Investigating Iron-Sulfur Proteins in Infectious Diseases: A Review of Characterization Techniques
Abstract
1. Introduction
2. Types of [Fe-S] Clusters
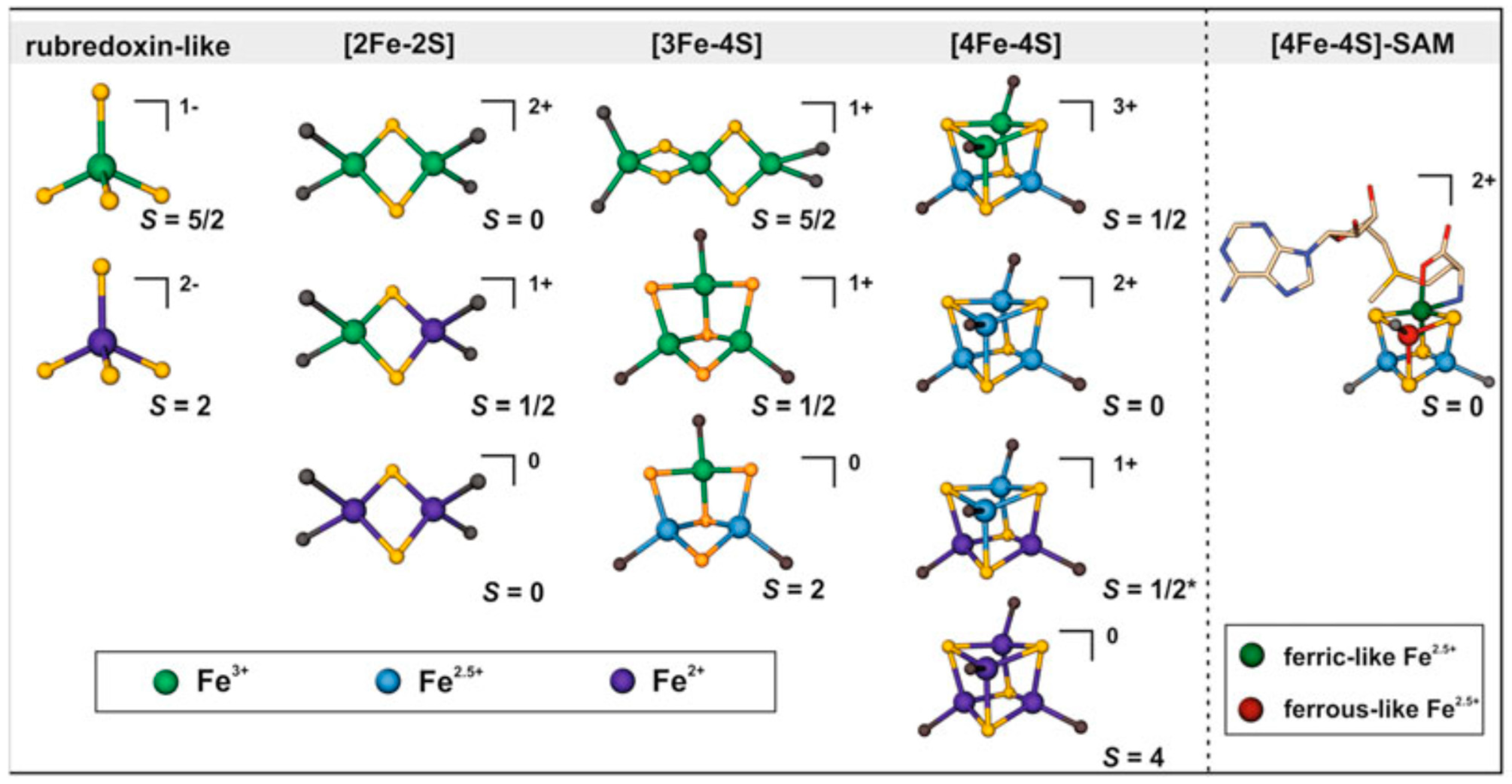
- [2Fe-2S] cluster: Two iron and two sulfur atoms combine to form one of the most basic [Fe-S] cluster formations. Usually, the sulfur atoms form a cluster with a diamond-like structure by tetrahedral coordinating with the iron atoms. Proteins involved in electron transport, such as ferredoxins, frequently have this cluster type (Figure 1) [25].
- [3Fe-4S] cluster: Three iron atoms coordinate four sulfur atoms. The artificial models of [3Fe-4S] clusters have a linear structure of three iron atoms, whereas protein-imposed structural constraints promote the development of a cuboidal shape for the [3Fe-4S]1+ cluster. They are vital enzymes required for metabolic processes, including nitrogen fixation and the citric acid cycle. (Figure 1) [26].
- [4Fe-4S] cluster: Proteins involved in redox reactions and electron transport pathways frequently include it. This more significant cluster has four sulfur and iron atoms. Sulfur atoms act as the coordinators of the iron atoms in a mixture of cubane and tetrahedral geometries (Figure 1). Aconitase and succinate dehydrogenase are enzymes and proteins that frequently include [4Fe-4S] clusters in redox processes [27]. Apo-aconitase is essential for cellular iron sensing, especially when the cell has an iron shortage [28]. When there is a shortage of iron in the cells, apo-aconitase acts as an iron sensor without the iron-sulfur cluster. This protein changes structurally to sense when iron is becoming less available. Because of this alteration, the protein responds to the iron status of the cell by adjusting its activity. Apo-aconitase is critical in preserving iron homeostasis within the cellular environment by employing its [4Fe-4S] cluster to enhance the cell’s sensitivity to changes in iron levels. In contrast, the radical S-adenosyl methionine (SAM) enzyme catalytic [4Fe-4S]1+ cluster coordinated three cysteines with either a solvent ligand or an unidentified small molecule, forming a gated cluster [29].
3. Different Characterization Techniques of [Fe-S] Clusters
3.1. Ultraviolet-Visible and Infrared Methods to Determine the [Fe-S] Cluster’s Structure and Function
3.2. Structural Characterizations
3.3. Biophysical Characterizations
3.4. Electrochemical Techniques
4. [Fe-S] Cluster Functions in Viral Polymerases
4.1. Viperin—A [Fe-S] Cluster Containing Nucleotide Dehydratase
4.2. Nsp5—A [Fe-S] Cluster Containing Protein That Modulates RNA Binding in Rotavirus
4.3. Tumor Antigen from Merkel Cell Polyomavirus (MCPyV) Consists of [Fe-S] Cluster
4.4. HBx—A [Fe-S] Cluster Containing Viral Replication Protein from Hepatitis B Virus (HBV)
4.5. GciS—A Glycine and Cysteine Rich Protein from Mimivirus That Contains [Fe-S] Cluster
4.6. Nsp12 and nsp13—A [Fe-S] Cluster Containing Protein That Regulates RNA Polymerase Activity in Coronaviruses
5. [Fe-S] Clusters Functions in Pathogenic Bacteria
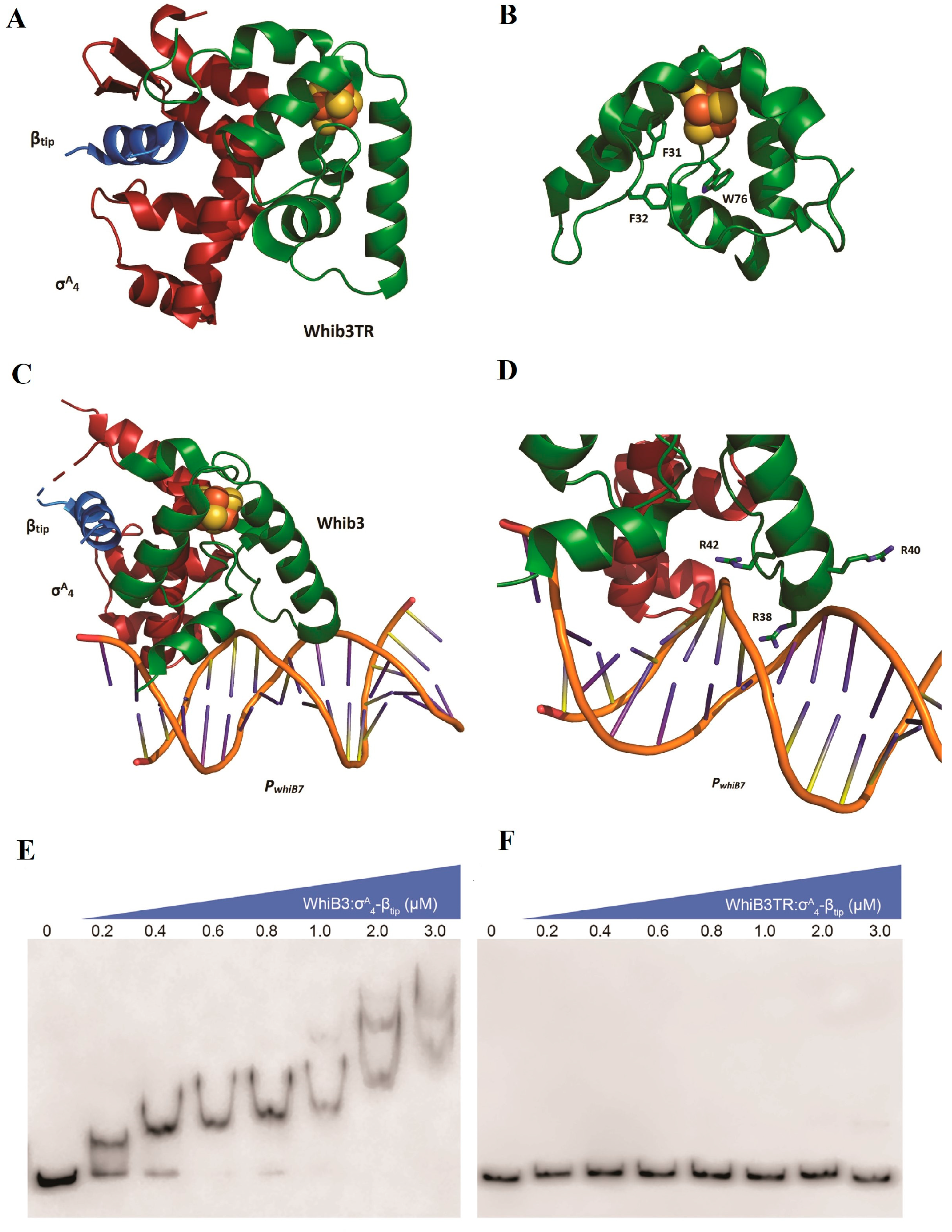
5.1. WhiB3—A Fe-S Cluster Containing Transcriptional Regulator from M Tuberculosis
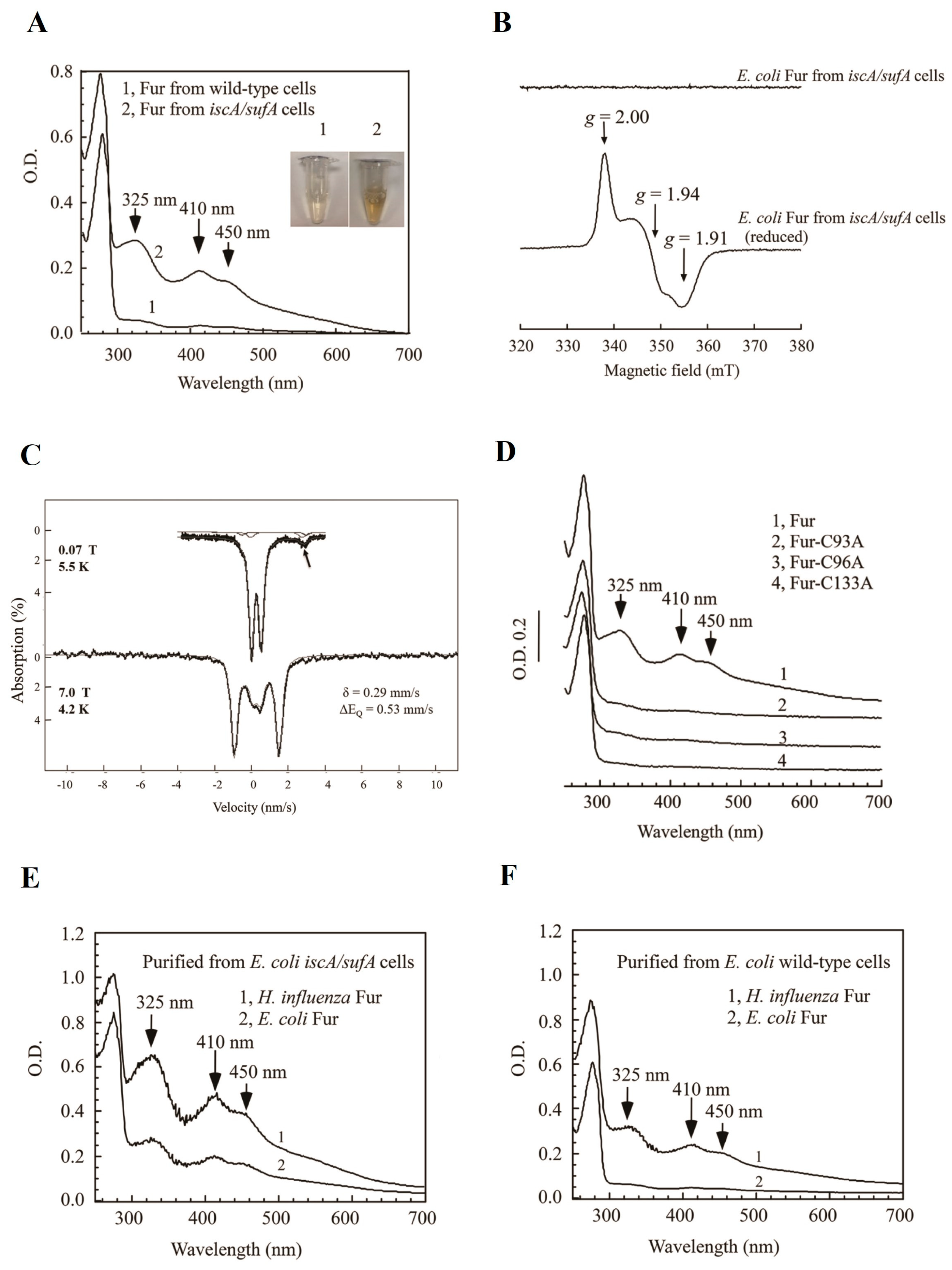
5.2. Ferric Uptake Regulators (Fur): A Transcription Factor That Regulates Intracellular Iron Homeostasis
5.3. Ferrous Iron Transport Protein C (FeoC): A Fe-S Cluster Containing Protein That Regulates the Ferrous Iron Channel
6. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Garcia, P.S.; D’Angelo, F.; Ollagnier de Choudens, S.; Dussouchaud, M.; Bouveret, E.; Gribaldo, S.; Barras, F. An Early Origin of Iron-Sulfur Cluster Biosynthesis Machineries before Earth Oxygenation. Nat. Ecol. Evol. 2022, 6, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef]
- Beinert, H.; Holm, R.H.; Münck, E. Iron-Sulfur Clusters: Nature’s Modular, Multipurpose Structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Schulz, V.; Basu, S.; Freibert, S.A.; Webert, H.; Boss, L.; Mühlenhoff, U.; Pierrel, F.; Essen, L.O.; Warui, D.M.; Booker, S.J.; et al. Functional spectrum and specificity of mitochondrial ferredoxins FDX1 and FDX2. Nat. Chem. Biol. 2023, 19, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Khodour, Y.; Kaguni, L.S.; Stiban, J. Iron-sulfur clusters in nucleic acid metabolism: Varying roles of ancient cofactors. Enzymes 2019, 45, 225–256. [Google Scholar] [PubMed]
- Rouault, T.A.; Tong, W.H. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008, 24, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Raza, M.K.; Karges, J. Iron-Sulfur Clusters in Viral Diseases. In Encyclopedia of Inorganic and Bioinorganic Chemistry; Scott, R.A., Ed.; Wiley: Hoboken, NJ, USA, 2024. [Google Scholar] [CrossRef]
- Honarmand, E.K.; Ciofi-Baffoni, S.; Hagedoorn, P.L.; Nicolet, Y.; Le Brun, N.E.; Hagen, W.R.; Armstrong, F.A. Iron-sulfur clusters as inhibitors and catalysts of viral replication. Nat. Chem. 2022, 14, 253–266. [Google Scholar] [CrossRef]
- Elchennawi, I.; Ollagnier de Choudens, S. Iron-Sulfur Clusters toward Stresses: Implication for Understanding and Fighting Tuberculosis. Inorganics 2022, 10, 174. [Google Scholar] [CrossRef]
- Fuss, J.O.; Tsai, C.-L.; Ishida, J.P.; Tainer, J.A. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochim. Biophys. Acta 2015, 1853, 1253–1271. [Google Scholar] [CrossRef]
- Barton, J.K.; Silva, R.M.B.; O’Brien, E. Redox Chemistry in the Genome: Emergence of the [4Fe4S] Cofactor in Repair and Replication. Annu. Rev. Biochem. 2019, 88, 163–190. [Google Scholar] [CrossRef]
- Boal, A.K.; Genereux, J.C.; Sontz, P.A.; Gralnick, J.A.; Newman, D.K.; Barton, J.K. Redox signaling between DNA repair proteins for efficient lesion detection. Proc. Natl. Acad. Sci. USA 2009, 106, 15237–15342. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.N.; Ter Beek, J.; Ekanger, L.A.; Johansson, E.; Barton, J.K. The [4Fe4S] cluster of yeast DNA polymerase ε is redox active and can undergo DNA-mediated signaling. J. Am. Chem. Soc. 2021, 143, 16147–16153. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.; Holt, M.E.; Thompson, M.K.; Salay, L.E.; Ehlinger, A.C.; Chazin, W.J.; Barton, J.K. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 2017, 355, eaag1789. [Google Scholar] [CrossRef] [PubMed]
- Salay, L.E.; Blee, A.M.; Raza, M.K.; Gallagher, K.S.; Chen, H.; Dorfeuille, A.J.; Barton, J.K.; Chazin, W.J. Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase. Biochemistry 2022, 61, 1113–1123. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar] [CrossRef]
- Pritts, J.D.; Michel, S.L.J. Fe-S clusters masquerading as zinc finger proteins. J. Inorg. Biochem. 2022, 230, 111756. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Calderone, L.A.; Pan, L.; Quist, T.; Pandelia, M.-E. The Fe and Zn cofactor dilemma. Biochim. Biophys. Acta-Proteins Proteom. 2023, 1871, 140931. [Google Scholar]
- Wiley, S.E.; Paddock, M.L.; Abresch, E.C.; Gross, L.; van der Geer, P.; Nechushtai, R.; Murphy, A.N.; Jennings, P.A.; Dixon, J.E. The outer mitochondrial membrane protein mitoNEET contains a novel redox-active 2Fe-2S cluster. J. Biol. Chem. 2007, 282, 23745–23749. [Google Scholar] [CrossRef]
- Ye, H.; Rouault, T.A. Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry 2010, 49, 4945–4956. [Google Scholar] [CrossRef]
- Ueda, C.; Langton, M.; Chen, J.; Pandelia, M.E. The HBx protein from hepatitis B virus coordinates a redox-active Fe-S cluster. J. Biol. Chem. 2022, 298, 101698. [Google Scholar] [CrossRef]
- Maio, N.; Lafont, B.A.P.; Sil, D.; Li, Y.; Bollinger, J.M., Jr.; Krebs, C.; Pierson, T.C.; Linehan, W.M.; Rouault, T.A. Fe-S cofactors in the SARS-CoV-2 RNA-dependent RNA polymerase are potential antiviral targets. Science 2021, 373, 236–241. [Google Scholar] [CrossRef]
- Maio, N.; Raza, M.K.; Li, Y.; Zhang, D.L.; Bollinger, J.M., Jr.; Krebs, C.; Rouault, T.A. An iron-sulfur cluster in the zinc-binding domain of the SARS-CoV-2 helicase modulates its RNA-binding and -unwinding activities. Proc. Natl. Acad. Sci. USA 2023, 120, e2303860120. [Google Scholar] [CrossRef]
- Pain, D.; Dancis, A. Roles of Fe-S proteins: From cofactor synthesis to iron homeostasis to protein synthesis. Curr. Opin. Genet. Dev. 2016, 38, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Schulz, V.; Freibert, S.A.; Boss, L.; Mühlenhoff, U.; Stehling, O.; Lill, R. Mitochondrial [2Fe-2S] ferredoxins: New functions for old dogs. FEBS Lett. 2023, 597, 102–121. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.M.; Heffron, K.; Kotlyar, V.; Sher, Y.; Maklashina, E.; Cecchini, G.; Armstrong, F.A. Electron transfer and catalytic control by the iron-sulfur clusters in a respiratory enzyme, E. coli fumarate reductase. J. Am. Chem. Soc. 2005, 127, 6977–6989. [Google Scholar] [CrossRef] [PubMed]
- Castro, L.; Tórtora, V.; Mansilla, S.; Radi, R. Aconitases: Non-Redox Iron-Sulfur Proteins Sensitive to Reactive Species. Acc. Chem. Res. 2019, 52, 2609–2619. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, O.V.; Piroddi, M.; Galli, F.; Lushchak, V.I. Aconitase post-translational modification as a key in linkage between Krebs cycle, iron homeostasis, redox signaling, and metabolism of reactive oxygen species. Redox Rep. 2014, 19, 8–15. [Google Scholar] [CrossRef]
- Broderick, J.B.; Duffus, B.R.; Duschene, K.S.; Shepard, E.M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014, 114, 4229–4317. [Google Scholar] [CrossRef]
- Pandelia, M.E.; Lanz, N.D.; Booker, S.J.; Krebs, C. Mössbauer spectroscopy of Fe/S proteins. Biochim Biophys Acta. 2015, 1853, 1395–1405. [Google Scholar] [CrossRef]
- Ueda, C.; Langton, M.; Pandelia, M.E. Characterization of Fe-S Clusters in Proteins by Mössbauer Spectroscopy. Methods Mol. Biol. 2021, 2353, 281–305. [Google Scholar]
- Valer, L.; Rossetto, D.; Scintilla, S.; Hu, Y.J.; Tomar, A.; Nader, S.; Betinol, I.O.; Mansy, S.S. Methods to identify and characterize iron-sulfur oligopeptides in water. Can. J. Chem. 2022, 100, 475–483. [Google Scholar] [CrossRef]
- Camponeschi, F.; Piccioli, M.; Banci, L. The Intriguing mitoNEET: Functional and Spectroscopic Properties of a Unique [2Fe-2S] Cluster Coordination Geometry. Molecules 2022, 27, 8218. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.C.; Kent, T.; Emptage, M.; Merkle, H.; Beinert, H.; Münck, E. Evidence for the formation of a linear [3Fe-4S] cluster in partially unfolded aconitase. J. Biol. Chem. 1984, 259, 14463–14471. [Google Scholar] [CrossRef] [PubMed]
- Crack, J.C.; Le Brun, N.E. Native Mass Spectrometry of Iron-Sulfur Proteins. Methods Mol. Biol. 2021, 2353, 231–258. [Google Scholar] [PubMed]
- Cai, K.; Markley, J.L. NMR as a Tool to Investigate the Processes of Mitochondrial and Cytosolic Iron-Sulfur Cluster Biosynthesis. Molecules 2018, 23, 2213. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Camponeschi, F.; Ciofi-Baffoni, S.; Piccioli, M. The NMR contribution to protein-protein networking in Fe-S protein maturation. J. Biol. Inorg. Chem. 2018, 23, 665–685. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.L.; Tainer, J.A. Robust Production, Crystallization, Structure Determination, and Analysis of [Fe-S] Proteins: Uncovering Control of Electron Shuttling and Gating in the Respiratory Metabolism of Molybdopterin Guanine Dinucleotide Enzymes. Methods Enzymol. 2018, 599, 157–196. [Google Scholar] [PubMed]
- Booker, S.J.; Lloyd, C.T. TwentyYears of Radical SAM! The Genesis of the Superfamily. ACS Bio Med Chem Au 2022, 2, 538–547. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, M.I.; Lanz, N.D.; Goldman, P.J.; Lee, K.-H.; Booker, S.J.; Drennan, C.L. Crystallographic Snapshots of Sulfur Insertion by Lipoyl Synthase. Proc. Natl. Acad. Sci. USA 2016, 113, 9446–9450. [Google Scholar] [CrossRef]
- Jeyachandran, V.R.; Pendyala, J.V.; McCarthy, E.L.; Boal, A.K.; Booker, S.J. Biochemical Approaches to Probe the Role of the Auxiliary Iron-Sulfur Cluster of Lipoyl Synthase from Mycobacterium Tuberculosis. Methods Mol. Biol. 2021, 2353, 307–332. [Google Scholar]
- Stich, T.A. Characterization of Paramagnetic Iron-Sulfur Clusters Using Electron Paramagnetic Resonance Spectroscopy. Methods Mol. Biol. 2021, 2353, 259–280. [Google Scholar]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yu, Y.; Tian, S.; Petrik, I.; Bhagi, A.; Lu, Y. Metalloproteins Containing Cytochrome, Iron–Sulfur, or Copper Redox Centers. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [CrossRef] [PubMed]
- Villalta, A.; Srour, B.; Lartigue, A.; Clémancey, M.; Byrne, D.; Chaspoul, F.; Loquet, A.; Guigliarelli, B.; Blondin, G.; Abergel, C.; et al. Evidence for [2Fe-2S]2+ and Linear [3Fe-4S]1+ Clusters in a Unique Family of Glycine/Cysteine-Rich Fe-S Proteins from Megavirinae Giant Viruses. J. Am. Chem. Soc. 2023, 145, 2733–2738. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.T.; Linkous, R.O.; Max, N.J.; Sestok, A.E.; Szalai, V.A.; Chacón, K.N. The FeoC [4Fe-4S] cluster is redox-active and rapidly oxygen-sensitive. Biochemistry 2019, 58, 4935–4949. [Google Scholar] [CrossRef]
- Lillig, C.H.; Berndt, C.; Vergnolle, O.; Lönn, M.E.; Hudemann, C.; Bill, E.; Holmgren, A. Characterization of human glutaredoxin 2 as iron-sulfur protein: A possible role as redox sensor. Proc. Natl. Acad. Sci. USA 2005, 102, 8168–8173. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.C.; Suess, D.L.M. An Open-Cuboidal [Fe3S4] Cluster Characterized in Both Biologically Relevant Redox States. J. Am. Chem. Soc. 2023, 145, 2075–2080. [Google Scholar] [CrossRef] [PubMed]
- Elgrishi, N.; Rountree, K.J.; McCarthy, B.D.; Rountree, E.S.; Eisenhart, T.T.; Dempsey, J.L. A Practical Beginner’s Guide to Cyclic Voltammetry. J. Chem. Educ. 2018, 95, 197–206. [Google Scholar] [CrossRef]
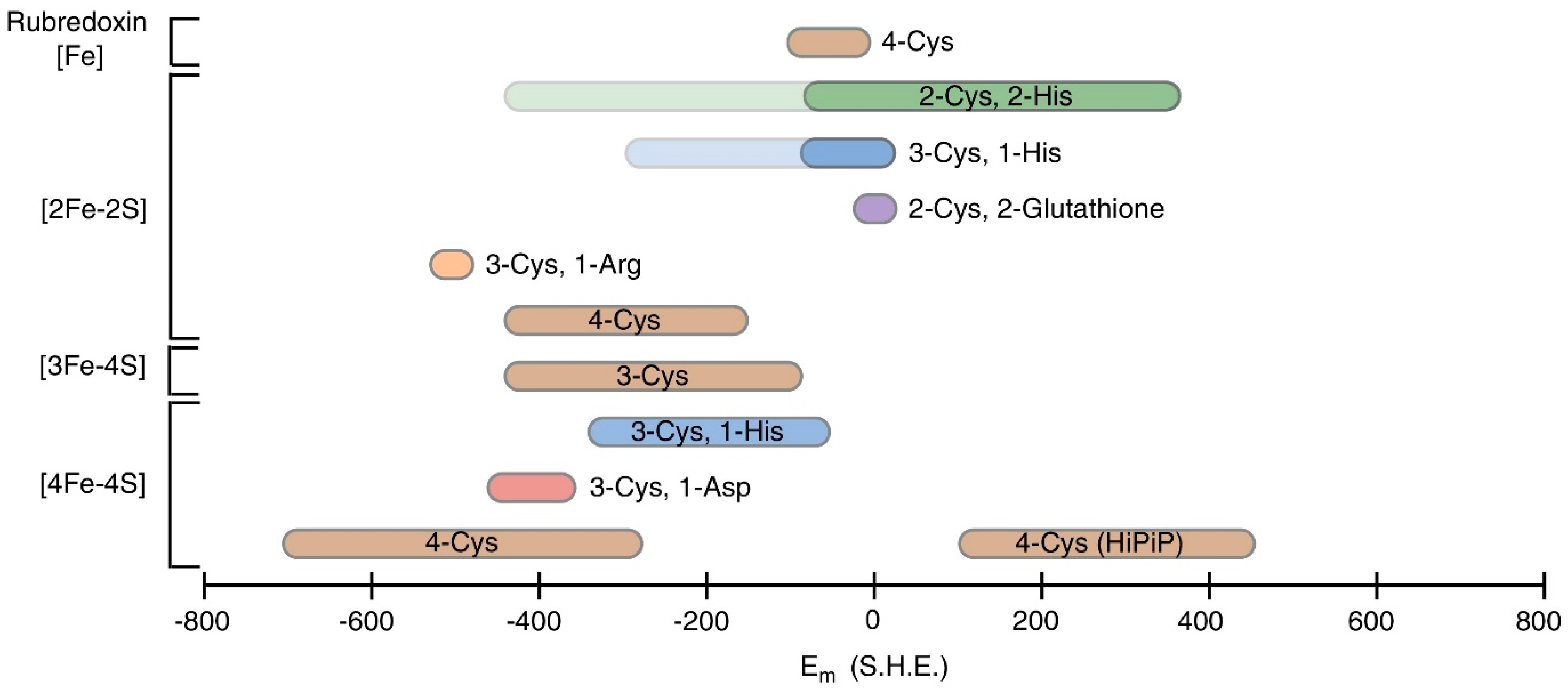
- Bak, D.W.; Elliott, S.J. Alternative FeS cluster ligands: Tuning redox potentials and chemistry. Curr. Opin. Chem. Biol. 2014, 19, 50–58. [Google Scholar] [CrossRef]
- Zanello, P. The competition between chemistry and biology in assembling iron-sulfur derivatives. Molecular structures and electrochemistry. Part II. {[Fe2S2](SγCys)4} proteins. Coord. Chem. Rev. 2014, 280, 54–83. [Google Scholar] [CrossRef]
- Nano, A.; Furst, A.L.; Hill, M.G.; Barton, J.K. DNA electrochemistry: Charge-transport pathways through DNA films on gold. J. Am. Chem. Soc. 2021, 143, 11631–11640. [Google Scholar] [CrossRef]
- Jones, J.E.; Le Sage, V.; Lakdawala, S.S. Viral and host heterogeneity and their effects on the viral life cycle. Nat. Rev. Microbiol. 2021, 19, 272–282. [Google Scholar] [CrossRef]
- Dixit, H.; Kulharia, M.; Verma, S.K. Metalloproteome of human-infective RNA viruses: A study towards understanding the role of metal ions in virology. Pathog. Dis. 2023, 81, ftad020. [Google Scholar] [CrossRef]
- Chen, A.Y.; Adamek, R.N.; Dick, B.L.; Credille, C.V.; Morrison, C.N.; Cohen, S.M. Targeting Metalloenzymes for Therapeutic Intervention. Chem. Rev. 2019, 119, 1323–1455. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Cong, J.P.; Shenk, T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: Induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 1997, 94, 13985–13990. [Google Scholar] [CrossRef] [PubMed]
- Gizzi, A.S.; Grove, T.L.; Arnold, J.J.; Jose, J.; Jangra, R.K.; Garforth, S.J.; Du, Q.; Cahill, S.M.; Dulyaninova, N.G.; Love, J.D.; et al. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 2018, 558, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Shaveta, G.; Shi, J.; Chow, V.T.K.; Song, J. Structural characterization reveals that viperin is a radical S-adenosyl-l-methionine (SAM) enzyme. J. Biochem. Biophys. Res. Commun. 2010, 391, 1390–1395. [Google Scholar] [CrossRef] [PubMed]
- Duschene, K.S.; Broderick, J.B. The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 2010, 584, 1263–1267. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Crawford, S.E.; Hyser, J.M.; Estes, M.K.; Prasad, B.V. Rotavirus non-structural proteins: Structure and function. Curr. Opin. Virol. 2012, 2, 380–388. [Google Scholar] [CrossRef]
- Martin, D.; Charpilienne, A.; Parent, A.; Boussac, A.; D’Autreaux, B.; Poupon, J.; Poncet, D. The rotavirus nonstructural protein NSP5 coordinates a [2Fe-2S] iron-sulfur cluster that modulates interaction to RNA. FASEB J. 2013, 27, 1074–1083. [Google Scholar] [CrossRef]
- Boothpur, R.; Brennan, D.C. Human polyoma viruses and disease with emphasis on clinical BK and JC. J. Clin. Virol. 2010, 47, 306–312. [Google Scholar] [CrossRef]
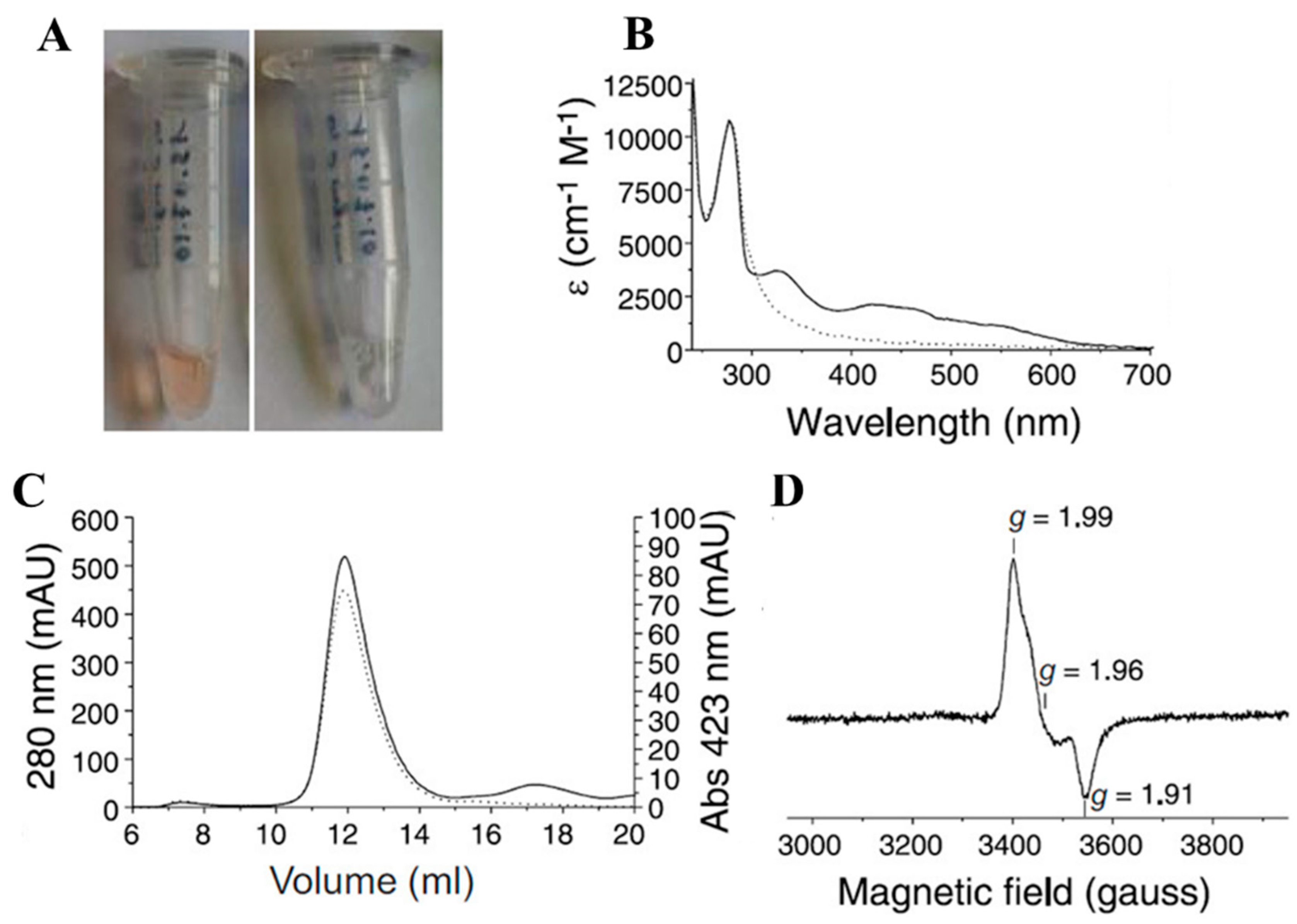
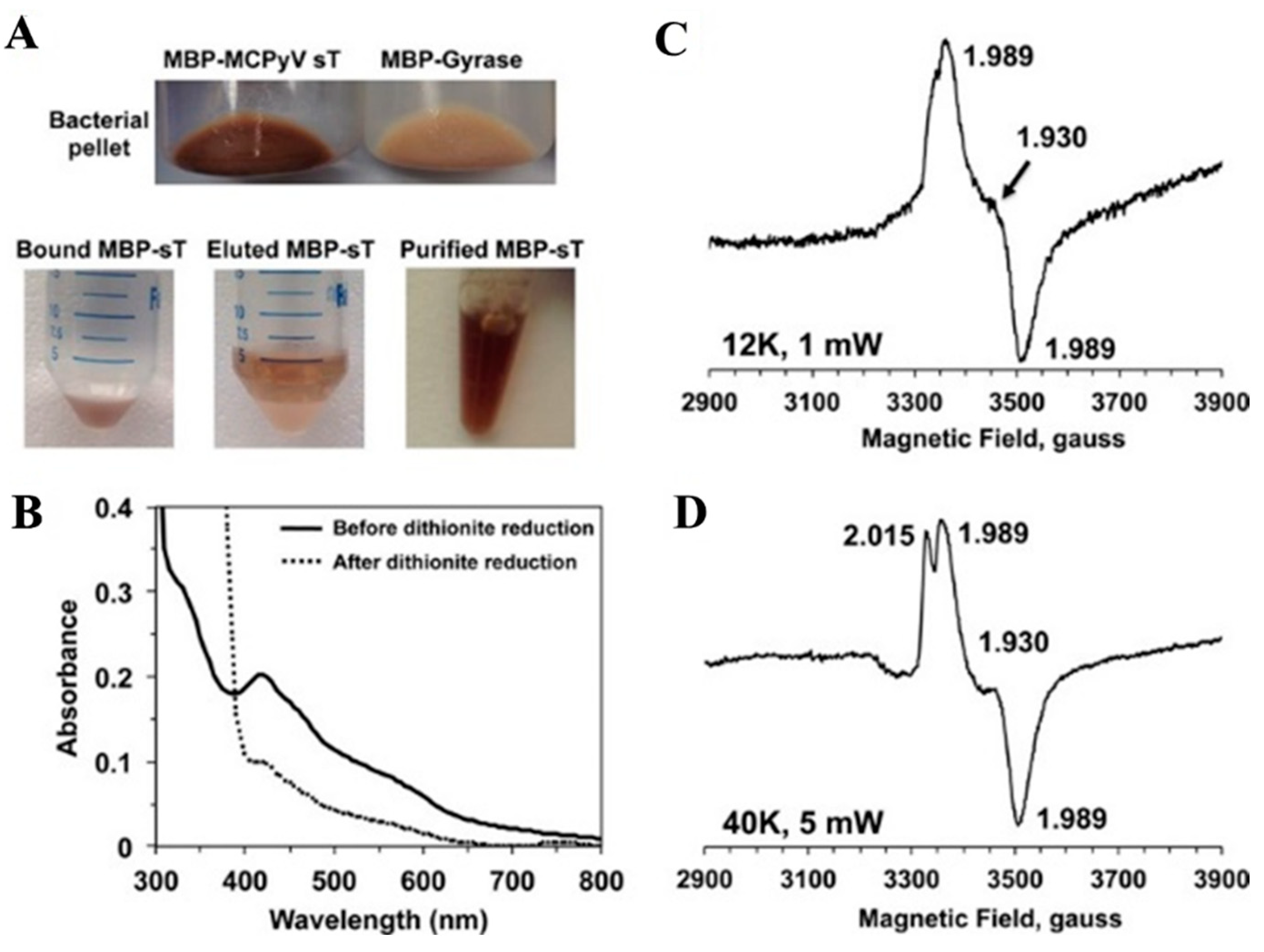
- Tsang, S.H.; Wang, R.; Nakamaru-Ogiso, E.; Knight, S.A.; Buck, C.B.; You, J. The Oncogenic Small Tumor Antigen of Merkel Cell Polyomavirus Is an Iron-Sulfur Cluster Protein That Enhances Viral DNA Replication. J. Virol. 2015, 90, 1544–1556. [Google Scholar] [CrossRef]
- Tan, M.; Bhadoria, A.S.; Cui, F.; Tan, A.; Van Holten, J.; Easterbrook, P.; Ford, N.; Han, Q.; Lu, Y.; Bulterys, M.; et al. Estimating the proportion of people with chronic hepatitis B virus infection eligible for hepatitis B antiviral treatment worldwide: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2021, 6, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Liu, M.; Wu, J.; Shi, Y. Structural and biochemical analysis of Bcl-2 interaction with the hepatitis B virus protein HBx. Proc. Natl. Acad. Sci. USA 2016, 113, 2074–2079. [Google Scholar] [CrossRef]
- Alvarez, S.W.; Sviderskiy, V.O.; Terzi, E.M.; Papagiannakopoulos, T.; Moreira, A.L.; Adams, S.; Sabatini, D.M.; Birsoy, K.; Possemato, R. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature 2017, 551, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Petronek, M.S.; Spitz, D.R.; Allen, B.G. Iron-Sulfur Cluster Biogenesis as a Critical Target in Cancer. Antioxidants 2021, 10, 1458. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Audic, S.; Robert, C.; Jungang, L.; de Lamballerie, X.; Drancourt, M.; Birtles, R.; Claverie, J.M.; Raoult, D. A giant virus in amoebae. Science 2003, 299, 2033. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.; Lai, S.T.; Poon, L.L.; Guan, Y.; Yam, L.Y.; Lim, W.; Nicholls, J.; Yee, W.K.; Yan, W.W.; Cheung, M.T.; et al. SARS study group. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Guan, Y.J.; et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef]
- Assiri, A.; McGeer, A.; Perl, T.M.; Price, C.S.; Al Rabeeah, A.A.; Cummings, D.A.; Alabdullatif, Z.N.; Assad, M.; Almulhim, A.; Makhdoom, H.; et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013, 369, 407–416. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Vernis, L.; El Banna, N.; Baïlle, D.; Hatem, E.; Heneman, A.; Huang, M.E. Fe-S Clusters Emerging as Targets of Therapeutic Drugs. Oxid. Med. Cell. Longev. 2017, 2017, 3647657. [Google Scholar] [CrossRef]
- Davis, N.K.; Chater, K.F. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol. Gen. Genet. 1992, 232, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.P.; Nguyen, L.; Gatfield, J.; Visconti, K.; Nguyen, K.; Schnappinger, D.; Ehrt, S.; Liu, Y.; Heifets, L.; Pieters, J.; et al. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2005, 102, 12200–12205. [Google Scholar] [CrossRef] [PubMed]
- Wan, T.; Horová, M.; Khetrapal, V.; Li, S.; Jones, C.; Schacht, A.; Sun, X.; Zhang, L. Structural basis of DNA binding by the WhiB-like transcription factor WhiB3 in Mycobacterium tuberculosis. J. Biol. Chem. 2023, 299, 104777. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Kim, D.; Latif, H.; O’Brien, E.J.; Szubin, R.; Palsson, B.O. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 2014, 5, 4910. [Google Scholar] [CrossRef] [PubMed]
- Fillat, M.F. The FUR (ferric uptake regulator) superfamily: Diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 2014, 546, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, C.R.; Tasnim, H.; Valdes, K.A.; Popescu, C.V.; Ding, H. Ferric uptake regulator (Fur) reversibly binds a [2Fe-2S] cluster to sense intracellular iron homeostasis in Escherichia coli. J. Biol. Chem. 2020, 295, 15454–15463. [Google Scholar] [CrossRef] [PubMed]
- Cartron, M.L.; Maddocks, S.; Gillingham, P.; Craven, C.J.; Andrews, S.C. Feo—Transport of ferrous iron into bacteria. Biometals 2006, 19, 143–157. [Google Scholar] [CrossRef]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003, 27, 215–337. [Google Scholar] [CrossRef]
- Cornelis, P.; Dingemans, J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 2013, 3, 75. [Google Scholar] [CrossRef]
- Hsueh, K.L.; Yu, L.K.; Chen, Y.H.; Cheng, Y.H.; Hsieh, Y.C.; Ke, S.C.; Hung, K.W.; Chen, C.J.; Huang, T.H. FeoC from Klebsiella pneumoniae contains a [4Fe-4S] cluster. J. Bacteriol. 2013, 195, 4726–4734. [Google Scholar] [CrossRef]
- Hsueh, K.L.; Yu, L.K.; Hsieh, Y.C.; Hsiao, Y.Y.; Chen, C.J. FeoC from Klebsiella pneumoniae uses its iron sulfur cluster to regulate the GTPase activity of the ferrous iron channel. Biochim. Biophys. Acta Proteins Proteom. 2023, 1871, 140855. [Google Scholar] [CrossRef] [PubMed]




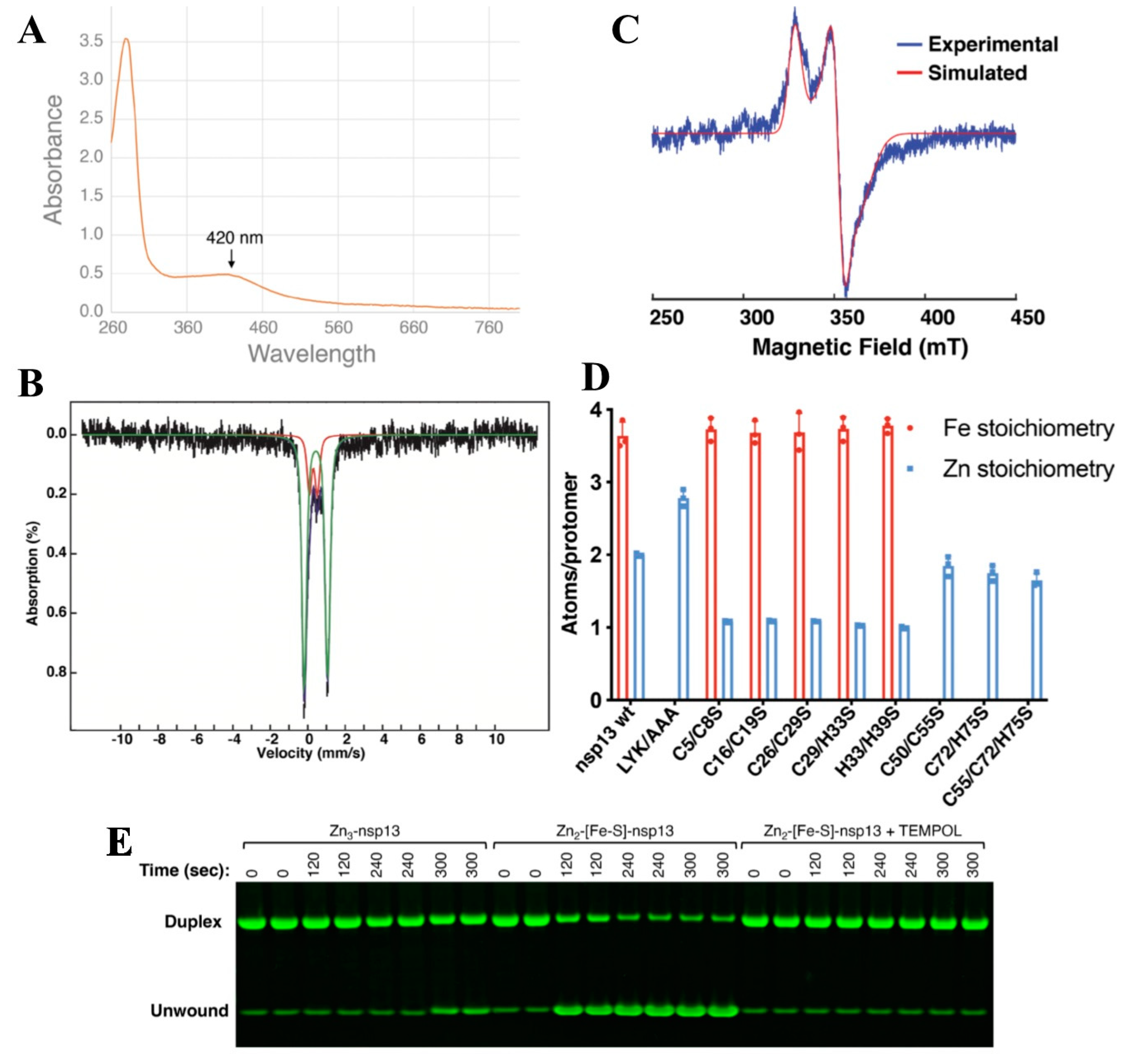

| Types of [Fe-S] Cluster | Stotal | EPRg (avg) | Mössbauer Parameters |
|---|---|---|---|
| a [2Fe-2S]-Oxidized | 1/2 | 1.95 | δ = 0.28 and ΔEQ = 0.61 |
| b [2Fe-2S]-Reduced | 0 | NA | δ = 0.30, δ = 0.72 and ΔEQ = 1.06, ΔEQ = 3.15 |
| c [3Fe-4S]-Cuboidal | 1/2 | 2.0 | δ = 0.27 and ΔEQ = 0.62 |
| d [3Fe-4S]-Linear | 5/2 | 5.85 | δ = 0.28 and ΔEQ = NA |
| e [4Fe-4S]-Native | NA | NA | δ = 0.44, ΔEQ = 1.25 |
| f [4Fe-4S]-Reduced | 1/2 | 1.96 | δ = 0.28, δ = 0.40 and ΔEQ = 0.80, ΔEQ = 1 |
| g [4Fe-4S]-Oxidized | 1/2 | 2.06 | δ = 0.48, δ = 0.60 and ΔEQ = 1, ΔEQ = 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raza, M.K.; Jeyachandran, V.R.; Bashir, S. Investigating Iron-Sulfur Proteins in Infectious Diseases: A Review of Characterization Techniques. Inorganics 2024, 12, 25. https://doi.org/10.3390/inorganics12010025
Raza MK, Jeyachandran VR, Bashir S. Investigating Iron-Sulfur Proteins in Infectious Diseases: A Review of Characterization Techniques. Inorganics. 2024; 12(1):25. https://doi.org/10.3390/inorganics12010025
Chicago/Turabian StyleRaza, Md Kausar, Vivian Robert Jeyachandran, and Sania Bashir. 2024. "Investigating Iron-Sulfur Proteins in Infectious Diseases: A Review of Characterization Techniques" Inorganics 12, no. 1: 25. https://doi.org/10.3390/inorganics12010025
APA StyleRaza, M. K., Jeyachandran, V. R., & Bashir, S. (2024). Investigating Iron-Sulfur Proteins in Infectious Diseases: A Review of Characterization Techniques. Inorganics, 12(1), 25. https://doi.org/10.3390/inorganics12010025







