Feature Papers in Autophagy
A topical collection in Cells (ISSN 2073-4409). This collection belongs to the section "Autophagy".
Viewed by 19358
Share This Topical Collection
Editor
Topical Collection Information
Dear Colleagues,
This Topical Collection “Feature Papers in Autophagy” aims to collect high-quality research articles, review articles, and communications (including opinion and hypothesis papers) in all fields of autophagy, with a focus on molecular and cell biological research. Since the aim of this Topical Collection is to illustrate, through selected works, frontier research in autophagy, we encourage Editorial Board Members of Cells or relevant experts and colleagues to contribute papers reflecting the latest progress in their particular research field.
Topics include, without being limited to:
- Molecular machinery and regulation of autophagic pathways
- Cellular functions of autophagy-related proteins and autophagy receptors (including non-autophagic functions)
- Regulation of autophagy by Intra- and Extracellular Signaling
- Molecular mechanisms of selective autophagy
- Methods to monitor autophagy
Role, interplay, involvement or function of autophagy in:
- Cell Physiology
- Cellular Metabolism
- Cellular Immunology
- Intra- and Extracellular Signaling
- Cell Movement and Motility
- Apoptosis
- Cell Aging, Senescence
- Cell Growth and Differentiation
- Stem Cells
- Disease, Genetic Disorders and Therapy
- Plant, Algae and Fungi Cell Biology
- Neurodegeneration, Neuroinflammation
Dr. Nikolai Engedal
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Published Papers (6 papers)
Open AccessArticle
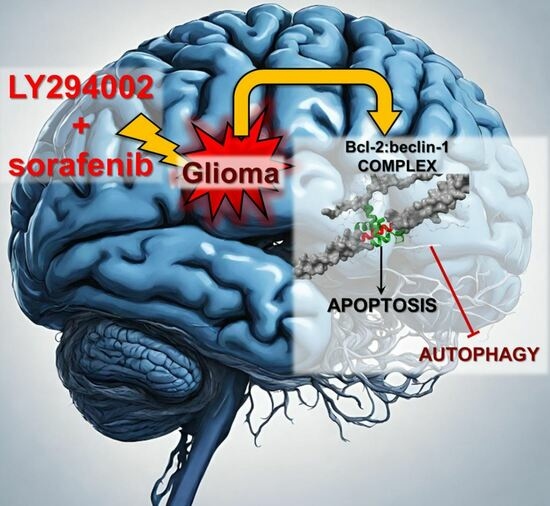
The Role of Bcl-2 and Beclin-1 Complex in “Switching” between Apoptosis and Autophagy in Human Glioma Cells upon LY294002 and Sorafenib Treatment
by
Adrian Zając, Aleksandra Maciejczyk, Joanna Sumorek-Wiadro, Kamil Filipek, Kamil Deryło, Ewa Langner, Jarosław Pawelec, Magdalena Wasiak, Mateusz Ścibiorski, Wojciech Rzeski, Marek Tchórzewski, Michał Reichert and Joanna Jakubowicz-Gil
Viewed by 1255
Abstract
Background: Gliomas are the most malignant tumors of the central nervous system. One of the factors in their high drug resistance is avoiding programmed death (PCD) induction. This is related to the overexpression of intracellular survival pathways: PI3K-Akt/PKB-mTOR and Ras-Raf-MEK-ERK. Apoptosis and autophagy
[...] Read more.
Background: Gliomas are the most malignant tumors of the central nervous system. One of the factors in their high drug resistance is avoiding programmed death (PCD) induction. This is related to the overexpression of intracellular survival pathways: PI3K-Akt/PKB-mTOR and Ras-Raf-MEK-ERK. Apoptosis and autophagy are co-existing processes due to the interactions between Bcl-2 and beclin-1 proteins. Their complex may be a molecular “toggle-switch” between PCD types. The aim of this research was to investigate the role of Bcl-2:beclin-1 complex in glioma cell elimination through the combined action of LY294002 and sorafenib. Methods: Drug cytotoxicity was estimated with an MTT test. The type of cell death was evaluated using variant microscopy techniques (fluorochrome staining, immunocytochemistry, and transmission electron microscopy), as well as the Bcl-2:beclin-1 complex formation and protein localization. Molecular analysis of PCD indicators was conducted through immunoblotting, immunoprecipitation, and ELISA testing. SiRNA was used to block Bcl-2 and beclin-1 expression. Results: The results showed the inhibitors used in simultaneous application resulted in Bcl-2:beclin-1 complex formation and apoptosis becoming dominant. This was accompanied by changes in the location of the tested proteins. Conclusions: “Switching” between apoptosis and autophagy using PI3K and Raf inhibitors with Bcl-2:beclin-1 complex formation opens new therapeutic perspectives against gliomas.
Full article
►▼
Show Figures
Open AccessArticle
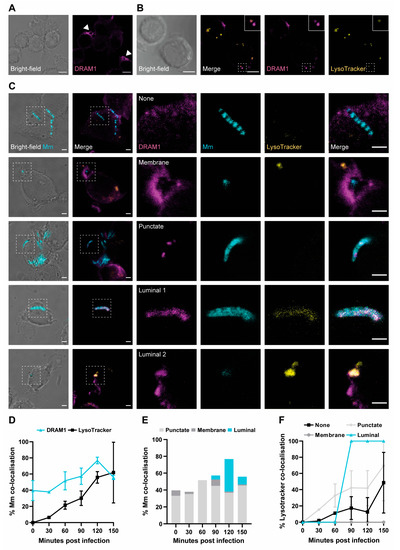
DRAM1 Promotes Lysosomal Delivery of Mycobacterium marinum in Macrophages
by
Adrianna Banducci-Karp, Jiajun Xie, Sem A. G. Engels, Christos Sarantaris, Patrick van Hage, Monica Varela, Annemarie H. Meijer and Michiel van der Vaart
Cited by 2 | Viewed by 1738
Abstract
Damage-Regulated Autophagy Modulator 1 (DRAM1) is an infection-inducible membrane protein, whose function in the immune response is incompletely understood. Based on previous results in a zebrafish infection model, we have proposed that DRAM1 is a host resistance factor against intracellular mycobacterial infection. To
[...] Read more.
Damage-Regulated Autophagy Modulator 1 (DRAM1) is an infection-inducible membrane protein, whose function in the immune response is incompletely understood. Based on previous results in a zebrafish infection model, we have proposed that DRAM1 is a host resistance factor against intracellular mycobacterial infection. To gain insight into the cellular processes underlying DRAM1-mediated host defence, here we studied the interaction of DRAM1 with
Mycobacterium marinum in murine RAW264.7 macrophages. We found that, shortly after phagocytosis, DRAM1 localised in a punctate pattern to mycobacteria, which gradually progressed to full DRAM1 envelopment of the bacteria. Within the same time frame, DRAM1-positive mycobacteria colocalised with the LC3 marker for autophagosomes and LysoTracker and LAMP1 markers for (endo)lysosomes. Knockdown analysis revealed that DRAM1 is required for the recruitment of LC3 and for the acidification of mycobacteria-containing vesicles. A reduction in the presence of LAMP1 further suggested reduced fusion of lysosomes with mycobacteria-containing vesicles. Finally, we show that DRAM1 knockdown impairs the ability of macrophages to defend against mycobacterial infection. Together, these results support that DRAM1 promotes the trafficking of mycobacteria through the degradative (auto)phagolysosomal pathway. Considering its prominent effect on host resistance to intracellular infection, DRAM1 is a promising target for therapeutic modulation of the microbicidal capacity of macrophages.
Full article
►▼
Show Figures
Open AccessReview
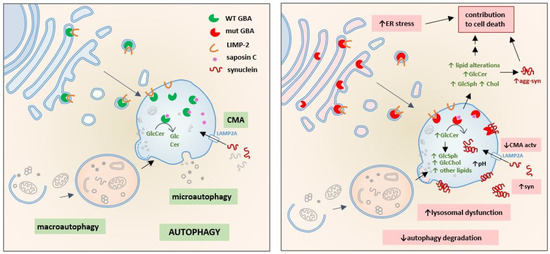
The Consequences of GBA Deficiency in the Autophagy–Lysosome System in Parkinson’s Disease Associated with GBA
by
Eddie Pradas and Marta Martinez-Vicente
Cited by 9 | Viewed by 3833
Abstract
GBA gene variants were the first genetic risk factor for Parkinson’s disease.
GBA encodes the lysosomal enzyme glucocerebrosidase (GBA), which is involved in sphingolipid metabolism. GBA exhibits a complex physiological function that includes not only the degradation of its substrate glucosylceramide but also
[...] Read more.
GBA gene variants were the first genetic risk factor for Parkinson’s disease.
GBA encodes the lysosomal enzyme glucocerebrosidase (GBA), which is involved in sphingolipid metabolism. GBA exhibits a complex physiological function that includes not only the degradation of its substrate glucosylceramide but also the metabolism of other sphingolipids and additional lipids such as cholesterol, particularly when glucocerebrosidase activity is deficient. In the context of Parkinson’s disease associated with GBA, the loss of GBA activity has been associated with the accumulation of α-synuclein species. In recent years, several hypotheses have proposed alternative and complementary pathological mechanisms to explain why lysosomal enzyme mutations lead to α-synuclein accumulation and become important risk factors in Parkinson’s disease etiology. Classically, loss of GBA activity has been linked to a dysfunctional autophagy–lysosome system and to a subsequent decrease in autophagy-dependent α-synuclein turnover; however, several other pathological mechanisms underlying GBA-associated parkinsonism have been proposed. This review summarizes and discusses the different hypotheses with a special focus on autophagy-dependent mechanisms, as well as autophagy-independent mechanisms, where the role of other players such as sphingolipids, cholesterol and other GBA-related proteins make important contributions to Parkinson’s disease pathogenesis.
Full article
►▼
Show Figures
Open AccessReview
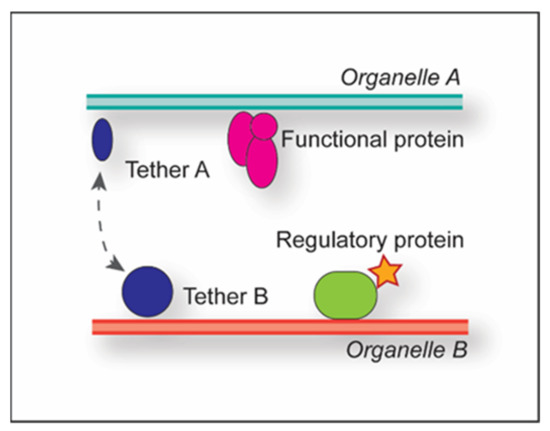
Membrane Contact Sites in Autophagy
by
Emma Zwilling and Fulvio Reggiori
Cited by 3 | Viewed by 3689
Abstract
Eukaryotes utilize different communication strategies to coordinate processes between different cellular compartments either indirectly, through vesicular transport, or directly, via membrane contact sites (MCSs). MCSs have been implicated in lipid metabolism, calcium signaling and the regulation of organelle biogenesis in various cell types.
[...] Read more.
Eukaryotes utilize different communication strategies to coordinate processes between different cellular compartments either indirectly, through vesicular transport, or directly, via membrane contact sites (MCSs). MCSs have been implicated in lipid metabolism, calcium signaling and the regulation of organelle biogenesis in various cell types. Several studies have shown that MCSs play a crucial role in the regulation of macroautophagy, an intracellular catabolic transport route that is characterized by the delivery of cargoes (proteins, protein complexes or aggregates, organelles and pathogens) to yeast and plant vacuoles or mammalian lysosomes, for their degradation and recycling into basic metabolites. Macroautophagy is characterized by the de novo formation of double-membrane vesicles called autophagosomes, and their biogenesis requires an enormous amount of lipids. MCSs appear to have a central role in this supply, as well as in the organization of the autophagy-related (ATG) machinery. In this review, we will summarize the evidence for the participation of specific MCSs in autophagosome formation, with a focus on the budding yeast and mammalian systems.
Full article
►▼
Show Figures
Open AccessArticle
Morphology of Phagophore Precursors by Correlative Light-Electron Microscopy
by
Sigurdur Runar Gudmundsson, Katri A. Kallio, Helena Vihinen, Eija Jokitalo, Nicholas Ktistakis and Eeva-Liisa Eskelinen
Cited by 10 | Viewed by 2659
Abstract
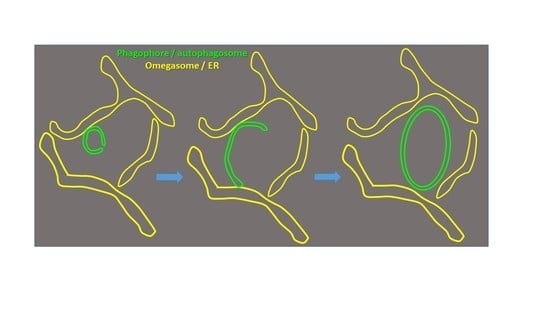
Autophagosome biogenesis occurs in the transient subdomains of the endoplasmic reticulum that are called omegasomes, which, in fluorescence microscopy, appear as small puncta, which then grow in diameter and finally shrink and disappear once the autophagosome is complete. Autophagosomes are formed by phagophores,
[...] Read more.
Autophagosome biogenesis occurs in the transient subdomains of the endoplasmic reticulum that are called omegasomes, which, in fluorescence microscopy, appear as small puncta, which then grow in diameter and finally shrink and disappear once the autophagosome is complete. Autophagosomes are formed by phagophores, which are membrane cisterns that elongate and close to form the double membrane that limits autophagosomes. Earlier electron-microscopy studies showed that, during elongation, phagophores are lined by the endoplasmic reticulum on both sides. However, the morphology of the very early phagophore precursors has not been studied at the electron-microscopy level. We used live-cell imaging of cells expressing markers of phagophore biogenesis combined with correlative light-electron microscopy, as well as electron tomography of ATG2A/B-double-deficient cells, to reveal the high-resolution morphology of phagophore precursors in three dimensions. We showed that phagophores are closed or nearly closed into autophagosomes already at the stage when the omegasome diameter is still large. We further observed that phagophore precursors emerge next to the endoplasmic reticulum as bud-like highly curved membrane cisterns with a small opening to the cytosol. The phagophore precursors then open to form more flat cisterns that elongate and curve to form the classically described crescent-shaped phagophores.
Full article
►▼
Show Figures
Open AccessArticle
Spermidine and Rapamycin Reveal Distinct Autophagy Flux Response and Cargo Receptor Clearance Profile
by
Sholto de Wet, Andre Du Toit and Ben Loos
Cited by 9 | Viewed by 4869
Abstract
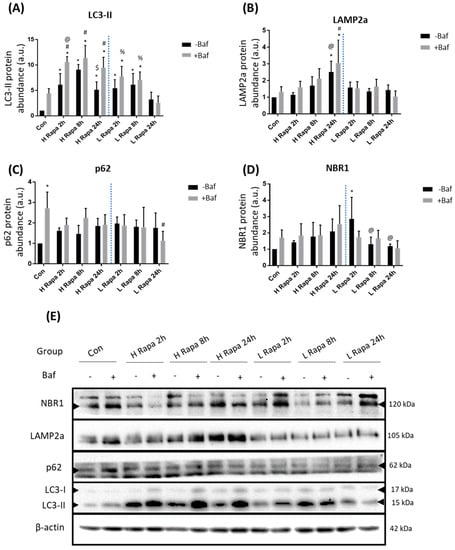
Autophagy flux is the rate at which cytoplasmic components are degraded through the entire autophagy pathway and is often measured by monitoring the clearance rate of autophagosomes. The specific means by which autophagy targets specific cargo has recently gained major attention due to
[...] Read more.
Autophagy flux is the rate at which cytoplasmic components are degraded through the entire autophagy pathway and is often measured by monitoring the clearance rate of autophagosomes. The specific means by which autophagy targets specific cargo has recently gained major attention due to the role of autophagy in human pathologies, where specific proteinaceous cargo is insufficiently recruited to the autophagosome compartment, albeit functional autophagy activity. In this context, the dynamic interplay between receptor proteins such as p62/Sequestosome-1 and neighbour of BRCA1 gene 1 (NBR1) has gained attention. However, the extent of receptor protein recruitment and subsequent clearance alongside autophagosomes under different autophagy activities remains unclear. Here, we dissect the concentration-dependent and temporal impact of rapamycin and spermidine exposure on receptor recruitment, clearance and autophagosome turnover over time, employing micropatterning. Our results reveal a distinct autophagy activity response profile, where the extent of autophagosome and receptor co-localisation does not involve the total pool of either entities and does not operate in similar fashion. These results suggest that autophagosome turnover and specific cargo clearance are distinct entities with inherent properties, distinctively contributing towards total functional autophagy activity. These findings are of significance for future studies where disease specific protein aggregates require clearance to preserve cellular proteostasis and viability and highlight the need of discerning and better tuning autophagy machinery activity and cargo clearance.
Full article
►▼
Show Figures