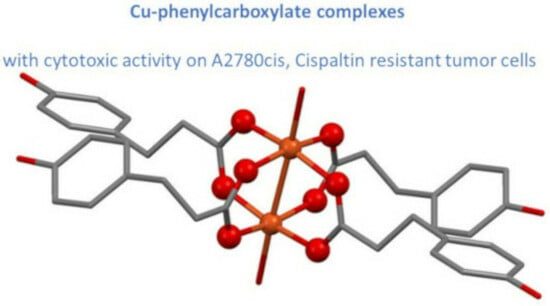
Synthesis, Characterization, DNA Binding and Cytotoxicity of Copper(II) Phenylcarboxylate Complexes
Abstract
:1. Introduction
2. Results
2.1. Crystal Structures
2.1.1. [Cu2(3-(4-Hydroxyphenyl)propanoate)4(H2O)2]·2H2O
2.1.2. [Cu2(Phenylacetate)4]·2H2O and [Cu2(Phenylpropanoate)4(H2O)2]
2.2. Infrared Spectra
2.3. Solution Studies
Major Species in Solution Characterization Using UV-Visible Spectra and Lipophilicity
2.4. Complex–DNA Binding Studies
2.4.1. Kb Determination (UV-Visible Spectra)
2.4.2. Mode of Binding (Relative Viscosity)
2.5. Cytotoxicity of the Compounds
3. Discussion
4. Materials and Methods
4.1. Synthetic Procedures
[Cu2(phenylcarboxylate)4] Complexes
4.2. Physical Methods
4.2.1. Characterization—General
4.2.2. Crystal Structure Determination
4.3. Lipophility Assessment
4.4. DNA Interaction
4.4.1. DNA Binding Constant: UV Absorption Titration Experiments
4.4.2. DNA Binding Mode: Variation of Viscosity Experiments
4.5. Cytotoxicity Studies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romani, A.M.P. Cisplatin in cancer treatment. Biochem. Pharmacol. 2022, 206, 115323. [Google Scholar] [CrossRef]
- Kellett, A.; Molphy, Z.; McKee, V.; Slator, C. Recent Advances in Anticancer Copper Compounds. In Metal-Based Anticancer Agents; Royal Society of Chemistry: London, UK, 2019; pp. 91–119. [Google Scholar]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in Copper Complexes as Anticancer Agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef] [PubMed]
- Zehra, S.; Tabassum, S.; Arjmand, F. Biochemical pathways of copper complexes: Progress over the past 5 years. Drug Discov. Today 2021, 26, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Krasnovskaya, O.; Naumov, A.; Guk, D.; Gorelkin, P.; Erofeev, A.; Beloglazkina, E.; Majouga, A. Copper Coordination Compounds as Biologically Active Agents. Int. J. Mol. Sci. 2020, 21, 3965. [Google Scholar] [CrossRef] [PubMed]
- McGivern, T.; Afsharpour, S.; Marmion, C. Copper Complexes as Artificial DNA Metallonucleases: From Sigman’s Reagent to Next Generation Anti-Cancer Agent? Inorg. Chim. Acta 2018, 472, 12–39. [Google Scholar] [CrossRef]
- Shi, X.; Chen, Z.; Wang, Y.; Guo, Z.; Wang, X. Hypotoxic copper complexes with potent anti-metastatic and anti-angiogenic activities against cancer cells. Dalton Trans. 2018, 47, 5049–5054. [Google Scholar] [CrossRef] [PubMed]
- Laws, K.; Suntharalingam, K. The next generation of anticancer metallopharmaceuticals: Cancer stem cell-active inorganics. ChemBioChem 2018, 19, 2246–2253. [Google Scholar] [CrossRef]
- Kaur, P.; Johnson, A.; Northcote-Smith, J.; Lu, C.; Suntharalingam, K. Immunogenic Cell Death of Breast Cancer Stem Cells Induced by an Endoplasmic Reticulum-Targeting Copper(II) Complex. ChemBioChem 2020, 21, 3618–3624. [Google Scholar] [CrossRef]
- Shi, X.; Fang, H.; Guo, Y.; Yuan, H.; Guo, Z.; Wang, X. Anticancer copper complex with nucleus, mitochondrion and cyclooxygenase-2 as multiple targets. J. Inorg. Biochem. 2019, 190, 38–44. [Google Scholar] [CrossRef]
- Chang, M.R.; Rusanov, D.A.; Arakelyan, J.; Alshehri, M.; Asaturova, A.V.; Kireeva, G.S.; Babak, M.V.; Ang, W.H. Targeting emerging cancer hallmarks by transition metal complexes: Cancer stem cells and tumor microbiome. Part I. Coord. Chem. Rev. 2023, 477, 214923. [Google Scholar] [CrossRef]
- Balsa, L.M.; Ruiz, M.C.; Santa Maria de la Parra, L.; Baran, E.J.; León, I.E. Anticancer and antimetastatic activity of copper(II)-tropolone complex against human breast cancer cells, breast multicellular spheroids and mammospheres. J. Inorg. Biochem. 2020, 204, 110975. [Google Scholar] [CrossRef] [PubMed]
- da Silva, D.A.; De Luca, A.; Squitti, R.; Rongioletti, M.; Rossi, L.; Machado, C.M.L.; Cerchiaro, G. Copper in tumors and the use of copper-based compounds in cancer treatment. J. Inorg. Biochem. 2022, 226, 111634. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.C.; Perelmulter, K.; Levín, P.; Romo, A.I.B.; Lemus, L.; Fogolín, M.B.; León, I.E.; Di Virgilio, A.L. Antiproliferative activity of two copper (II) complexes on colorectal cancer cell models: Impact on ROS production, apoptosis induction and NF-κB inhibition. Eur. J. Pharm. Sci. 2022, 169, 106092. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-DePaz, Y.; Resendiz-Acevedo, K.; Davila-Manzanilla, S.G.; Garcia-Ramos, J.C.; Ortiz-Frade, L.; Serment-Guerrero, J.; Ruiz-Azuara, L. DNA, a target of mixed chelate copper(II) compounds (Casiopeinas(R)) studied by electrophoresis, UV-vis and circular dichroism techniques. J. Inorg. Biochem. 2022, 231, 111772. [Google Scholar] [CrossRef]
- Peña, Q.; Sciortino, G.; Maréchal, J.-D.; Bertaina, S.; Simaan, A.J.; Lorenzo, J.; Capdevila, M.; Bayón, P.; Iranzo, O.; Palacios, Ò. Copper (II) N, N, O-Chelating Complexes as Potential Anticancer Agents. Inorg. Chem. 2021, 60, 2939–2952. [Google Scholar] [CrossRef]
- Fernández, C.Y.; Alvarez, N.; Rocha, A.; Ellena, J.; Costa-Filho, A.J.; Batista, A.A.; Facchin, G. New Copper(II)-L-Dipeptide-Bathophenanthroline Complexes as Potential Anticancer Agents—Synthesis, Characterization and Cytotoxicity Studies—And Comparative DNA-Binding Study of Related Phen Complexes. Molecules 2023, 28, 896. [Google Scholar]
- Alvarez, N.; Rocha, A.; Collazo, V.; Ellena, J.; Costa-Filho, A.J.; Batista, A.A.; Facchin, G. Development of Copper Complexes with Diimines and Dipicolinate as Anticancer Cytotoxic Agents. Pharmaceutics 2023, 15, 1345. [Google Scholar] [CrossRef]
- Alvarez, N.; Leite, C.M.; Napoleone, A.; Mendes, L.F.S.; Fernandez, C.Y.; Ribeiro, R.R.; Ellena, J.; Batista, A.A.; Costa-Filho, A.J.; Facchin, G. Tetramethyl-phenanthroline copper complexes in the development of drugs to treat cancer: Synthesis, characterization and cytotoxicity studies of a series of copper(II)-L-dipeptide-3,4,7,8-tetramethyl-phenanthroline complexes. J. Biol. Inorg. Chem. 2022, 27, 431–441. [Google Scholar] [CrossRef]
- Alvarez, N.; Viña, D.; Leite, C.M.; Mendes, L.F.; Batista, A.A.; Ellena, J.; Costa-Filho, A.J.; Facchin, G. Synthesis and structural characterization of a series of ternary copper (II)-L-dipeptide-neocuproine complexes. Study of their cytotoxicity against cancer cells including MDA-MB-231, triple negative breast cancer cells. J. Inorg. Biochem. 2020, 203, 110930. [Google Scholar] [CrossRef]
- Alvarez, N.; Mendes, L.F.; Kramer, M.G.; Torre, M.H.; Costa-Filho, A.J.; Ellena, J.; Facchin, G. Development of Copper (II)-diimine-iminodiacetate mixed ligand complexes as potential antitumor agents. Inorg. Chim. Acta 2018, 483, 61–70. [Google Scholar] [CrossRef]
- Alvarez, N.; Noble, C.; Torre, M.H.; Kremer, E.; Ellena, J.; Peres de Araujo, M.; Costa-Filho, A.J.; Mendes, L.F.; Kramer, M.G.; Facchin, G. Synthesis, structural characterization and cytotoxic activity against tumor cells of heteroleptic copper (I) complexes with aromatic diimines and phosphines. Inorg. Chim. Acta 2017, 466, 559–564. [Google Scholar] [CrossRef]
- Iglesias, S.; Alvarez, N.; Kramer, G.; Torre, M.H.; Kremer, E.; Ellena, J.; Costa-Filho, A.J.; Facchin, G. Structural Characterization and Cytotoxic Activity of Heteroleptic Copper (II) Complexes with L-Dipeptides and 5-NO2-Phenanthroline.: Crystal Structure of [Cu (Phe-Ala)(5-NO2-Phen)]·4H2O. Struct. Chem. Crystallogr. Commun. 2015, 1, 1–7. [Google Scholar]
- Iglesias, S.; Alvarez, N.; Torre, M.H.; Kremer, E.; Ellena, J.; Ribeiro, R.R.; Barroso, R.P.; Costa-Filho, A.J.; Kramer, M.G.; Facchin, G. Synthesis, structural characterization and cytotoxic activity of ternary copper (II)–dipeptide–phenanthroline complexes. A step towards the development of new copper compounds for the treatment of cancer. J. Inorg. Biochem. 2014, 139, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Costa-Filho, A.J.; Nascimento, O.R.; Calvo, R. Electron paramagnetic resonance study of weak exchange interactions between metal ions in a model system: CuIIGly-Trp. J. Phys. Chem. B 2004, 108, 9549–9555. [Google Scholar] [CrossRef]
- Balzano, T. Active Clinical Trials in Hepatic Encephalopathy: Something Old, Something New and Something Borrowed. Neurochem. Res. 2023, 48, 2309–2319. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.; Ali, S.; Tahir, M.N. Polymeric Copper(II) Paddlewheel Carboxylate: Structural Description, Electrochemistry, and DNA-binding Studies. Z. Für Anorg. Und Allg. Chem. 2018, 644, 172–179. [Google Scholar] [CrossRef]
- Benslimane, M.; Redjel, Y.K.; Merazig, H.; Daran, J.-C. catena-Poly[bis([mu]3-2-phenylacetato-[kappa]3O,O′:O)bis([mu]2-2-phenylacetato-[kappa]2O:O′)dicopper(II)(Cu-Cu)]. Acta Crystallogr. Sect. E 2013, 69, m397. [Google Scholar] [CrossRef]
- Rap, V.M.; Manohar, H. Synthesis and crystal structure of methanol and acetic acid adducts of copper acetate. Predominance of σ-interaction between the two copper atoms in the dimer. Inorganica Chim. Acta 1979, 34, L213–L214. [Google Scholar] [CrossRef]
- Kanazawa, Y.; Mitsudome, T.; Takaya, H.; Hirano, M. Pd/Cu-Catalyzed Dehydrogenative Coupling of Dimethyl Phthalate: Synchrotron Radiation Sheds Light on the Cu Cycle Mechanism. ACS Catal. 2020, 10, 5909–5919. [Google Scholar] [CrossRef]
- Kendin, M.; Nikiforov, A.; Svetogorov, R.; Degtyarenko, P.; Tsymbarenko, D. A 3D-Coordination Polymer Assembled from Copper Propionate Paddlewheels and Potassium Propionate 1D-Polymeric Rods Possessing a Temperature-Driven Single-Crystal-to-Single-Crystal Phase Transition. Cryst. Growth Des. 2021, 21, 6183–6194. [Google Scholar] [CrossRef]
- Jassal, A.K.; Sharma, S.; Hundal, G.; Hundal, M.S. Structural Diversity, Thermal Studies, and Luminescent Properties of Metal Complexes of Dinitrobenzoates: A Single Crystal to Single Crystal Transformation from Dimeric to Polymeric Complex of Copper(II). Cryst. Growth Des. 2015, 15, 79–93. [Google Scholar] [CrossRef]
- Udupa, M.R.; Krebs, B. Crystal and molecular structure of tetra-μ-N-acetylglycinatodiaquodicopper(II). Inorganica Chim. Acta 1979, 37, 1–4. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sect. B 2016, 72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Bruno, I.J.; Cole, J.C.; Edgington, P.R.; Kessler, M.; Macrae, C.F.; McCabe, P.; Pearson, J.; Taylor, R. New software for searching the Cambridge Structural Database and visualizing crystal structures. Acta Crystallogr. Sect. B 2002, 58, 389–397. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- Vaughan, G.B.M.; Schmidt, S.; Poulsen, H.F. Multicrystal approach to crystal structure solution and refinement. Z. Für Krist. Cryst. Mater. 2004, 219, 813–825. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Shi, Q.; Shi, Q.Z.; Gao, Y.C. Self-assembly of porous two-dimensional copper(II) α, β-unsaturated carboxylate complexes with trimethyl phosphate. Transit. Met. Chem. 2000, 25, 382–387. [Google Scholar] [CrossRef]
- Pathak, S.; Biswas, N.; Jana, B.; Ghorai, T.K. Synthesis and characterization of a nano Cu2 cluster. Adv. Mater. Proc. 2017, 2, 275–279. [Google Scholar]
- Li, J.-R.; Yu, J.; Lu, W.; Sun, L.-B.; Sculley, J.; Balbuena, P.B.; Zhou, H.-C. Porous materials with pre-designed single-molecule traps for CO2 selective adsorption. Nat. Commun. 2013, 4, 1538. [Google Scholar] [CrossRef]
- Kristiansson, O.; Tergenius, L.-E. Structure and host–guest properties of the nanoporous diaquatetrakis(p-nitrobenzoato)dicopper(II) framework. J. Chem. Soc. Dalton Trans. 2001, 9, 1415–1420. [Google Scholar] [CrossRef]
- Desiraju, G.R. A Bond by Any Other Name. Angew. Chem. Int. Ed. 2011, 50, 52–59. [Google Scholar] [CrossRef] [PubMed]
- de la Flor, G.; Orobengoa, D.; Tasci, E.; Perez-Mato, J.M.; Aroyo, M.I. Comparison of structures applying the tools available at the Bilbao Crystallographic Server. J. Appl. Crystallogr. 2016, 49, 653–664. [Google Scholar] [CrossRef]
- Doyle, A.; Felcman, J.; Gambardella, M.T.d.P.; Verani, C.N.; Tristão, M.L.B. Anhydrous copper(II) hexanoate from cuprous and cupric oxides. The crystal and molecular structure of Cu2(O2CC5H11)4. Polyhedron 2000, 19, 2621–2627. [Google Scholar] [CrossRef]
- Katzsch, F.; Münch, A.S.; Mertens, F.O.R.L.; Weber, E. Copper(II) benzoate dimers coordinated by different linear alcohols—A systematic study of crystal structures. J. Mol. Struct. 2014, 1064, 122–129. [Google Scholar] [CrossRef]
- Krause, L.; Herbst-Irmer, R.; Stalke, D. An empirical correction for the influence of low-energy contamination. J. Appl. Crystallogr. 2015, 48, 1907–1913. [Google Scholar] [CrossRef]
- Sheng, G.-H.; Zhou, Q.-C.; Sun, J.; Cheng, X.-S.; Qian, S.-S.; Zhang, C.-Y.; You, Z.-L.; Zhu, H.-L. Synthesis, structure, and urease inhibitory activities of three binuclear copper(II) complexes with protocatechuic acid derivative. J. Coord. Chem. 2014, 67, 1265–1278. [Google Scholar] [CrossRef]
- Massignani, S.; Scatena, R.; Lanza, A.; Monari, M.; Condello, F.; Nestola, F.; Pettinari, C.; Zorzi, F.; Pandolfo, L. Coordination polymers from mild condition reactions of copper(II) carboxylates with pyrazole (Hpz). Influence of carboxylate basicity on the self-assembly of the [Cu3(μ3-OH)(μ-pz)3]2+ secondary building unit. Inorg. Chim. Acta 2017, 455, 618–626. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds, Applications in Coordination, Organometallic, and Bioinorganic Chemistry, 6th ed.; Wiley-Interscience: Hoboken, NJ, USA, 2009. [Google Scholar]
- Prenesti, E.; Daniele, P.; Prencipe, M.; Ostacoli, G. Spectrum–structure correlation for visible absorption spectra of copper (II) complexes in aqueous solution. Polyhedron 1999, 18, 3233–3241. [Google Scholar] [CrossRef]
- Prenesti, E.; Daniele, P.G.; Berto, S.; Toso, S. Spectrum–structure correlation for visible absorption spectra of copper (II) complexes showing axial co-ordination in aqueous solution. Polyhedron 2006, 25, 2815–2823. [Google Scholar] [CrossRef]
- Karaliota, A.; Kretsi, O.; Tzougraki, C. Synthesis and characterization of a binuclear coumarin-3-carboxylate copper(II) complex. J. Inorg. Biochem. 2001, 84, 33–37. [Google Scholar] [CrossRef]
- Bhirud, R.G.; Srivastava, T.S. Synthesis, characterization and superoxide dismutase activity of some ternary copper (II) dipeptide-2, 2′-bipyridine, 1, 10-phenanthroline and 2, 9-dimethyl-1, 10-phenanthroline complexes. Inorg. Chim. Acta 1991, 179, 125–131. [Google Scholar] [CrossRef]
- Muhammad, N.; Ikram, M.; Perveen, F.; Ibrahim, M.; Ibrahim, M.; Abel; Viola; Rehman, S.; Shujah, S.; Khan, W.; et al. Syntheses, crystal structures and DNA binding potential of copper(II) carboxylates. J. Mol. Struct. 2019, 1196, 771–782. [Google Scholar] [CrossRef]
- Iqbal, M.; Ali, S.; Tahir, M.N.; Haleem, M.A.; Gulab, H.; Shah, N.A. A binary copper(II) complex having a stepped polymeric structure: Synthesis, characterization, DNA-binding and anti-fungal studies. J. Serb. Chem. Soc. 2020, 85, 203–214. [Google Scholar] [CrossRef]
- Suh, D.; Chaires, J.B. Criteria for the mode of binding of DNA binding agents. Bioorg. Med. Chem. 1995, 3, 723–728. [Google Scholar] [CrossRef]
- Sheldrick, G. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Benesi, H.A.; Hildebrand, J.H. A Spectrophotometric Investigation of the Interaction of Iodine with Aromatic Hydrocarbons. J. Am. Chem. Soc. 2002, 71, 2703–2707. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. DRUG-DNA Interactions and their study by UV-visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Scruggs, R.L.; Ross, P.D. Viscosity study of DNA. Biopolymers 1964, 2, 593–609. [Google Scholar] [CrossRef]



| Complex | C1 | C3 |
|---|---|---|
| Formula | C36H44Cu2O16 | C16H14CuO4 |
| Dcalc./g cm−3 | 1.594 | 1.618 |
| μ/mm−1 | 2.130 | 2.374 |
| Formula Weight | 859.832 | 333.81 |
| Color | Blue | Blue |
| Shape | Prism | Plate |
| Size/mm3 | 0.15 × 0.10 × 0.10 | 0.30 × 0.15 × 0.08 |
| Crystal System | Triclinic | monoclinic |
| Space Group | P | P21/c |
| a/Å | 8.6810(2) | 5.17356(6) |
| b/Å | 10.6746(3) | 26.2143(3) |
| c/Å | 11.3849(3) | 10.20173(12) |
| α/° | 66.930(3) | 90 |
| β/° | 70.661(2) | 97.8378(11) |
| γ/° | 71.814(2) | 90 |
| V/Å3 | 895.43(5) | 1370.64(3) |
| Z | 1 | 4 |
| Θmin/° | 4.347 | 3.372 |
| Θmax/° | 80.066 | 79.397 |
| Measured Refl. | 15,912 | 13,429 |
| Independent Refl. | 3875 | 2965 |
| Reflections with I > 2σ(I) | 3820 | 2743 |
| Rint | 0.0193 | 0.0451 |
| Parameters | 251 | 191 |
| Restraints | 0 | 0 |
| Largest Peak | 0.622 | 0.476 |
| Deepest Hole | −0.737 | −0.597 |
| GooF | 1.040 | 1.027 |
| wR2 (all data) | 0.0708 | 0.0855 |
| wR2 | 0.0706 | 0.0835 |
| R1 (all data) | 0.0278 | 0.0349 |
| R1 | 0.0275 | 0.0325 |
| CCDC deposition number | 2,288,430 | 2,288,436 |
| Bond Lengths (Å) | Angles (°) | ||
|---|---|---|---|
| Cu1-Cu2 | 2.6075(4) | O1-Cu1-O4 | 90.98(5) |
| Cu1-O4 | 1.9649(11) | O5′-Cu1-O4 | 169.16(4) |
| Cu1-O1 | 1.9604(10) | O5′-Cu1-O1 | 88.48(4) |
| Cu1-O5′ | 1.9751(10) | O2′-Cu1-O4 | 91.39(5) |
| Cu1-O2′ | 1.9628(11) | O2′-Cu1-O1 | 168.36(5) |
| O5′-Cu1-O2′ | 87.05(5) | ||
| Compound | ν(O-H) | ν(C=O) + ν(COO)as | ν(COO)s | ν(Cu-O) |
|---|---|---|---|---|
| [Cu2(3-(4-hydroxyphenyl)propanoate)4(H2O)2]·2H2O | 3330 sh | 1582 s, 1516 w | 1425 m | 532 w |
| [Cu2(phenylpropanoate)4(H2O)2] | 3500–3200 sh | 1588 s, 1516 w | 1431 m | 480 w |
| [Cu2(phenylacetate)4] | 3500–3200 sh | 1594 s, 1514 s | 1438 m | 532 w |
| Compound | λmax/ε * | n * | λmax/ε ** | n ** | P |
|---|---|---|---|---|---|
| [Cu2(3-(4-hydroxyphenyl)propanoate)4(H2O)2]·2H2O | 710/388 | 43 | 726/134 | 65 | 0.10 |
| [Cu2(phenylpropanoate)4(H2O)2] | 715/313 | 44 | 713/288 | 73 | 0.24 |
| [Cu2(phenylacetate)4] | 711/404 | 48 | 736/150 | 72 | 0.47 |
| Compound | C1 | C2 | C3 | L1 | L2 | L3 |
|---|---|---|---|---|---|---|
| Kb (M-1) | 5.2 × 102 | 2.0 × 102 | 8.7 × 102 | ND * | ND * | ND * |
| Cytotoxicity, IC50 (µM) | |||||
|---|---|---|---|---|---|
| Compound | MCF-7 | MDA-MB-231 | A549 | A278cis | MRC-5 |
| C1 | >50 | >50 | >50 | 26.80 ± 4.50 | >50 |
| C2 | 20.20 ± 0.78 | >50 | >50 | 13.50 ± 0.57 | >50 |
| C3 | >50 | >50 | >50 | 7.85 ± 0.86 | >50 |
| Cisplatin | 8.91 ± 2.60 | 24.90 ± 3.40 | 14.40 ± 1.40 | 26.90 ± 0.60 | 29.09 ± 0.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández, C.Y.; Rocha, A.; Azam, M.; Alvarez, N.; Min, K.; Batista, A.A.; Costa-Filho, A.J.; Ellena, J.; Facchin, G. Synthesis, Characterization, DNA Binding and Cytotoxicity of Copper(II) Phenylcarboxylate Complexes. Inorganics 2023, 11, 398. https://doi.org/10.3390/inorganics11100398
Fernández CY, Rocha A, Azam M, Alvarez N, Min K, Batista AA, Costa-Filho AJ, Ellena J, Facchin G. Synthesis, Characterization, DNA Binding and Cytotoxicity of Copper(II) Phenylcarboxylate Complexes. Inorganics. 2023; 11(10):398. https://doi.org/10.3390/inorganics11100398
Chicago/Turabian StyleFernández, Carlos Y., Analu Rocha, Mohammad Azam, Natalia Alvarez, Kim Min, Alzir A. Batista, Antonio J. Costa-Filho, Javier Ellena, and Gianella Facchin. 2023. "Synthesis, Characterization, DNA Binding and Cytotoxicity of Copper(II) Phenylcarboxylate Complexes" Inorganics 11, no. 10: 398. https://doi.org/10.3390/inorganics11100398