Real-Time Authentication of Camellia Oil by Rapid Evaporative Ionization Mass Spectrometry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Sample Preparation
2.3. Analytical Conditions
2.4. Optimization of REIMS Parameters
2.5. Data Processing
3. Results and Discussion
3.1. Mass Spectra Identification and Comparison
3.2. Model Building and Validation
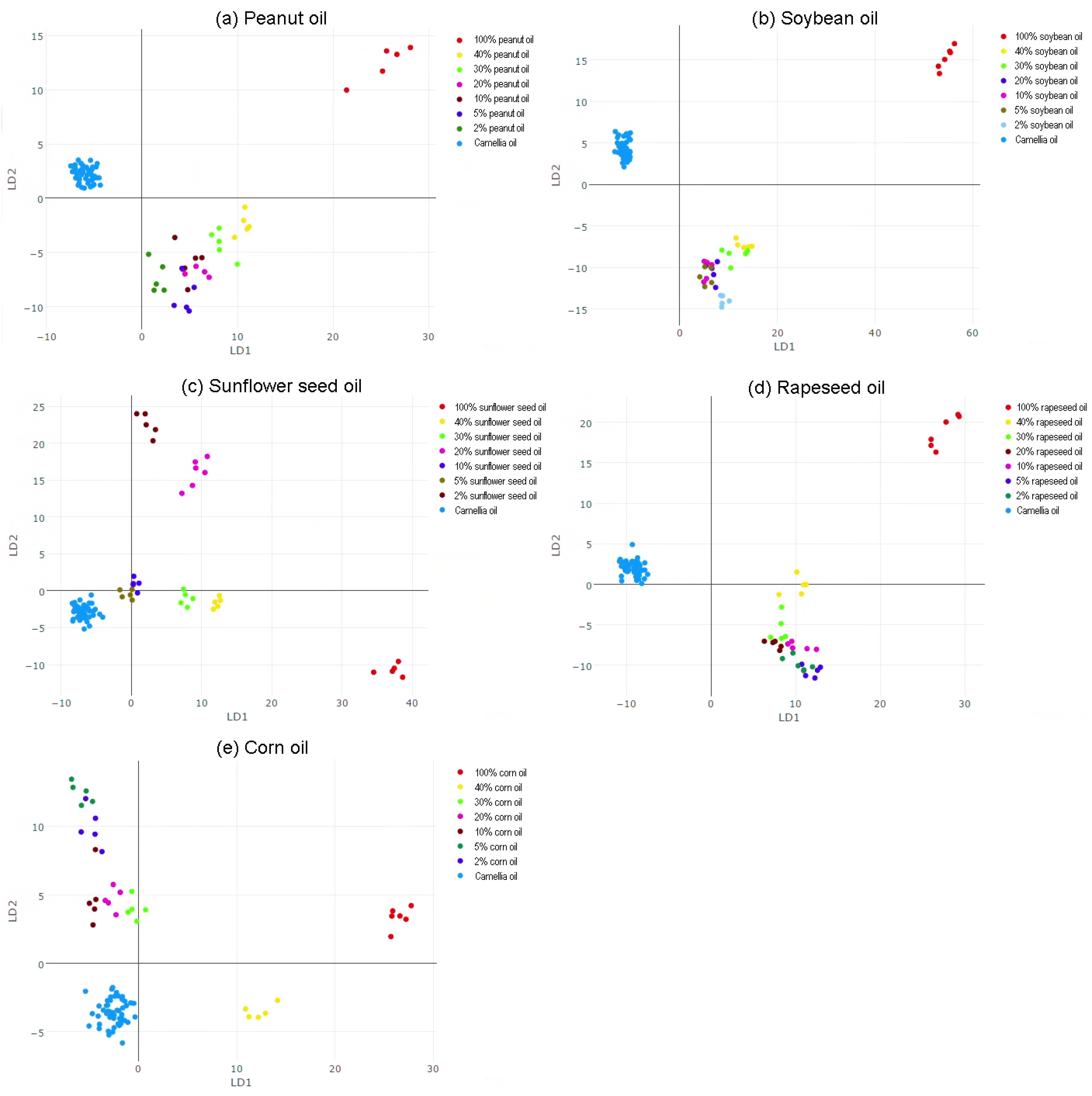
3.3. Discrimination of Pure and Doped Camellia Oil
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cheng, X.; Yang, T.; Wang, Y.; Zhou, B.; Yan, L.; Teng, L.; Wang, F.; Chen, L.; He, Y.; Guo, K.; et al. New Method for Effective Identification of Adulterated Camellia Oil Basing on Camellia Oleifera-Specific DNA. Arab. J. Chem. 2018, 11, 815–826. [Google Scholar] [CrossRef]
- Su, M.H.; Shih, M.C.; Lin, K.-H. Chemical Composition of Seed Oils in Native Taiwanese Camellia Species. Food Chem. 2014, 156, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Jin, L.; Liu, Q.; Zhao, K.; Lin, L.; Zheng, J.; Li, C.; Chen, B.; Shen, Y. Recent Advances in the Extraction, Composition Analysis and Bioactivity of Camellia (Camellia Oleifera Abel.) Oil. Trends Food Sci. Technol. 2024, 143, 104211. [Google Scholar] [CrossRef]
- Yu, J.; Yan, H.; Wu, Y.; Wang, Y.; Xia, P. Quality Evaluation of the Oil of Camellia Spp. Foods 2022, 11, 2221. [Google Scholar] [CrossRef]
- He, W.; Lei, T. Identification of Camellia Oil Using FT-IR Spectroscopy and Chemometrics Based on Both Isolated Unsaponifiables and Vegetable Oils. Spectrochim. Acta A. Mol. Biomol. Spectrosc. 2020, 228, 117839. [Google Scholar] [CrossRef]
- Dou, X.; Mao, J.; Zhang, L.; Xie, H.; Chen, L.; Yu, L.; Ma, F.; Wang, X.; Zhang, Q.; Li, P. Multispecies Adulteration Detection of Camellia Oil by Chemical Markers. Molecules 2018, 23, 241. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, C.; Chen, H.; Ye, J.; Zhou, H. Identification and Detection of Adulterated Camellia Oleifera Abel. Oils by Near Infrared Transmittance Spectroscopy. Int. J. Food Prop. 2016, 19, 300–313. [Google Scholar] [CrossRef]
- Du, Q.; Zhu, M.; Shi, T.; Luo, X.; Gan, B.; Tang, L.; Chen, Y. Adulteration Detection of Corn Oil, Rapeseed Oil and Sunflower Oil in Camellia Oil by in Situ Diffuse Reflectance near-Infrared Spectroscopy and Chemometrics. Food Control 2021, 121, 107577. [Google Scholar] [CrossRef]
- Li, Y.; Fang, T.; Zhu, S.; Huang, F.; Chen, Z.; Wang, Y. Detection of Olive Oil Adulteration with Waste Cooking Oil via Raman Spectroscopy Combined with iPLS and SiPLS. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 189, 37–43. [Google Scholar] [CrossRef]
- Wang, T.; Wu, H.-L.; Long, W.-J.; Hu, Y.; Cheng, L.; Chen, A.-Q.; Yu, R.-Q. Rapid Identification and Quantification of Cheaper Vegetable Oil Adulteration in Camellia Oil by Using Excitation-Emission Matrix Fluorescence Spectroscopy Combined with Chemometrics. Food Chem. 2019, 293, 348–357. [Google Scholar] [CrossRef]
- Shi, T.; Wu, G.; Jin, Q.; Wang, X. Detection of Camellia Oil Adulteration Using Chemometrics Based on Fatty Acids GC Fingerprints and Phytosterols GC–MS Fingerprints. Food Chem. 2021, 352, 129422. [Google Scholar] [CrossRef]
- Hu, Q.; Zhang, J.; Xing, R.; Yu, N.; Chen, Y. Integration of Lipidomics and Metabolomics for the Authentication of Camellia Oil by Ultra-Performance Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry Coupled with Chemometrics. Food Chem. 2022, 373, 131534. [Google Scholar] [CrossRef]
- Zhu, M.; Shi, T.; Chen, Y.; Luo, S.; Leng, T.; Wang, Y.; Guo, C.; Xie, M. Prediction of Fatty Acid Composition in Camellia Oil by 1H NMR Combined with PLS Regression. Food Chem. 2019, 279, 339–346. [Google Scholar] [CrossRef]
- Hai, Z.; Wang, J. Detection of Adulteration in Camellia Seed Oil and Sesame Oil Using an Electronic Nose. Eur. J. Lipid Sci. Technol. 2006, 108, 116–124. [Google Scholar] [CrossRef]
- Duan, D.; Huang, Y.; Zou, Y.; He, B.; Tang, R.; Yang, L.; Zhang, Z.; Su, S.; Wang, G.; Zhang, D.; et al. Discrimination of Camellia Seed Oils Extracted by Supercritical CO2 Using Electronic Tongue Technology. Food Sci. Biotechnol. 2021, 30, 1303–1312. [Google Scholar] [CrossRef]
- Majchrzak, T.; Wojnowski, W.; Dymerski, T.; Gębicki, J.; Namieśnik, J. Electronic Noses in Classification and Quality Control of Edible Oils: A Review. Food Chem. 2018, 246, 192–201. [Google Scholar] [CrossRef]
- Li, S.; Zhu, X.; Zhang, J.; Li, G.; Su, D.; Shan, Y. Authentication of Pure Camellia Oil by Using Near Infrared Spectroscopy and Pattern Recognition Techniques. J. Food Sci. 2012, 77, C374–C380. [Google Scholar] [CrossRef] [PubMed]
- Guitton, Y.; Dervilly-Pinel, G.; Jandova, R.; Stead, S.; Takats, Z.; Le Bizec, B. Rapid Evaporative Ionisation Mass Spectrometry and Chemometrics for High-Throughput Screening of Growth Promoters in Meat Producing Animals. Food Addit. Contam. Part A 2018, 35, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Tzafetas, M.; Mitra, A.; Paraskevaidi, M.; Bodai, Z.; Kalliala, I.; Bowden, S.; Lathouras, K.; Rosini, F.; Szasz, M.; Savage, A.; et al. The Intelligent Knife (iKnife) and Its Intraoperative Diagnostic Advantage for the Treatment of Cervical Disease. Proc. Natl. Acad. Sci. USA 2020, 117, 7338–7346. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Chen, K.; Wang, H.; Zhang, M.; Yu, X.; Wang, J.; Shen, Q. In Situ and Real-Time Authentication of Thunnus Species by iKnife Rapid Evaporative Ionization Mass Spectrometry Based Lipidomics without Sample Pretreatment. Food Chem. 2020, 318, 126504. [Google Scholar] [CrossRef]
- Jones, E.A.; Simon, D.; Karancsi, T.; Balog, J.; Pringle, S.D.; Takats, Z. Matrix Assisted Rapid Evaporative Ionization Mass Spectrometry. Anal. Chem. 2019, 91, 9784–9791. [Google Scholar] [CrossRef] [PubMed]
- Barlow, R.S.; Fitzgerald, A.G.; Hughes, J.M.; McMillan, K.E.; Moore, S.C.; Sikes, A.L.; Tobin, A.B.; Watkins, P.J. Rapid Evaporative Ionization Mass Spectrometry: A Review on Its Application to the Red Meat Industry with an Australian Context. Metabolites 2021, 11, 171. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.; Brunius, C.; Chevallier, O.; Dervilly, G.; Elliott, C.; Guitton, Y.; Prenni, J.; Savolainen, O.; Hemeryck, L.; Vidkjær, N.; et al. Making Complex Measurements of Meat Composition Fast: Application of Rapid Evaporative Ionisation Mass Spectrometry to Measuring Meat Quality and Fraud. Meat Sci. 2021, 181, 108333. [Google Scholar] [CrossRef] [PubMed]
- Strittmatter, N.; Jones, E.; Veselkov, K.; Rebec, M.; Bundy, J.; Takats, Z. Analysis of Intact Bacteria Using Rapid Evaporative Ionisation Mass Spectrometry. Chem. Commun. 2013, 49, 6188–6190. [Google Scholar] [CrossRef]
- He, Q.; Yang, M.; Chen, X.; Yan, X.; Li, Y.; He, M.; Liu, T.; Chen, F.; Zhang, F. Differentiation between Fresh and Frozen-Thawed Meat Using Rapid Evaporative Ionization Mass Spectrometry: The Case of Beef Muscle. J. Agric. Food Chem. 2021, 69, 5709–5724. [Google Scholar] [CrossRef]
- Shen, Q.; Song, G.; Zhao, Q.; Wang, P.; Yang, H.; Xue, J.; Wang, H.; Cui, Y.; Wang, H. Detection of Lipidomics Characterization of Tuna Meat during Different Wet-Aging Stages Using iKnife Rapid Evaporative Ionization Mass Spectrometry. Food Res. Int. 2022, 156, 111307. [Google Scholar] [CrossRef]
- Lu, W.; Wang, P.; Ge, L.; Chen, X.; Guo, S.; Zhao, Q.; Zhu, X.; Cui, Y.; Zhang, M.; Chen, K.; et al. Real-Time Authentication of Minced Shrimp by Rapid Evaporative Ionization Mass Spectrometry. Food Chem. 2022, 383, 132432. [Google Scholar] [CrossRef]
- Gao, H.; Lin, J.; Jia, X.; Zhao, Y.; Wang, S.; Bai, H.; Ma, Q. Real-Time Authentication of Animal Species Origin of Leather Products Using Rapid Evaporative Ionization Mass Spectrometry and Chemometric Analysis. Talanta 2021, 225, 122069. [Google Scholar] [CrossRef]
- Shen, Q.; Wang, J.; Li, S.; Rao, W.; Wang, Y.; Wang, H. In Situ Rapid Evaporative Ionization Mass Spectrometry Method for Real-Time Discrimination of Pelodiscus Sinensis in Different Culturing Modes without Sample Preparation. Food Anal. Methods 2019, 12, 2699–2708. [Google Scholar] [CrossRef]
- Cardoso, V.; Sabin, G.; Hantao, L. Rapid Evaporative Ionization Mass Spectrometry (REIMS) Combined with Chemometrics for Real-Time Beer Analysis. Anal. Methods 2022, 14, 1540–1546. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cao, X.; Han, T.; Pei, H.; Ren, H.; Stead, S. A Novel Methodology for Real-Time Identification of the Botanical Origins and Adulteration of Honey by Rapid Evaporative Ionization Mass Spectrometry. Food Control 2019, 106, 106753. [Google Scholar] [CrossRef]
- Robson, K.; Birse, N.; Chevallier, O.; Elliott, C. Metabolomic Profiling to Detect Different Forms of Beef Fraud Using Rapid Evaporative Ionisation Mass Spectrometry (REIMS). NPJ Sci. Food 2022, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Black, C.; Chevallier, O.P.; Haughey, S.A.; Balog, J.; Stead, S.; Pringle, S.D.; Riina, M.V.; Martucci, F.; Acutis, P.L.; Morris, M.; et al. A Real Time Metabolomic Profiling Approach to Detecting Fish Fraud Using Rapid Evaporative Ionisation Mass Spectrometry. Metabolomics 2017, 13, 153. [Google Scholar] [CrossRef]
- Song, G.; Li, L.; Wang, H.; Zhang, M.; Yu, X.; Wang, J.; Shen, Q. Electric Soldering Iron Ionization Mass Spectrometry Based Lipidomics for in Situ Monitoring Fish Oil Oxidation Characteristics during Storage. J. Agric. Food Chem. 2020, 68, 2240–2248. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Wang, H.; Zhang, M.; Zhang, Y.; Wang, H.; Yu, X.; Wang, J.; Shen, Q. Real-Time Monitoring of the Oxidation Characteristics of Antarctic Krill Oil (Euphausia superba) during Storage by Electric Soldering Iron Ionization Mass Spectrometry-Based Lipidomics. J. Agric. Food Chem. 2020, 68, 1457–1467. [Google Scholar] [CrossRef]
- Strittmatter, N.; Lovrics, A.; Sessler, J.; McKenzie, J.S.; Bodai, Z.; Doria, M.L.; Kucsma, N.; Szakacs, G.; Takats, Z. Shotgun Lipidomic Profiling of the NCI60 Cell Line Panel Using Rapid Evaporative Ionization Mass Spectrometry. Anal. Chem. 2016, 88, 7507–7514. [Google Scholar] [CrossRef] [PubMed]
- LaFranchi, B.W.; Petrucci, G.A. A Comprehensive Characterization of Photoelectron Resonance Capture Ionization Aerosol Mass Spectrometry for the Quantitative and Qualitative Analysis of Organic Particulate Matter. Int. J. Mass Spectrom. 2006, 258, 120–133. [Google Scholar] [CrossRef]
- Liu, T.; Wang, W.; He, M.; Chen, F.; Liu, J.; Yang, M.; Guo, W.; Zhang, F. Real-Time Traceability of Sorghum Origin by Soldering Iron-Based Rapid Evaporative Ionization Mass Spectrometry and Chemometrics. Electrophoresis 2022, 43, 1841–1849. [Google Scholar] [CrossRef]
- Balog, J.; Kumar, S.; Alexander, J.; Golf, O.; Huang, J.; Wiggins, T.; Abbassi-Ghadi, N.; Enyedi, A.; Kacska, S.; Kinross, J.; et al. In Vivo Endoscopic Tissue Identification by Rapid Evaporative Ionization Mass Spectrometry (REIMS). Angew. Chem. Int. Ed. 2015, 54, 11059–11062. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, H.; Rao, W.; Cui, Y.; Yu, X.; Dai, Z.; Shen, Q. Rapid Evaporative Ionization Mass Spectrometry-Based Lipidomics Tracking of Grass Carp (Ctenopharyngodon idellus) during In Vitro Multiple-Stage Digestion. J. Agric. Food Chem. 2018, 66, 6246–6253. [Google Scholar] [CrossRef]
- Song, G.; Zhang, M.; Zhang, Y.; Wang, H.; Li, S.; Dai, Z.; Shen, Q. In Situ Method for Real-Time Discriminating Salmon and Rainbow Trout without Sample Preparation Using iKnife and Rapid Evaporative Ionization Mass Spectrometry-Based Lipidomics. J. Agric. Food Chem. 2019, 67, 4679–4688. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Zhao, X.; Wang, H.; Cao, X. A Real-Time Metabolomic Profiling Approach to Identify Virgin Olive Oil Adulteration by Rapid Evaporative Ionization Mass Spectrometry. Food Anal. Methods 2023, 16, 985–996. [Google Scholar] [CrossRef]
- Rigano, F.; Stead, S.; Mangraviti, D.; Jandova, R.; Petit, D.; Marino, N.; Mondello, L. Use of an “Intelligent Knife” (Iknife), Based on the Rapid Evaporative Ionization Mass Spectrometry Technology, for Authenticity Assessment of Pistachio Samples. Food Anal. Methods 2019, 12, 558–568. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, H.; Zhao, Q.; Zhu, X.; Wang, P.; Xue, J.; Chen, K.; Shen, Q. Real-Time Detection of Authenticity and Adulteration of Krill Phospholipids with Soybean Phospholipids Using Rapid Evaporative Ionization Mass Spectrometry: Application on Commercial Samples. Food Control 2021, 121, 107680. [Google Scholar] [CrossRef]
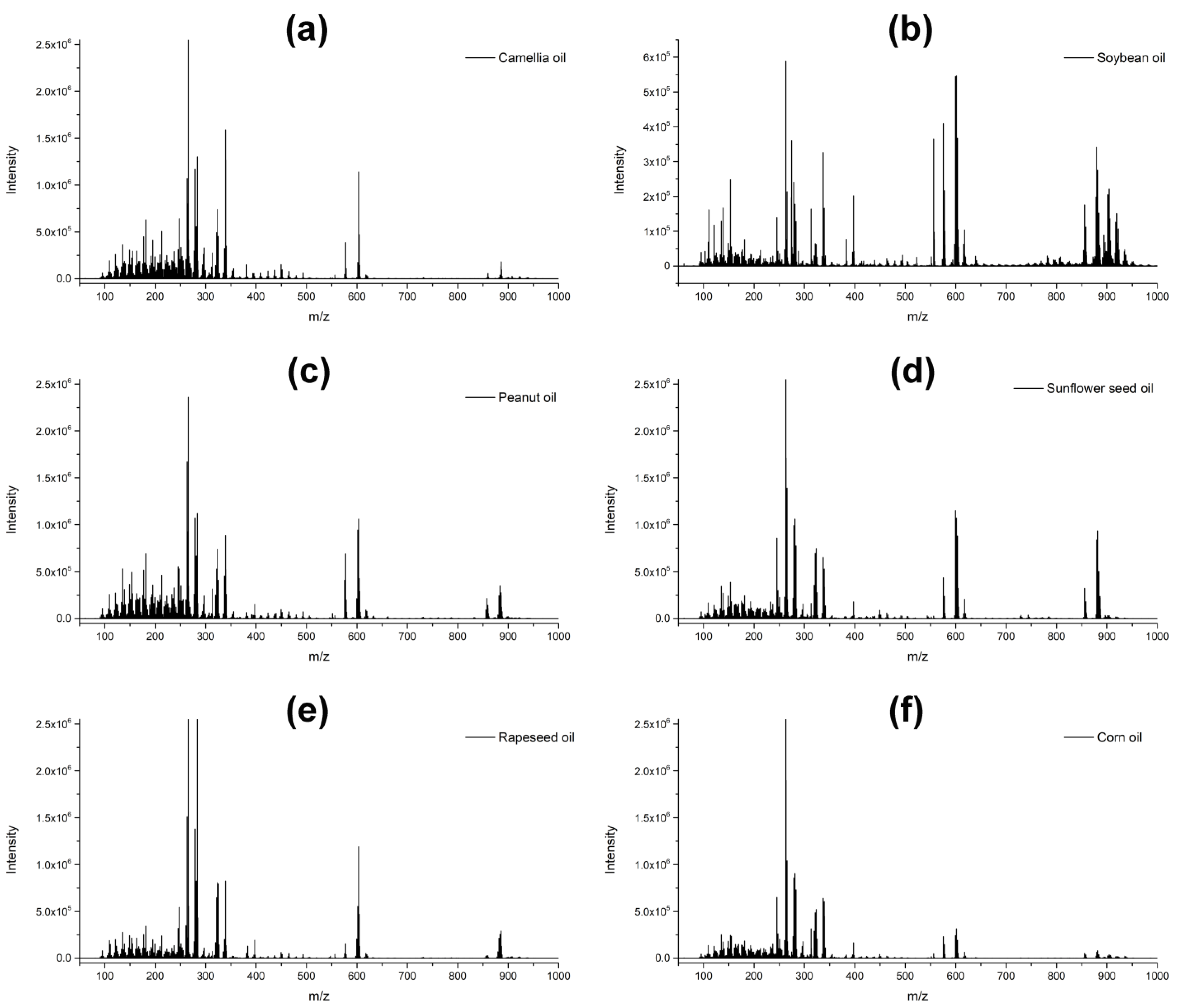

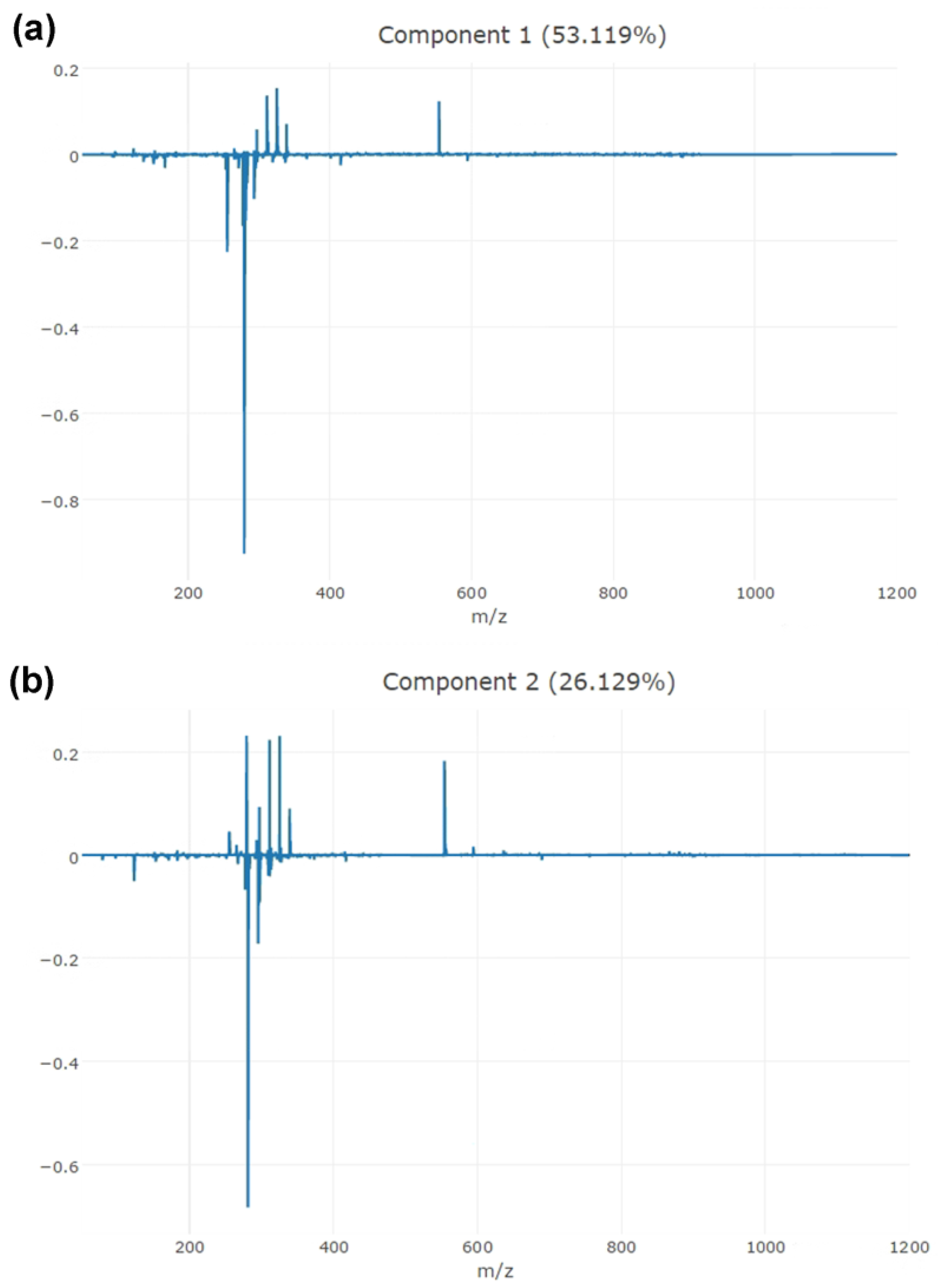
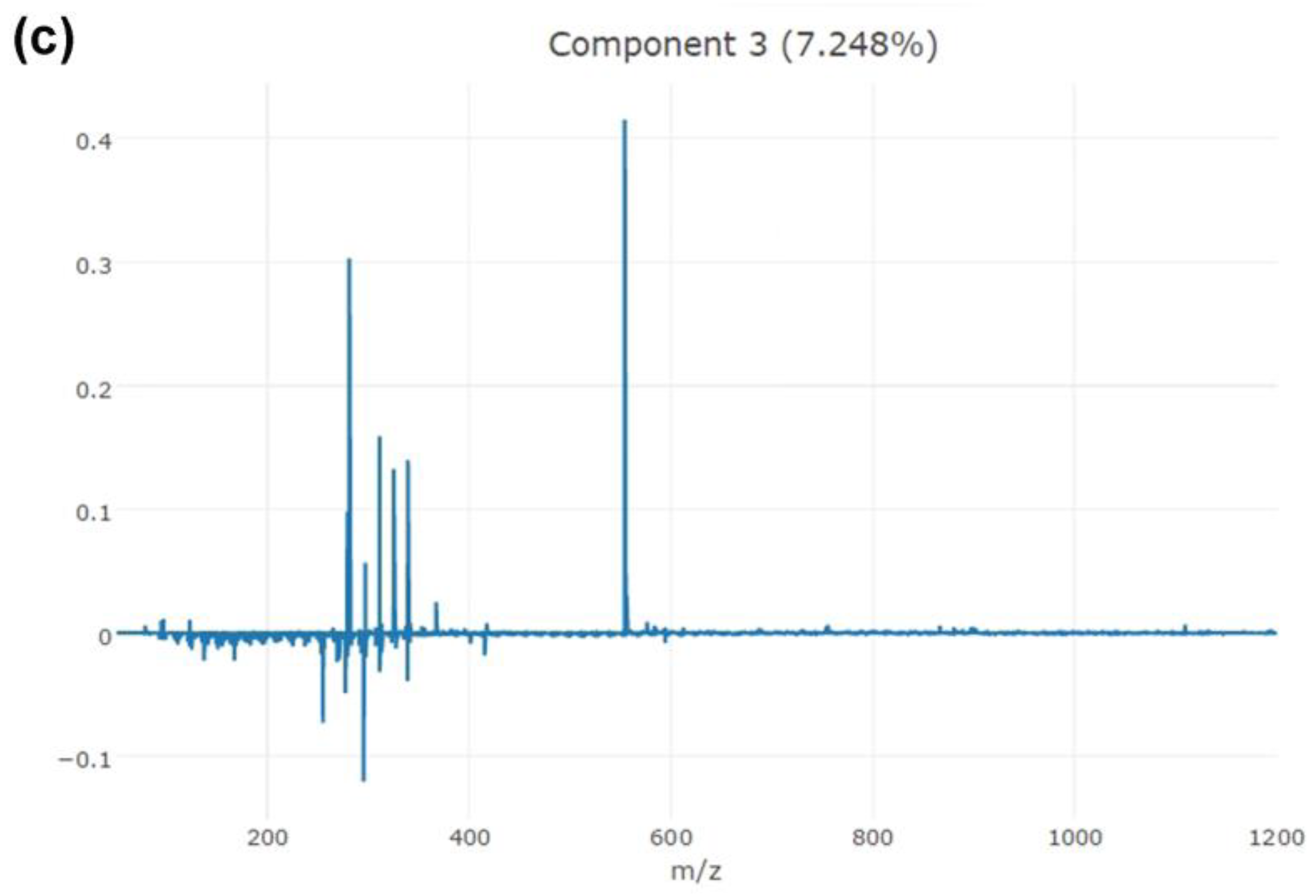
| Significant Ion (m/z) | Tentative Assignment | Main Class | Formula |
|---|---|---|---|
| 271.2259 | Juniperic acid | Fatty acids and conjugates | C16H32O3 |
| 275.1991 | Parinaric acid | Fatty acids and conjugates | C18H28O2 |
| 277.2156 | Pinolenic acid | Fatty acids and conjugates | C18H30O2 |
| 305.2462 | Sciadonic acid | Fatty acids and conjugates | C20H34O2 |
| 307.2616 | 20:2 (7Z,14Z) | Fatty acids and conjugates | C20H36O2 |
| 321.2767 | 21:2 (5Z,14Z) | Fatty acids and conjugates | C21H38O2 |
| 337.309 | 22:1 (7Z) | Fatty acids and conjugates | C22H42O2 |
| 339.3248 | Behenic acid | Fatty acids and conjugates | C22H44O2 |
| 367.3553 | Lignoceric acid | Fatty acids and conjugates | C24H48O2 |
| 395.3858 | Cerotic acid | Fatty acids and conjugates | C26H52O2 |
| 727.4859 | PG (P-16:0/18:3) | Phosphatidylglycerols | C40H73O9P |
| 729.5021 | PG (O-16:0/18:3) | Phosphatidylglycerols | C40H75O9P |
| 753.501 | PG (P-18:0/18:4) | Phosphatidylglycerols | C42H75O9P |
| 755.5164 | PG (O-18:0/18:4) | Phosphatidylglycerols | C42H77O9P |
| 853.7217 | TG (15:0/18:4/19:0) | Triacylglycerols | C55H98O6 |
| 881.752 | TG (18:1/18:1/18:2) | Triacylglycerols | C57H102O6 |
| 883.7704 | TG (18:1/18:1/18:1) | Triacylglycerols | C57H=O6 |
| 887.7182 | TG (15:1/18:4/22:3) | Triacylglycerols | C58H96O6 |
| 889.7326 | TG (18:3/18:3/19:1) | Triacylglycerols | C58H98O6 |
| Camellia Oil | Peanut Oil | Sunflower Seed Oil | Rapeseed Oil | Corn Oil | Soybean Oil | Outlier | Total | |
|---|---|---|---|---|---|---|---|---|
| Camellia oil | 137 | 0 | 0 | 0 | 0 | 0 | 4 | 141 |
| Peanut oil | 0 | 110 | 0 | 0 | 0 | 0 | 5 | 115 |
| Sunflower seed oil | 0 | 0 | 57 | 0 | 0 | 0 | 8 | 65 |
| Rapeseed oil | 0 | 0 | 0 | 92 | 0 | 0 | 8 | 100 |
| Corn oil | 0 | 0 | 0 | 0 | 124 | 0 | 6 | 130 |
| Soybean oil | 0 | 0 | 0 | 0 | 0 | 86 | 9 | 95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, J.; Liu, Q.; Jing, H.; Chen, X. Real-Time Authentication of Camellia Oil by Rapid Evaporative Ionization Mass Spectrometry. Separations 2024, 11, 68. https://doi.org/10.3390/separations11030068
Xiang J, Liu Q, Jing H, Chen X. Real-Time Authentication of Camellia Oil by Rapid Evaporative Ionization Mass Spectrometry. Separations. 2024; 11(3):68. https://doi.org/10.3390/separations11030068
Chicago/Turabian StyleXiang, Jun, Qi Liu, Huihua Jing, and Xiaoqing Chen. 2024. "Real-Time Authentication of Camellia Oil by Rapid Evaporative Ionization Mass Spectrometry" Separations 11, no. 3: 68. https://doi.org/10.3390/separations11030068





