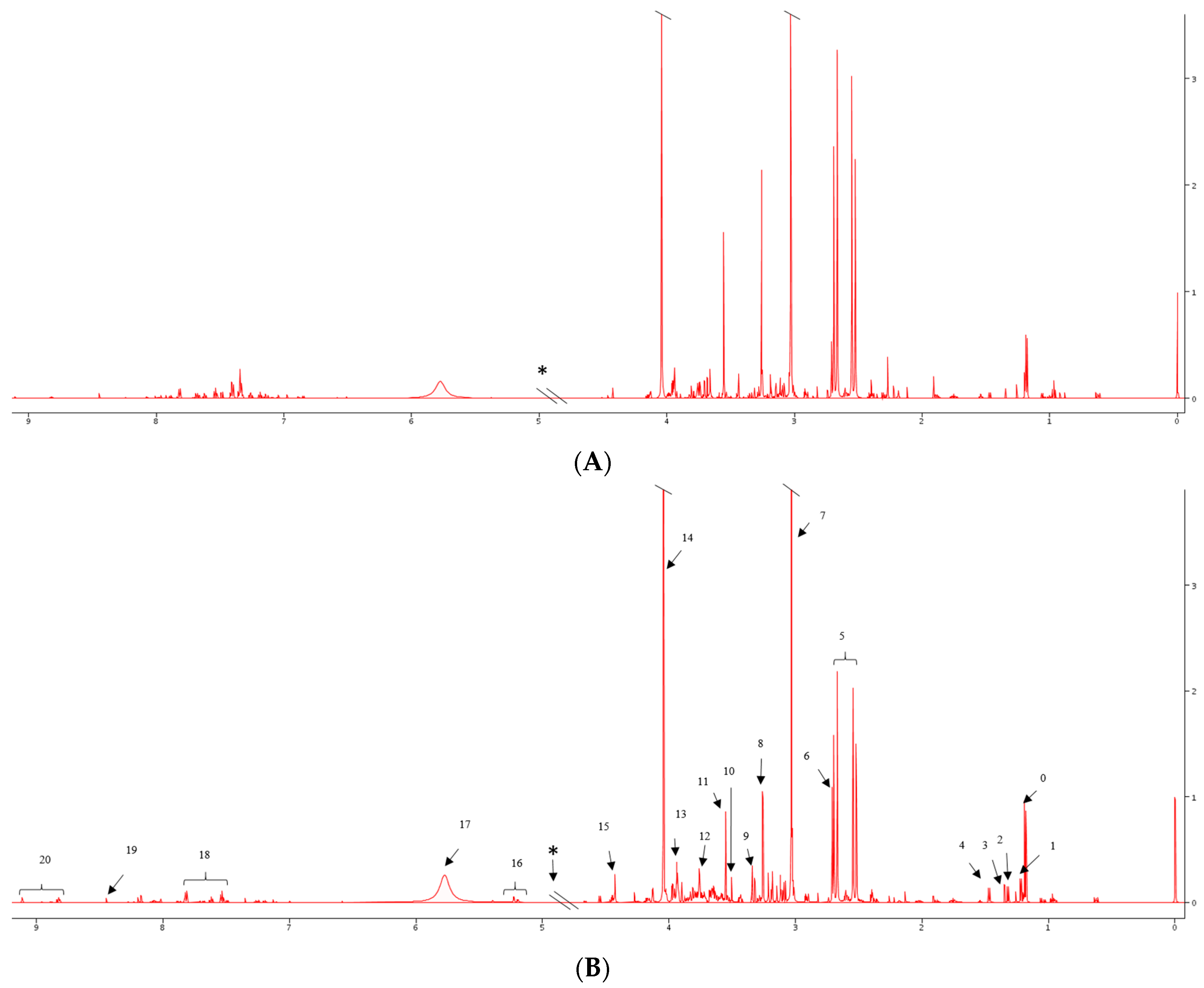
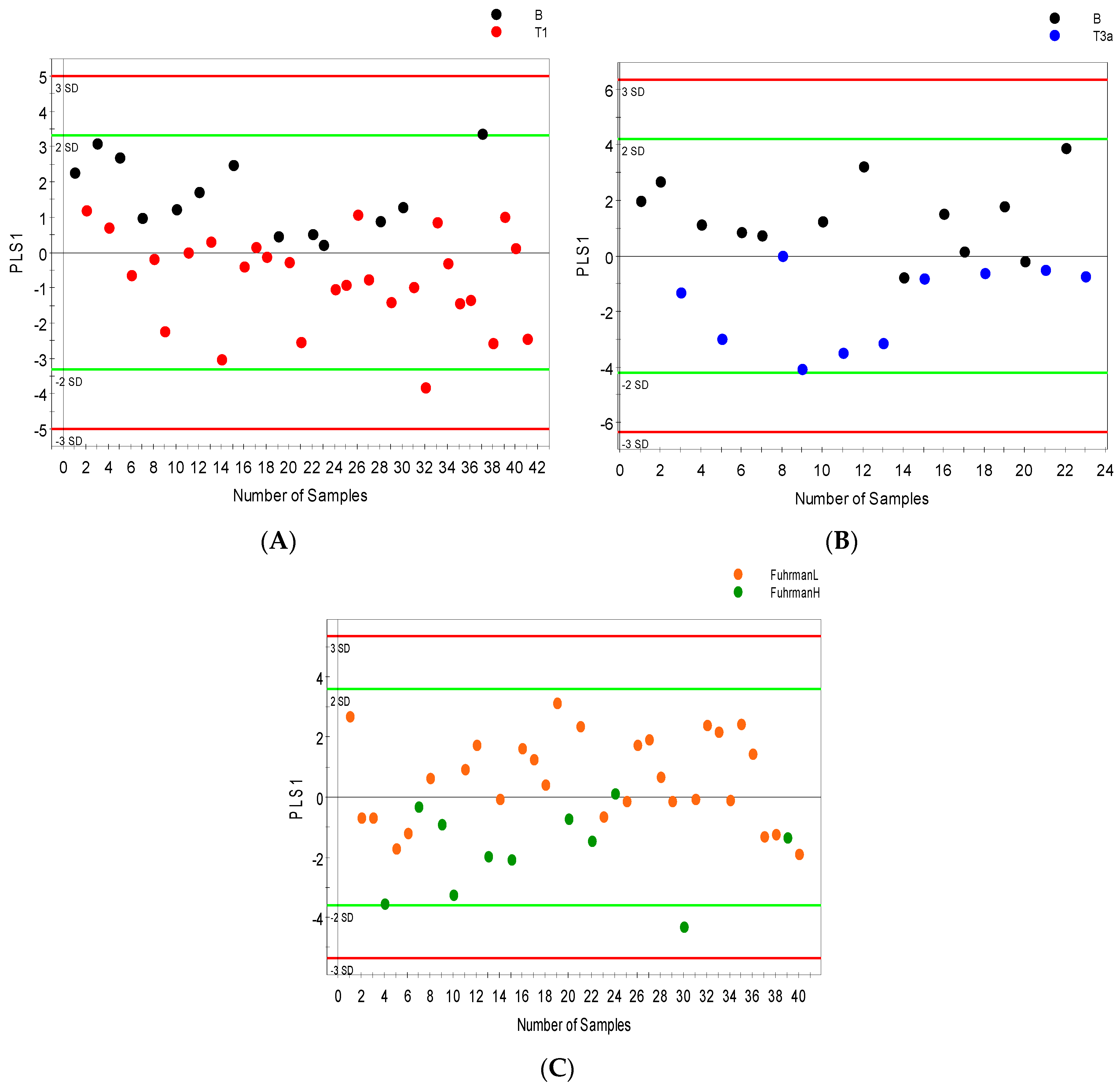
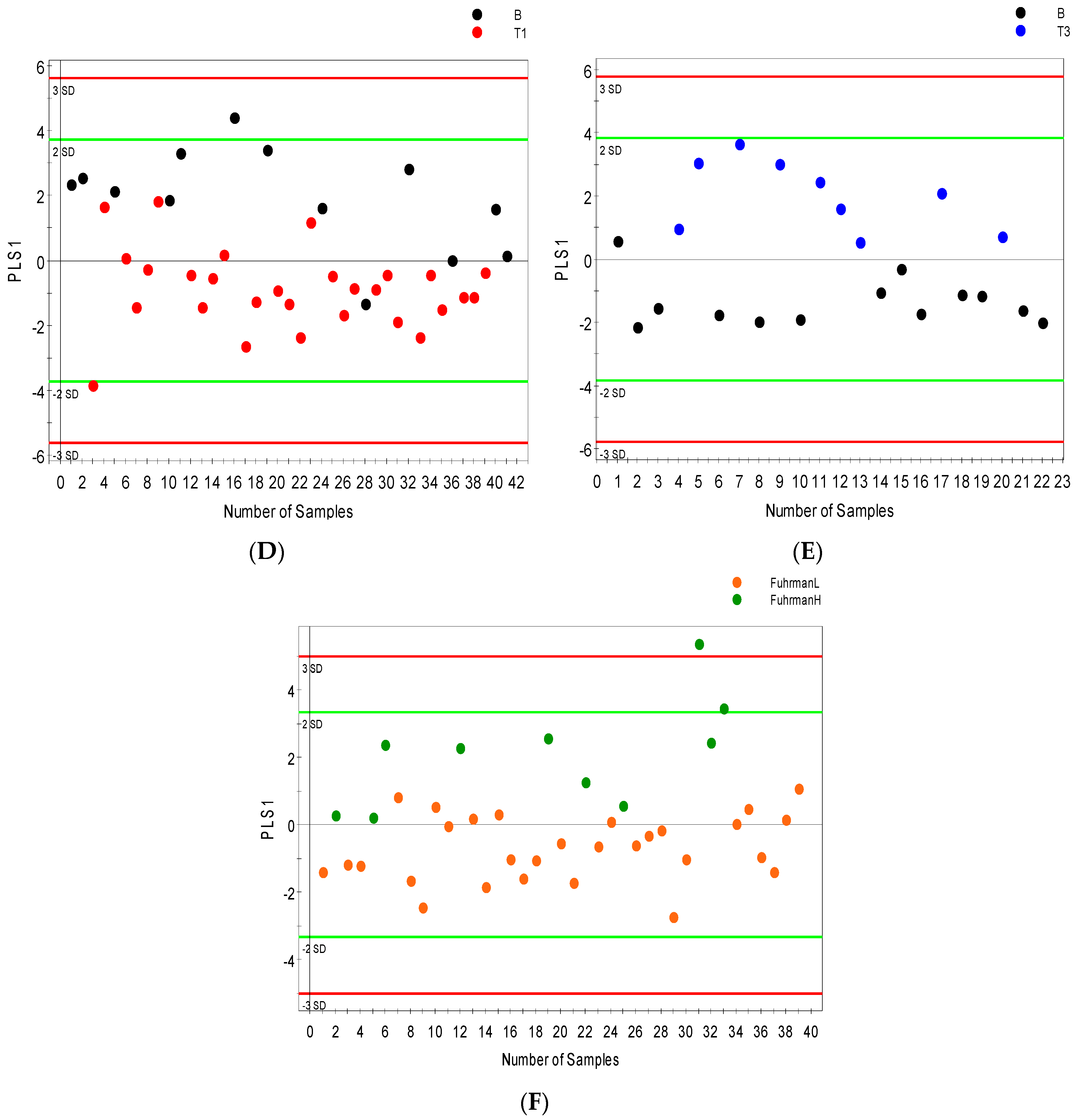
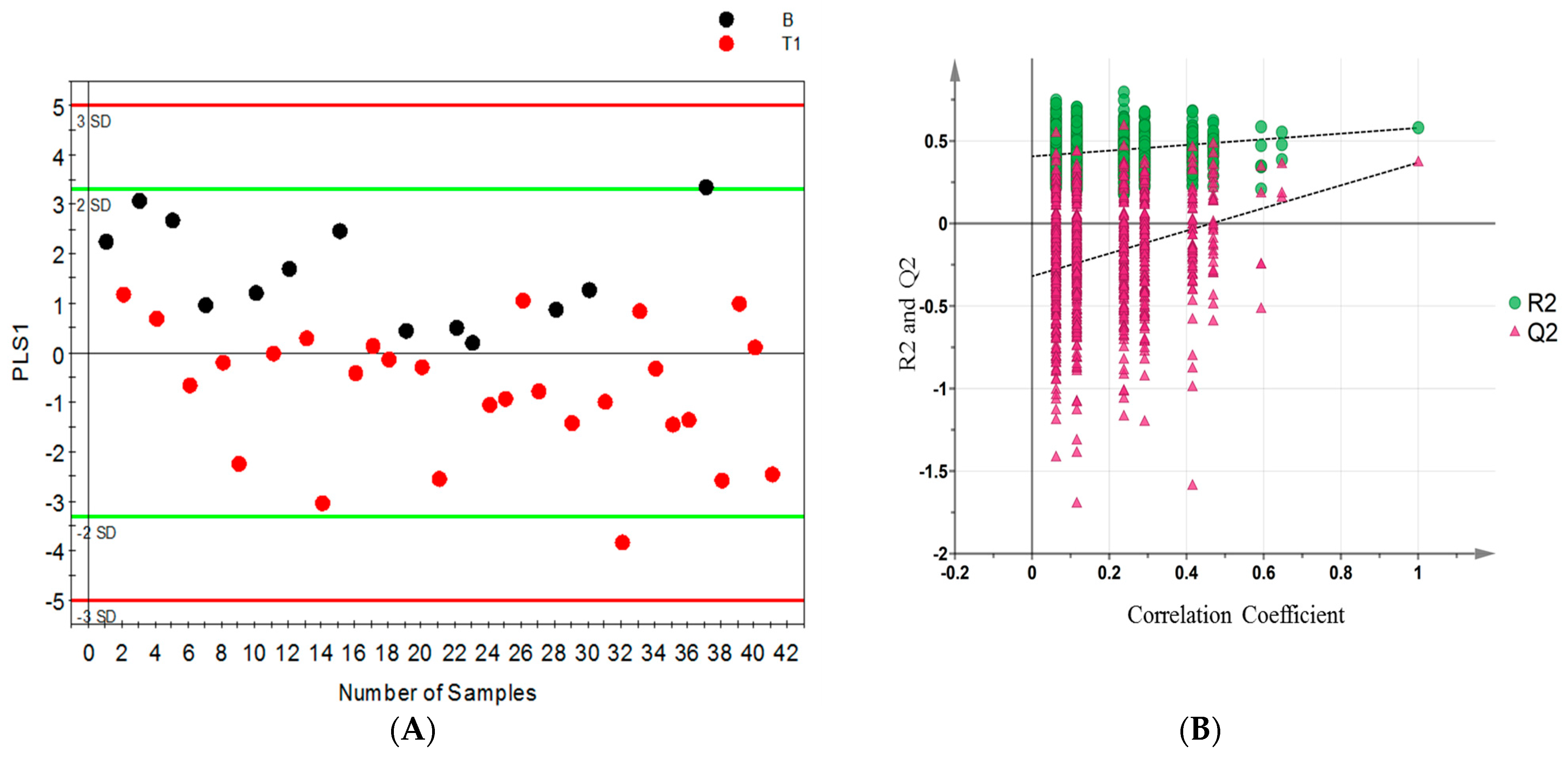
| Sample Group | Number of Sample (Urine and Serum) | Age at Surgery Range (Years) | Mean Age (Years) | Number of Men | Number of Women |
|---|---|---|---|---|---|
| Controls | 13 | 39–69 | 53.67 | 9 | 4 |
| All RCC | 40 | 36–84 | 61.89 | 23 | 17 |
| ccRCC | 37 | 36–84 | 62.49 | 22 | 15 |
| Papillary | 2 | 37–72 | 53.91 | 1 | 1 |
| Unclassified | 1 | 56 | 55.52 | - | 1 |
| Stage I (T1a: 22; T1b: 6) | 28 | 36–84 | 60.88 | 16 | 12 |
| Stage II | 1 | 46 | 45.52 | 1 | - |
| Stage III | 10 | 59–80 | 66.35 | 6 | 4 |
| Stage IV | 1 | 57 | 56.87 | - | 1 |
| Smokers | 20 | 50–84 | 62.91 | 14 | 6 |
| Non-smokers | 28 | 36–82 | 57.31 | 13 | 15 |
| Unknown smoking status | 5 | 62-69 | 65.30 | 4 | 1 |
| BMI 19–25 | 13 | 39–84 | 61.02 | 4 | 9 |
| BMI above 25 | 28 | 36–74 | 58.02 | 21 | 7 |
| BMI unknown | 12 | 45–80 | 63.94 | 7 | 5 |
| Model Type | R2 | Q2 | CV Anova p-Value | Q2 Intercept |
|---|---|---|---|---|
| NMR serum | ||||
| B vs. pT1 | 0.46 | 0.28 | 3. 4 × 10−3 | −0.27 |
| B vs. pT3 | 0.58 | 0.37 | 1.6 × 10−2 | −0.32 |
| FuhrmanL vs. FuhrmanH | 0.37 | 0.23 | 8.7 × 10−3 | −0.24 |
| NMR urine | ||||
| B vs. pT1 | 0.50 | 0.37 | 4.9 × 10−4 | −0.28 |
| B vs. pT3 | 0.72 | 0.68 | 2.6 × 10−5 | −0.41 |
| FuhrmanL vs. FuhrmanH | 0.54 | 0.36 | 8.6 × 10−4 | −0.29 |
| GCMS serum | ||||
| B vs. pT1 | 0.63 | 0.48 | 3.2 × 10−3 | −0.28 |
| pT1 vs. pT3 | 0.70 | 0.54 | 4.1 × 10−5 | −0.45 |
| FuhrmanL vs. FuhrmanH | 0.60 | 0.47 | 1.0 × 10−5 | −0.26 |
| GCMS urine | ||||
| B vs. pT3 | 0.87 | 0.70 | 3.4 × 10−4 | −0.43 |
| B vs. FuhrmanH | 0.84 | 0.62 | 1.1 × 10−3 | −0.46 |
| Metabolomics Platform | Decrease in Cancer vs. Benign | Increase in Cancer vs. Benign | ||||||
|---|---|---|---|---|---|---|---|---|
| Serum | p-Value (<0.05) | Urine | p-Value (<0.05) | Serum | p-Value (<0.05) | Urine | p-Value (<0.05) | |
| 1H NMR Metabolites | Citrate | Citrate | 0.039 | 2-oxoisocaproate | Pyruvate | 0.040 | ||
| Methanol | Succinate | 0.004 | Lactate | 0.049 | ||||
| Threonine Glycine | Glycine | 0.047 | Creatine | Oxypurinol | ||||
| Histidine Taurine | 3-hydroxybutyrate Creatinine | Isoleucine | 0.041 | Gluconate Hypoxanthine | ||||
| Glutamine | 2-aminoisobutyrate | 0.008 | Glutamate | 0.003 | Malonate | |||
| Phenylalanine | 0.025 | Ornithine | 0.048 | Betaine | ||||
| Tryptophan | ||||||||
| Methylhistidine | 0.017 | Trigonelline | ||||||
| Dimethylamine | ||||||||
| GCMS Metabolites | 5-methylcytosine | 0.049 | Acetate Threonine | Glutamate Tyrosine | Glucose | 0.001 | ||
| Eicosanoate | 0.003 | Gluconate | Octadecanoate | Erythritol | 0.011 | |||
| Thymine | Galactose | 0.030 | 2-oxoglutarate | 0.032 | ||||
| Mannitol | Pyruvate | 0.018 | Myo-inositol | 0.040 | ||||
| Citrate | Lactate | 0.018 | ||||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falegan, O.S.; Ball, M.W.; Shaykhutdinov, R.A.; Pieroraio, P.M.; Farshidfar, F.; Vogel, H.J.; Allaf, M.E.; Hyndman, M.E. Urine and Serum Metabolomics Analyses May Distinguish between Stages of Renal Cell Carcinoma. Metabolites 2017, 7, 6. https://doi.org/10.3390/metabo7010006
Falegan OS, Ball MW, Shaykhutdinov RA, Pieroraio PM, Farshidfar F, Vogel HJ, Allaf ME, Hyndman ME. Urine and Serum Metabolomics Analyses May Distinguish between Stages of Renal Cell Carcinoma. Metabolites. 2017; 7(1):6. https://doi.org/10.3390/metabo7010006
Chicago/Turabian StyleFalegan, Oluyemi S., Mark W. Ball, Rustem A. Shaykhutdinov, Phillip M. Pieroraio, Farshad Farshidfar, Hans J. Vogel, Mohamad E. Allaf, and Matthew E. Hyndman. 2017. "Urine and Serum Metabolomics Analyses May Distinguish between Stages of Renal Cell Carcinoma" Metabolites 7, no. 1: 6. https://doi.org/10.3390/metabo7010006
APA StyleFalegan, O. S., Ball, M. W., Shaykhutdinov, R. A., Pieroraio, P. M., Farshidfar, F., Vogel, H. J., Allaf, M. E., & Hyndman, M. E. (2017). Urine and Serum Metabolomics Analyses May Distinguish between Stages of Renal Cell Carcinoma. Metabolites, 7(1), 6. https://doi.org/10.3390/metabo7010006






