Plasma Rich in Growth Factors in Bone Regeneration: The Proximity to the Clot as a Differential Factor in Osteoblast Cell Behaviour
Abstract
:1. Introduction
2. Material and Methods
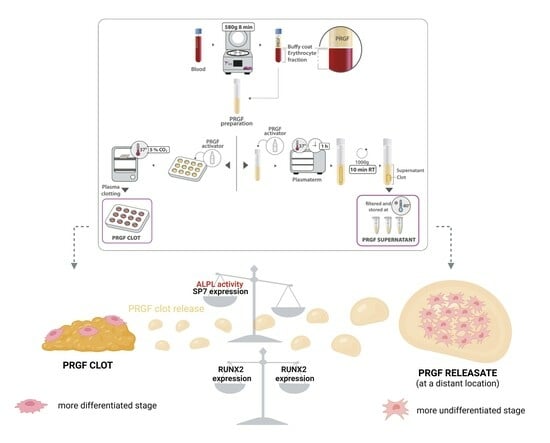
2.1. PRGF Preparation and Haematological Characterisation
2.2. Osteoblast Cell Isolation
2.3. Osteoblast Cell Characterisation
2.4. Osteoblast Proliferation
2.5. Osteoblast Gene Expression and Enzyme Activity Determination
2.5.1. Cell Culture
2.5.2. RNA Extraction and Quality Control (QC) Assessment
2.5.3. RT-qPCR Assays
2.5.4. Gene Expression Analyses
2.5.5. Enzymatic Activity Determination
3. Results
3.1. PRGF Preparation and Haematological Characterisation
3.2. Osteoblast Cell Characterisation
3.3. Osteoblast Proliferation
3.4. Osteoblast Gene Expression and ALPL Activity Quantification
3.4.1. Osteoblast Gene Expression
3.4.2. ALPL Activity Determination
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shen, F.; Shi, Y. Recent Advances in Single-Cell View of Mesenchymal Stem Cell in Osteogenesis. Front. Cell Dev. Biol. 2022, 9, 809918. [Google Scholar] [CrossRef] [PubMed]
- Mollentze, J.; Durandt, C.; Pepper, M.S. An in vitro and in vivo comparison of osteogenic differentiation of human mesenchymal stromal/stem cells. Stem Cells Int. 2021, 2021, 9919361. [Google Scholar] [CrossRef] [PubMed]
- Kirkham, G.; Cartmell, S. Genes and proteins involved in the regulation of osteogenesis. Top. Tissue Eng. 2007, 3, 1–22. [Google Scholar]
- Fung Ling Chau, J.; Fook Leong, W.; Li, B. Signaling pathways governing osteoblast proliferation, differentiation and function. Histol. Histopathol. 2009, 24, 1593–1606. [Google Scholar]
- Kulterer, B.; Friedl, G.; Jandrositz, A.; Sanchez-Cabo, F.; Prokesch, A.; Paar, C.; Scheideler, M.; Windhager, R.; Preisegger, K.-H.; Trajanoski, Z. Gene expression profiling of human mesenchymal stem cells derived from bone marrow during expansion and osteoblast differentiation. BMC Genom. 2007, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Dvorakova, J.; Wiesnerova, L.; Chocholata, P.; Kulda, V.; Landsmann, L.; Cedikova, M.; Kripnerova, M.; Eberlova, L.; Babuska, V. Human cells with osteogenic potential in bone tissue research. BioMedical Eng. OnLine 2023, 22, 33. [Google Scholar] [CrossRef] [PubMed]
- Rochira, A.; Siculella, L.; Damiano, F.; Palermo, A.; Ferrante, F.; Carluccio, M.A.; Calabriso, N.; Giannotti, L.; Stanca, E. Concentrated Growth Factors (CGF) Induce Osteogenic Differentiation in Human Bone Marrow Stem Cells. Biology 2020, 9, 370. [Google Scholar] [CrossRef]
- Huang, W.; Yang, S.; Shao, J.; Li, Y.-P. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front. Biosci. A J. Virtual Libr. 2007, 12, 3068. [Google Scholar] [CrossRef]
- Beederman, M.; Lamplot, J.D.; Nan, G.; Wang, J.; Liu, X.; Yin, L.; Li, R.; Shui, W.; Zhang, H.; Kim, S.H. BMP signaling in mesenchymal stem cell differentiation and bone formation. J. Biomed. Sci. Eng. 2013, 6, 32. [Google Scholar] [CrossRef]
- Park, J.-H.; Koh, E.-B.; Seo, Y.-J.; Oh, H.-S.; Byun, J.-H. BMP-9 Improves the Osteogenic Differentiation Ability over BMP-2 through p53 Signaling In Vitro in Human Periosteum-Derived Cells. Int. J. Mol. Sci. 2023, 24, 15252. [Google Scholar] [CrossRef]
- Celil, A.B.; Campbell, P.G. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J. Biol. Chem. 2005, 280, 31353–31359. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Zhang, X.; Chen, X.; Wang, Z.; Zheng, S.; Cheng, Y.; Liu, S.; Hao, L. The role of insulin-like growth factor-1 in bone remodeling: A review. Int. J. Biol. Macromol. 2023, 238, 124125. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Peel, S.A.; Ho, S.K.; Sándor, G.K.; Clokie, C.M. Platelet-rich plasma induces mRNA expression of VEGF and PDGF in rat bone marrow stromal cell differentiation. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2009, 107, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Kuriwaka-Kido, R.; Kondo, T.; Endo, I.; Kido, S. Regulation of osteoblast differentiation by interleukin-11 via AP-1 and Smad signaling. Endocr. J. 2012, 59, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Kespohl, B.; Schumertl, T.; Bertrand, J.; Lokau, J.; Garbers, C. The cytokine interleukin-11 crucially links bone formation, remodeling and resorption. Cytokine Growth Factor Rev. 2021, 60, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Stein, G.S.; Lian, J.B.; Van Wijnen, A.J.; Stein, J.L.; Montecino, M.; Javed, A.; Zaidi, S.K.; Young, D.W.; Choi, J.-Y.; Pockwinse, S.M. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 2004, 23, 4315–4329. [Google Scholar] [CrossRef] [PubMed]
- Mammoli, F.; Castiglioni, S.; Parenti, S.; Cappadone, C.; Farruggia, G.; Iotti, S.; Davalli, P.; Maier, J.A.M.; Grande, A.; Frassineti, C. Magnesium Is a Key Regulator of the Balance between Osteoclast and Osteoblast Differentiation in the Presence of Vitamin D3. Int. J. Mol. Sci. 2019, 20, 385. [Google Scholar] [CrossRef] [PubMed]
- Șelaru, A.; Samoilă, I.; Dinescu, S.; Costache, M. An overview on osteogenic differentiation process: Minimum essential information for bone tissue engineering. Rev. Biol. Biomed. Sci. 2018, 1, 9. [Google Scholar] [CrossRef]
- Arumugam, B.; Vishal, M.; Shreya, S.; Malavika, D.; Rajpriya, V.; He, Z.; Partridge, N.C.; Selvamurugan, N. Parathyroid hormone-stimulation of Runx2 during osteoblast differentiation via the regulation of lnc-SUPT3H-1:16 (RUNX2-AS1:32) and miR-6797-5p. Biochimie 2019, 158, 43–52. [Google Scholar] [CrossRef]
- Xia, H.; Tian, Y.; Lin, Y.; Huang, Q.; Xue, Y. Evaluating Osteogenic Differentiation of Osteoblastic Precursors upon Intermittent Administration of PTH/IGFBP7. Front. Pharmacol. 2022, 13, 839035. [Google Scholar] [CrossRef]
- Chan, W.C.W.; Tan, Z.; To, M.K.T.; Chan, D. Regulation and role of transcription factors in osteogenesis. Int. J. Mol. Sci. 2021, 22, 5445. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, W.; Zou, J.; Kratz, K.; Deng, Z.; Lendlein, A.; Ma, N. Histone Modification of Osteogenesis Related Genes Triggered by Substrate Topography Promotes Human Mesenchymal Stem Cell Differentiation. ACS Appl. Mater. Interfaces 2023, 15, 29752–29766. [Google Scholar] [CrossRef] [PubMed]
- Ponzetti, M.; Rucci, N. Osteoblast Differentiation and Signaling: Established Concepts and Emerging Topics. Int. J. Mol. Sci. 2021, 22, 6651. [Google Scholar] [CrossRef] [PubMed]
- O’brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Nardocci, G.; Carrasco, M.E.; Acevedo, E.; Hodar, C.; Meneses, C.; Montecino, M. Identification of a novel long noncoding RNA that promotes osteoblast differentiation. J. Cell. Biochem. 2018, 119, 7657–7666. [Google Scholar] [CrossRef] [PubMed]
- Aurilia, C.; Donati, S.; Palmini, G.; Miglietta, F.; Iantomasi, T.; Brandi, M.L. The Involvement of Long Non-Coding RNAs in Bone. Int. J. Mol. Sci. 2021, 22, 3909. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Guan, Z.; Yu, B.; Guo, J.; Shi, Y.; Hu, L. Circular RNA hsa_circ_0076906 competes with OGN for miR-1305 biding site to alleviate the progression of osteoporosis. Int. J. Biochem. Cell Biol. 2020, 122, 105719. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Veziroglu, E.M.; Mias, G.I. Characterizing Extracellular Vesicles and Their Diverse RNA Contents. Front. Genet. 2020, 11, 558262. [Google Scholar] [CrossRef]
- Li, Y.; Yin, P.; Guo, Z.; Lv, H.; Deng, Y.; Chen, M.; Gu, Y.; Tang, P.; Zhang, L. Bone-Derived Extracellular Vesicles: Novel Players of Interorgan Crosstalk. Front. Endocrinol. 2019, 10, 846. [Google Scholar] [CrossRef]
- Ren, J.; Yu, R.; Xue, J.; Tang, Y.; Su, S.; Liao, C.; Guo, Q.; Guo, W.; Zheng, J. How Do Extracellular Vesicles Play a Key Role in the Maintenance of Bone Homeostasis and Regeneration? A Comprehensive Review of Literature. Int. J. Nanomed. 2022, 17, 5375–5389. [Google Scholar] [CrossRef]
- Gao, M.; Gao, W.; Papadimitriou, J.M.; Zhang, C.; Gao, J.; Zheng, M. Exosomes—The enigmatic regulators of bone homeostasis. Bone Res. 2018, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Piao, Y.; Liu, Q.; Yang, X. Platelet-rich plasma-derived extracellular vesicles: A superior alternative in regenerative medicine? Cell Prolif. 2021, 54, e13123. [Google Scholar] [CrossRef] [PubMed]
- Antich-Rosselló, M.; Forteza-Genestra, M.A.; Monjo, M.; Ramis, J.M. Platelet-Derived Extracellular Vesicles for Regenerative Medicine. Int. J. Mol. Sci. 2021, 22, 8580. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Troya, M.; Falcon-Pérez, J.M.; López-Sarrio, S.; González, E.; Alkhraisat, M.H. Advances in Platelet Rich Plasma-Derived Extracellular Vesicles for Regenerative Medicine: A Systematic-Narrative Review. Int. J. Mol. Sci. 2023, 24, 13043. [Google Scholar] [CrossRef]
- Otahal, A.; Kuten-Pella, O.; Kramer, K.; Neubauer, M.; Lacza, Z.; Nehrer, S.; De Luna, A. Functional repertoire of EV-associated miRNA profiles after lipoprotein depletion via ultracentrifugation and size exclusion chromatography from autologous blood products. Sci. Rep. 2021, 11, 5823. [Google Scholar] [CrossRef]
- Nishiyama, K.; Okudera, T.; Watanabe, T.; Isobe, K.; Suzuki, M.; Masuki, H.; Okudera, H.; Uematsu, K.; Nakata, K.; Kawase, T. Basic characteristics of plasma rich in growth factors (PRGF): Blood cell components and biological effects. Clin. Exp. Dent. Res. 2016, 2, 96–103. [Google Scholar] [CrossRef]
- Anitua, E.; Zalduendo, M.; Troya, M.; Padilla, S.; Orive, G. Leukocyte inclusion within a platelet rich plasma-derived fibrin scaffold stimulates a more pro-inflammatory environment and alters fibrin properties. PLoS ONE 2015, 10, e0121713. [Google Scholar] [CrossRef]
- Anitua, E.; Zalduendo, M.; Troya, M. Autologous plasma rich in growth factors technology for isolation and ex vivo expansion of human dental pulp stem cells for clinical translation. Regen. Med. 2019, 14, 97–111. [Google Scholar] [CrossRef]
- Anitua, E.; Tejero, R.; Zalduendo, M.M.; Orive, G. Plasma rich in growth factors promotes bone tissue regeneration by stimulating proliferation, migration, and autocrine secretion in primary human osteoblasts. J. Periodontol. 2013, 84, 1180–1190. [Google Scholar] [CrossRef]
- Mokhtari, H.; Montaseri, A.; Mojaddadi, M.; Maleki Dizaj, S. Evaluation of the Effect of Platelet-Rich Plasma (Prp) on Osteoblast And Osteoclast Differentiation in the Presence of Polycaprolactone/Hydroxyapatite 3D Scaffold: An In Vitro Study. Stud. Med. Sci. 2020, 31, 725–734. [Google Scholar]
- Wang, J.; Li, W.; He, X.; Li, S.; Pan, H.; Yin, L. Injectable platelet-rich fibrin positively regulates osteogenic differentiation of stem cells from implant hole via the ERK1/2 pathway. Platelets 2023, 34, 2159020. [Google Scholar] [CrossRef]
- Gentile, P.; Garcovich, S. Systematic Review—The Potential Implications of Different Platelet-Rich Plasma (PRP) Concentrations in Regenerative Medicine for Tissue Repair. Int. J. Mol. Sci. 2020, 21, 5702. [Google Scholar] [CrossRef]
- Bacevich, B.M.; Smith, R.D.J.; Reihl, A.M.; Mazzocca, A.D.; Hutchinson, I.D. Advances with Platelet-Rich Plasma for Bone Healing. Biologics 2024, 18, 29–59. [Google Scholar] [CrossRef]
- Anitua, E.; Fernández-De-Retana, S.; Alkhraisat, M.H. Platelet rich plasma in oral and maxillofacial surgery from the perspective of composition. Platelets 2021, 32, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Allende, M.; Eguia, A.; Alkhraisat, M.H. Bone-Regenerative Ability of Platelet-Rich Plasma Following Sinus Augmentation with Anorganic Bovine Bone: A Systematic Review with Meta-Analysis. Bioengineering 2022, 9, 597. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E. Plasma rich in growth factors: Preliminary results of use in the preparation of future sites for implants. Int. J. Oral Maxillofac. Implant. 1999, 14, 529–535. [Google Scholar]
- Anitua, E.; Murias-Freijo, A.; Alkhraisat, M.H.; Orive, G. Clinical, radiographical, and histological outcomes of plasma rich in growth factors in extraction socket: A randomized controlled clinical trial. Clin. Oral Investig. 2015, 19, 589–600. [Google Scholar] [CrossRef]
- Stumbras, A.; Januzis, G.; Gervickas, A.; Kubilius, R.; Juodzbalys, G. Randomized and Controlled Clinical Trial of Bone Healing After Alveolar Ridge Preservation Using Xenografts and Allografts Versus Plasma Rich in Growth Factors. J. Oral Implant. 2020, 46, 515–525. [Google Scholar] [CrossRef]
- Janmey, P.A.; Miller, R.T. Mechanisms of mechanical signaling in development and disease. J. Cell Sci. 2011, 124 Pt 1, 9–18. [Google Scholar] [CrossRef]
- Thorne, R.G.; Hrabetová, S.; Nicholson, C. Diffusion of epidermal growth factor in rat brain extracellular space measured by integrative optical imaging. J. Neurophysiol. 2004, 92, 3471–3481. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Troya, M.; Pino, A. A novel protein-based autologous topical serum for skin regeneration. J. Cosmet. Dermatol. 2020, 19, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, L.A.; Ziaeian, B.; Palmer, T.; Schwartz, P.H. CHAPTER 9—Immunocytochemical Analysis of Stem Cells. In Human Stem Cell Manual; Loring, J.F., Wesselschmidt, R.L., Schwartz, P.H., Eds.; Academic Press: Oxford, UK, 2007; pp. 108–126. [Google Scholar]
- Van Acker, S.I.; Van Den Bogerd, B.; Haagdorens, M.; Koppen, C.; Pintelon, I. Immunocytochemical characterization of ex vivo cultured conjunctival explants; marker validation for the identification of squamous epithelial cells and goblet cells. Front. Med. 2023, 10, 1024926. [Google Scholar] [CrossRef] [PubMed]
- Quentmeier, H.; Osborn, M.; Reinhardt, J.; Zaborski, M.; Drexler, H.G. Immunocytochemical Analysis of Cell Lines Derived from Solid Tumors. J. Histochem. Cytochem. 2001, 49, 1369–1378. [Google Scholar] [CrossRef] [PubMed]
- Abuna, R.P.F.; Oliveira, F.S.; Ramos, J.I.R.; Lopes, H.B.; Freitas, G.P.; Souza, A.T.P.; Beloti, M.M.; Rosa, A.L. Selection of reference genes for quantitative real-time polymerase chain reaction studies in rat osteoblasts. J. Cell. Physiol. 2018, 234, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.0031. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons using rank sums. Technometrics 1964, 6, 241–252. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Team, R.D.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Pohlert, T. PMCMRplus: Calculate Pairwise Multiple Comparisons of Mean Rank Sums Extended; R package version 1.9.10; 2018; Volume 1, p. 147. Available online: https://CRAN.R-project.org/package=PMCMRplus (accessed on 12 December 2023).
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual: (Python Documentation Manual Part 2); CreateSpace: Scotts Valley, CA, USA, 2009. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Sa, B. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 4611–4622. [Google Scholar]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ (−delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71. [Google Scholar]
- Li, X.; Wang, Y.; Li, J.; Mei, X.; Liu, Y.; Huang, H. qPCRtools: An R package for qPCR data processing and visualization. Front. Genet. 2022, 13, 1002704. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, I.L.; Abril, N.; Zafra, R.; Morales-Prieto, N.; Hernández, V.M.; Ruiz, M.T.; Perez-Caballero, R.; Martínez-Moreno, A.; Pérez, J. Identification of reference genes for real-time PCR cytokine gene expression studies in sheep experimentally infected with Fasciola hepatica. Sci. Rep. 2019, 9, 1485. [Google Scholar] [CrossRef] [PubMed]
- Riedel, G.; Rüdrich, U.; Fekete-Drimusz, N.; Manns, M.P.; Vondran, F.W.; Bock, M. An extended ΔCT-method facilitating normalisation with multiple reference genes suited for quantitative RT-PCR analyses of human hepatocyte-like cells. PLoS ONE 2014, 9, e93031. [Google Scholar] [CrossRef]
- Hellemans, J.; Preobrazhenska, O.; Willaert, A.; Debeer, P.; Verdonk, P.C.; Costa, T.; Janssens, K.; Menten, B.; Roy, N.V.; Vermeulen, S.J.; et al. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat. Genet. 2004, 36, 1213–1218. [Google Scholar] [CrossRef]
- Poppe, B.; Vandesompele, J.; Schoch, C.; Lindvall, C.; Mrózek, K.; Bloomfield, C.D.; Beverloo, H.B.; Michaux, L.; Dastugue, N.; Herens, C.; et al. Expression analyses identify MLL as a prominent target of 11q23 amplification and support an etiologic role for MLL gain of function in myeloid malignancies. Blood 2004, 103, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Yang, J.; Li, X.; Zhou, C.; Chen, Y.; Wang, Z.; Qiu, X.; Liu, Y.; Zhang, H.; Greenbaum, J.; et al. A systematic dissection of human primary osteoblasts in vivo at single-cell resolution. bioRxiv 2020, 13, 20629–20650. [Google Scholar] [CrossRef]
- Kanno, T.; Takahashi, T.; Tsujisawa, T.; Ariyoshi, W.; Nishihara, T. Platelet-rich plasma enhances human osteoblast-like cell proliferation and differentiation. J. Oral Maxillofac. Surg. 2005, 63, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, H.; Orita, S.; Inage, K.; Fujimoto, K.; Shiga, Y.; Abe, K.; Inoue, M.; Norimoto, M.; Umimura, T.; Ishii, T.; et al. Freeze-Dried Platelet-Rich Plasma Induces Osteoblast Proliferation via Platelet-Derived Growth Factor Receptor-Mediated Signal Transduction. Asian Spine J. 2020, 14, 1–8. [Google Scholar] [CrossRef]
- Rutkovskiy, A.; Stensløkken, K.O.; Vaage, I.J. Osteoblast Differentiation at a Glance. Med. Sci. Monit. Basic Res. 2016, 22, 95–106. [Google Scholar] [CrossRef]
- Komori, T. Regulation of proliferation, differentiation and functions of osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef] [PubMed]
- Hojo, H.; Saito, T.; He, X.; Guo, Q.; Onodera, S.; Azuma, T.; Koebis, M.; Nakao, K.; Aiba, A.; Seki, M.; et al. Runx2 regulates chromatin accessibility to direct the osteoblast program at neonatal stages. Cell Rep. 2022, 40, 111315. [Google Scholar] [CrossRef]
- Amarasekara, D.S.; Kim, S.; Rho, J. Regulation of Osteoblast Differentiation by Cytokine Networks. Int. J. Mol. Sci. 2021, 22, 2851. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Reed, D.A.; Min, L.; Gopinathan, G.; Li, S.; Dangaria, S.J.; Li, L.; Geng, Y.; Galang, M.-T.; Gajendrareddy, P.; et al. Lyophilized platelet-rich fibrin (PRF) promotes craniofacial bone regeneration through Runx2. Int. J. Mol. Sci. 2014, 15, 8509–8525. [Google Scholar] [CrossRef]
- Sumida, R.; Maeda, T.; Kawahara, I.; Yusa, J.; Kato, Y. Platelet-rich fibrin increases the osteoprotegerin/receptor activator of nuclear factor-κB ligand ratio in osteoblasts. Exp. Ther. Med. 2019, 18, 358–365. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Choukroun, J.; Ghanaati, S.; Miron, R.J. Effects of an injectable platelet-rich fibrin on osteoblast behavior and bone tissue formation in comparison to platelet-rich plasma. Platelets 2018, 29, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.-C.; Wang, C.-Y.; Jang, J.S.-C.; Lee, C.-H.; Wu, J.-L. Large-pore platelet-rich fibrin with a mg ring to allow mc3t3-e1 preosteoblast migration and to improve osteogenic ability for bone defect repair. Int. J. Mol. Sci. 2021, 22, 4022. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, Y.; Liu, Y.; Yu, J.; Sun, X.; Wang, L.; Zhou, Y. Effects of platelet-rich fibrin on osteogenic differentiation of Schneiderian membrane derived mesenchymal stem cells and bone formation in maxillary sinus. Cell Commun. Signal. 2022, 20, 88. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Zheng, Z.; Zhang, M.; Long, L.; Zhang, X.; Tan, B.; Zhu, Z.; Liao, J.; Chen, W. Lyophilized Platelet-Rich Fibrin Exudate-Loaded Carboxymethyl Chitosan/GelMA Hydrogel for Efficient Bone Defect Repair. ACS Appl. Mater. Interfaces 2023, 15, 26349–26362. [Google Scholar] [CrossRef] [PubMed]
- Kargarpour, Z.; Nasirzade, J.; Panahipour, L.; Mitulović, G.; Miron, R.J.; Gruber, R. Platelet-rich fibrin increases BMP2 expression in oral fibroblasts via activation of TGF-β signaling. Int. J. Mol. Sci. 2021, 22, 7935. [Google Scholar] [CrossRef]
- Lee, K.-S.; Kim, H.-J.; Li, Q.-L.; Chi, X.-Z.; Ueta, C.; Komori, T.; Wozney, J.M.; Kim, E.-G.; Choi, J.-Y.; Ryoo, H.-M.; et al. Runx2 is a common target of transforming growth factor β1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol. Cell. Biol. 2000, 20, 8783–8792. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wu, S.; Chen, W.; Li, Y.-P. The roles and regulatory mechanisms of TGF-β and BMP signaling in bone and cartilage development, homeostasis and disease. Cell Res. 2024, 34, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Kawane, T.; Qin, X.; Jiang, Q.; Miyazaki, T.; Komori, H.; Yoshida, C.A.; Matsuura-Kawata, V.K.d.S.; Sakane, C.; Matsuo, Y.; Nagai, K.; et al. Runx2 is required for the proliferation of osteoblast progenitors and induces proliferation by regulating Fgfr2 and Fgfr3. Sci. Rep. 2018, 8, 13551. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, M.; Wang, S.; Xiao, Z.; Xiong, Y.; Wang, G. Recent Advances of Osterix Transcription Factor in Osteoblast Differentiation and Bone Formation. Front. Cell Dev. Biol. 2020, 8, 601224. [Google Scholar] [CrossRef]
- Yoshida, C.A.; Komori, H.; Maruyama, Z.; Miyazaki, T.; Kawasaki, K.; Furuichi, T.; Fukuyama, R.; Mori, M.; Yamana, K.; Nakamura, K.; et al. SP7 inhibits osteoblast differentiation at a late stage in mice. PLoS ONE 2012, 7, e32364. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, Z.; Feng, J.Q.; Dusevich, V.M.; Sinha, K.; Zhang, H.; Darnay, B.G.; de Crombrugghe, B. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc. Natl. Acad. Sci. USA 2010, 107, 12919–12924. [Google Scholar] [CrossRef]
- Bialek, P.; Kern, B.; Yang, X.; Schrock, M.; Sosic, D.; Hong, N.; Wu, H.; Yu, K.; Ornitz, D.M.; Olson, E.N.; et al. A twist code determines the onset of osteoblast differentiation. Dev. Cell 2004, 6, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Sinha, K.M.; Zhou, X. Genetic and molecular control of osterix in skeletal formation. J. Cell. Biochem. 2013, 114, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Matsuyama, T.; Miyamoto, M.; Yonamine, Y.; Izumi, Y. Platelet-rich plasma/osteoblasts complex induces bone formation via osteoblastic differentiation following subcutaneous transplantation. J. Periodontal Res. 2006, 41, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, H.; Montaseri, A.; Mojaddadi, A.; Mokhtari Zonouzi, H.R.; Karimiyan, N.; Arami, S. Effect of Platelet-Rich Plasma on Differentiation of Osteoblasts and Osteoclasts in the Presence of Three-Dimensional Scaffold. Pharm. Sci. 2018, 24, 124–130. [Google Scholar] [CrossRef]
- Li, X.; Yang, H.; Zhang, Z.; Yan, Z.; Lv, H.; Zhang, Y.; Wu, B. Platelet-rich fibrin exudate promotes the proliferation and osteogenic differentiation of human periodontal ligament cells in vitro. Mol. Med. Rep. 2018, 18, 4477–4485. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, A.a.-S.; Loty, S.; Isaac, J.; Bouchard, P.; Berdal, A.; Sautier, J.-M. Platelet-poor plasma stimulates the proliferation but inhibits the differentiation of rat osteoblastic cells in vitro. Clin. Oral Implant. Res. 2009, 20, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Hojo, H.; Ohba, S. Sp7 action in the skeleton: Its mode of action, functions, and relevance to skeletal diseases. Int. J. Mol. Sci. 2022, 23, 5647. [Google Scholar] [CrossRef] [PubMed]
- Rashid, H.; Ma, C.; Chen, H.; Wang, H.; Hassan, M.Q.; Sinha, K.; de Crombrugghe, B.; Javed, A. Sp7 and Runx2 molecular complex synergistically regulate expression of target genes. Connect. Tissue Res. 2014, 55 (Suppl. S1), 83–87. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Zalduendo, M.; Troya, M.; Tierno, R.; Alkhraisat, M.H. The inclusion of leukocytes into platelet rich plasma reduces scaffold stability and hinders extracellular matrix remodelling. Ann. Anat.-Anat. Anz. 2022, 240, 151853. [Google Scholar] [CrossRef]
- Yamada, Y.; Ito, K.; Nakamura, S.; Ueda, M.; Nagasaka, T. Promising cell-based therapy for bone regeneration using stem cells from deciduous teeth, dental pulp, and bone marrow. Cell Transplant. 2011, 20, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Ueda, M.; Naiki, T.; Takahashi, M.; Hata, K.-I.; Nagasaka, T. Autogenous injectable bone for regeneration with mesenchymal stem cells and platelet-rich plasma: Tissue-engineered bone regeneration. Tissue Eng. 2004, 10, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Niu, L.; Zhao, T.; Shi, Z.; Di, T.; Feng, G.; Li, J.; Huang, Z. Combining mesenchymal stem cell sheets with platelet-rich plasma gel/calcium phosphate particles: A novel strategy to promote bone regeneration. Stem Cell Res. Ther. 2015, 6, 256. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.J.; Choi, J.Y. Addition of mesenchymal stem cells to the scaffold of platelet-rich plasma is beneficial for the reduction of the consolidation period in mandibular distraction osteogenesis. J. Oral Maxillofac. Surg. 2010, 68, 1112–1124. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, J.; Yu, L.; Zhou, J.; Zheng, D.; Zhang, B. Effect of Concentrated Growth Factor (CGF) on the Promotion of Osteogenesis in Bone Marrow Stromal Cells (BMSC) In Vivo. Sci. Rep. 2018, 8, 5876. [Google Scholar] [CrossRef] [PubMed]
- Awadeen, M.A.; Al-Belasy, F.A.; Ameen, L.E.; Helal, M.E.; Grawish, M.E. Early therapeutic effect of platelet-rich fibrin combined with allogeneic bone marrow-derived stem cells on rats’ critical-sized mandibular defects. World J. Stem Cells 2020, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, E.; Fujioka-Kobayashi, M.; Sculean, A.; Chappuis, V.; Buser, D.; Schaller, B.; Dőri, F.; Miron, R.J. Effects of platelet rich plasma (PRP) on human gingival fibroblast, osteoblast and periodontal ligament cell behaviour. BMC Oral Health 2017, 17, 91. [Google Scholar]
- Blatt, S.; Thiem, D.G.; Kyyak, S.; Pabst, A.; Al-Nawas, B.; Kämmerer, P.W. Possible implications for improved osteogenesis? The combination of platelet-rich fibrin with different bone substitute materials. Front. Bioeng. Biotechnol. 2021, 9, 640053. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, X.; Liu, W.; Lu, W. Thrombin-activated platelet-rich plasma enhances osteogenic differentiation of human periodontal ligament stem cells by activating SIRT1-mediated autophagy. Eur. J. Med. Res. 2021, 26, 105. [Google Scholar] [CrossRef]
- Baca-Gonzalez, L.; Serrano Zamora, R.; Rancan, L.; González Fernández-Tresguerres, F.; Fernández-Tresguerres, I.; López-Pintor, R.M.; López-Quiles, J.; Leco, I.; Torres, J. Plasma rich in growth factors (PRGF) and leukocyte-platelet rich fibrin (L-PRF): Comparative release of growth factors and biological effect on osteoblasts. Int. J. Implant Dent. 2022, 8, 39. [Google Scholar] [CrossRef]
- Kosmidis, K.; Ehsan, K.; Pitzurra, L.; Loos, B.; Jansen, I. An in vitro study into three different PRF preparations for osteogenesis potential. J. Periodontal Res. 2023, 58, 483–492. [Google Scholar] [CrossRef]
- Sato, M.; Saitoh, I.; Kiyokawa, Y.; Iwase, Y.; Kubota, N.; Ibano, N.; Noguchi, H.; Yamasaki, Y.; Inada, E. Tissue-nonspecific alkaline phosphatase, a possible mediator of cell maturation: Towards a new paradigm. Cells 2021, 10, 3338. [Google Scholar] [CrossRef]
- Kyeyune-Nyombi, E.; Nicolas, V.; Strong, D.; Farley, J. Paradoxical effects of phosphate to directly regulate the level of skeletal alkaline phosphatase activity in human osteosarcoma (SaOS-2) cells and inversely regulate the level of skeletal alkaline phosphatase mRNA. Calcif. Tissue Int. 1995, 56, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Kim, M.G.; Byun, J.H.; Kim, G.C.; Ro, J.H.; Hwang, D.S.; Choi, B.B.; Park, G.C.; Kim, U.K. The effect of biomechanical stimulation on osteoblast differentiation of human jaw periosteum-derived stem cells. Maxillofac. Plast. Reconstr. Surg. 2017, 39, 7. [Google Scholar] [CrossRef] [PubMed]
- Nonoyama, S.; Karakida, T.; Chiba-Ohkuma, R.; Yamamoto, R.; Ujiie, Y.; Nagano, T.; Yamakoshi, Y.; Gomi, K. Development and Characterization of Alkaline Phosphatase-Positive Human Umbilical Cord Perivascular Cells. Cells 2021, 10, 3011. [Google Scholar] [CrossRef] [PubMed]
- Low, M.G. Glycosylphosphatidylinositol: An anchor for eukaryotic membrane proteins. Biochem. Soc. Trans. 1988, 16, 271–272. [Google Scholar] [CrossRef]
- Davitz, M.A.; Hereld, D.; Shak, S.; Krakow, J.; Englund, P.T.; Nussenzweig, V. A glycan-phosphatidylinositol-specific phospholipase D in human serum. Science 1987, 238, 81–84. [Google Scholar] [CrossRef]
- Anh, D.; Eden, A.; Farley, J. Quantitation of soluble and skeletal alkaline phosphatase, and insoluble alkaline phosphatase anchor-hydrolase activities in human serum. Clin. Chim. Acta 2001, 311, 137–148. [Google Scholar] [CrossRef]
- Fedde, K.N. Human osteosarcoma cells spontaneously release matrix-vesicle-like structures with the capacity to mineralize. Bone Miner. 1992, 17, 145–151. [Google Scholar] [CrossRef]
- Xie, M.; Low, M. Streptolysin-O induces release of glycosylphosphatidylinositol-anchored alkaline phosphatase from ROS cells by vesiculation independently of phospholipase action. Biochem. J. 1995, 305, 529–537. [Google Scholar] [CrossRef]
- Dean, D.; Schwartz, Z.; Bonewald, L.; Muniz, O.; Morales, S.; Gomez, R.; Brooks, B.; Qiao, M.; Howell, D.; Boyan, B. Matrix vesicles produced by osteoblast-like cells in culture become significantly enriched in proteoglycan-degrading metalloproteinases after addition of β-glycerophosphate and ascorbic acid. Calcif. Tissue Int. 1994, 54, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, D.; Hofmann, C.; Jakob, F.; Klopocki, E.; Graser, S. Tissue-Nonspecific Alkaline Phosphatase—A Gatekeeper of Physiological Conditions in Health and a Modulator of Biological Environments in Disease. Biomolecules 2020, 10, 1648. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.A.; Felice, B.; Sappia, L.D.; Moura, S.L.; Martí, M.; Pividori, M.I. Osteoblastic exosomes. A non-destructive quantitative approach of alkaline phosphatase to assess osteoconductive nanomaterials. Mater. Sci. Eng. C 2020, 115, 110931. [Google Scholar] [CrossRef] [PubMed]





| Sample | Platelet Count (×106/mL) | Leukocyte Count (×106/mL) | Erythrocyte Count (×109/mL) |
|---|---|---|---|
| Peripheral blood | 176 | 6.1 | 4.59 |
| PRGF | 391 (2.2 x) * | 0.2 (0.03 x) * | 0.01 (0.00 x) * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anitua, E.; Zalduendo, M.; Tierno, R.; Alkhraisat, M.H. Plasma Rich in Growth Factors in Bone Regeneration: The Proximity to the Clot as a Differential Factor in Osteoblast Cell Behaviour. Dent. J. 2024, 12, 122. https://doi.org/10.3390/dj12050122
Anitua E, Zalduendo M, Tierno R, Alkhraisat MH. Plasma Rich in Growth Factors in Bone Regeneration: The Proximity to the Clot as a Differential Factor in Osteoblast Cell Behaviour. Dentistry Journal. 2024; 12(5):122. https://doi.org/10.3390/dj12050122
Chicago/Turabian StyleAnitua, Eduardo, Mar Zalduendo, Roberto Tierno, and Mohammad Hamdan Alkhraisat. 2024. "Plasma Rich in Growth Factors in Bone Regeneration: The Proximity to the Clot as a Differential Factor in Osteoblast Cell Behaviour" Dentistry Journal 12, no. 5: 122. https://doi.org/10.3390/dj12050122