Rapid Bacterial Recognition over a Wide pH Range by Boronic Acid-Based Ditopic Dendrimer Probes for Gram-Positive Bacteria
Abstract
:1. Introduction
2. Results and Discussion
2.1. Intermolecular Interactions between Dendrimer Probes and Bacteria
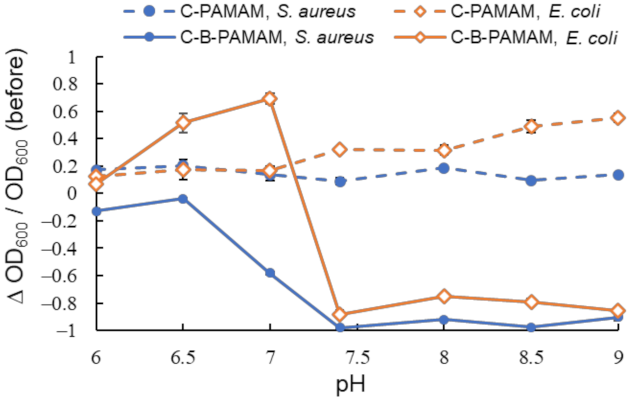
2.1.1. Electrostatic Interaction
2.1.2. Hydrophobic Interaction
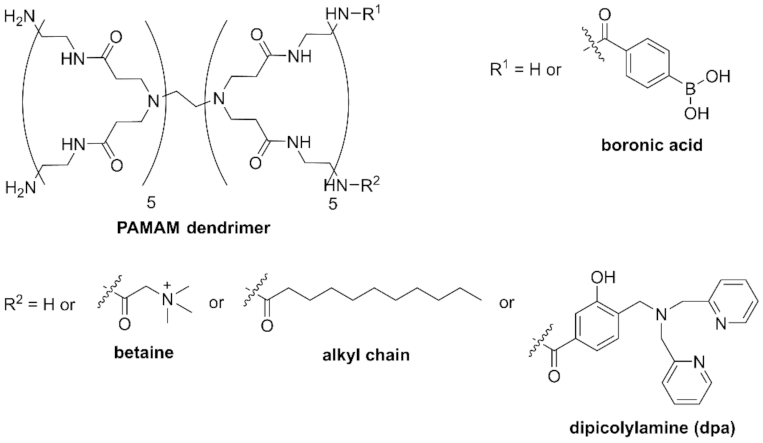
2.2. dpa-Modified Ditopic Dendrimer Probes
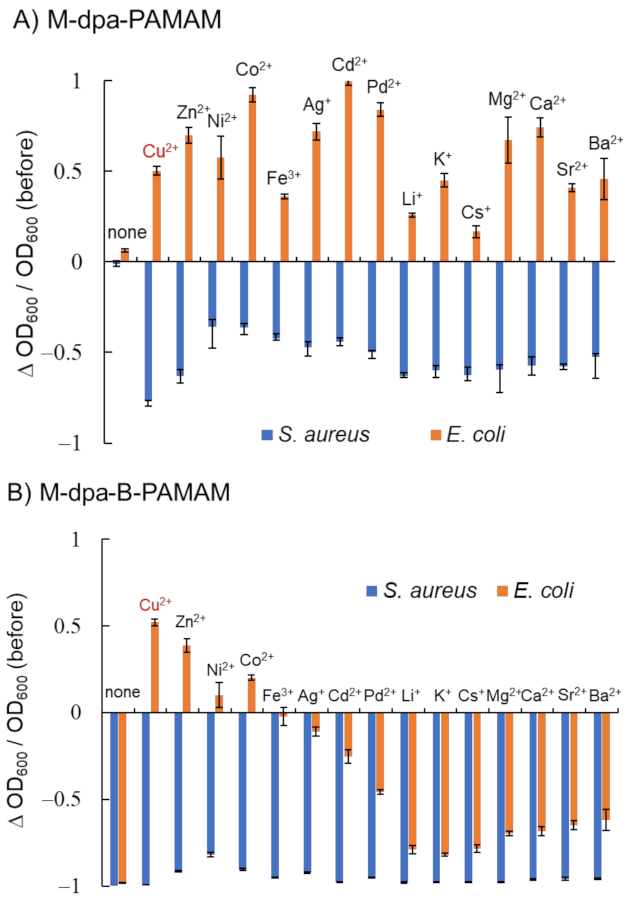
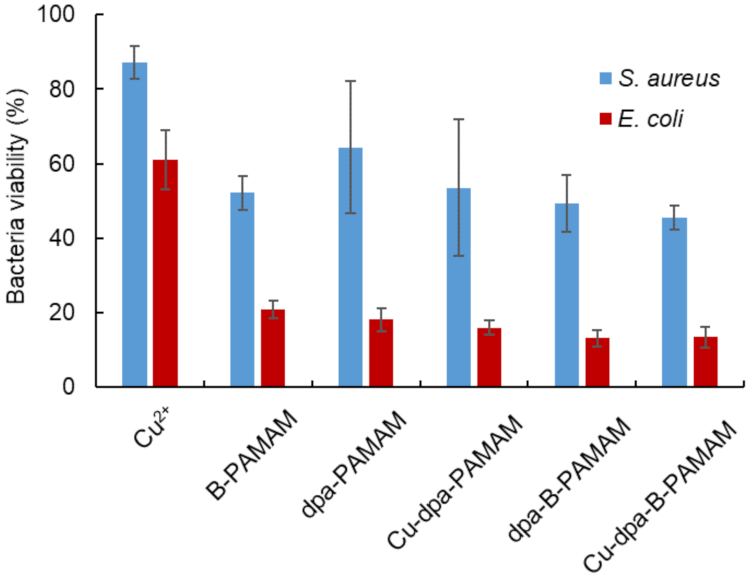
2.2.1. Bacterial Recognition by Cu-dpa-B-PAMAM
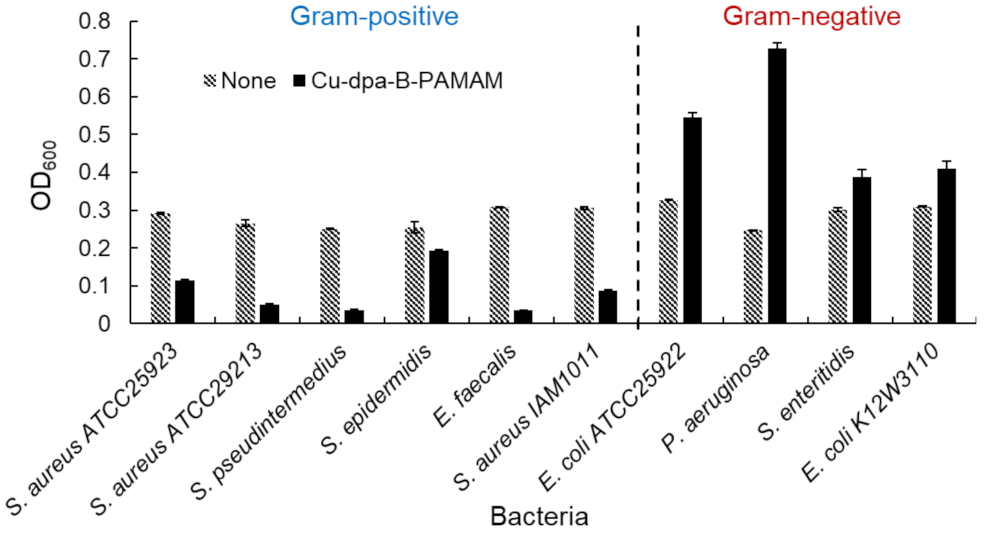
2.2.2. Selectivity toward Gram-Positive Bacteria and Assay for Viability
3. Materials and Methods
3.1. Reagents and Apparatus
3.1.1. Reagents
3.1.2. Apparatus
3.2. Chemical Syntheses
3.2.1. Synthesis of B-PAMAMs
3.2.2. Synthesis of Bt-B-PAMAM
3.2.3. Synthesis of C-PAMAM
3.2.4. Synthesis of C-B-PAMAM
3.2.5. Synthesis of dpa
3.2.6. Synthesis of dpa-PAMAM
3.2.7. Synthesis of dpa-B-PAMAM
3.3. Biological Experiments
3.3.1. Cell Culture
- S. aureus: OD600 = 1.0, CFU = 4.5 × 108 mL−1.
- E. coli: OD600 = 1.0, CFU = 1.0 × 109 mL−1.
3.3.2. Bacterial Detection
- ΔOD600 = OD600(after) − OD600(before or control).
- Turbidity change = ΔOD600/OD600(before or control).
3.3.3. MTT Assay
3.3.4. Statistical Analyses of Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ventola, C.L. The antibiotic resistance crisis. Pharmacol. Ther. 2015, 40, 277–283. [Google Scholar]
- Reardon, S. Antibiotic resistance sweeping developing world. Nature 2014, 509, 141–142. [Google Scholar] [CrossRef] [Green Version]
- Jacoby, G.A.; Archer, G.L.N. New mechanisms of bacterial resistance to antimicrobial agents. N. Engl. J. Med. 1991, 324, 601–612. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Swaminathan, B.; Feng, P. Rapid detection of food-borne pathogenic bacteria. Annu. Rev. Microbiol. 1994, 48, 401–426. [Google Scholar] [CrossRef]
- Lazcka, O.; Campo, F.J.D.; Muñoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef]
- Mandal, P.K.; Biswas, A.K.; Choi, K.; Pal, U.K. Methods for rapid detection of foodborne pathogens: An overview. Am. J. Food Technol. 2011, 6, 87–102. [Google Scholar] [CrossRef] [Green Version]
- Ray, P.C.; Khan, S.A.; Singh, A.K.; Senapati, D.; Fan, Z. Nanomaterials for targeted detection and photothermal killing of bacteria. Chem. Soc. Rev. 2012, 41, 3193–3209. [Google Scholar] [CrossRef]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef]
- Lqbal, S.S.; Mayo, M.W.; Bruno, J.G.; Bronk, B.V.; Batt, C.A.; Chambers, J.P. A review of molecular recognition technologies for detection of biological threat agents. Biosens. Bioelectron. 2000, 15, 549–578. [Google Scholar] [CrossRef]
- Lay, J.O., Jr. MALDI-TOF mass spectrometry of bacteria. Mass Spec. Rev. 2001, 20, 172–194. [Google Scholar] [CrossRef]
- Skottrup, P.D.; Nicolaisen, M.; Justesen, A.F. Towards on-site pathogen detection using antibody-based sensors. Biosens. Bioelectron. 2008, 24, 339–348. [Google Scholar] [CrossRef]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Gang, J.J. Challenges of microarray applications for microbial detection and gene expression profiling in food. J. Microb. Biochem. Technol. 2011, S2, 2. [Google Scholar] [CrossRef] [Green Version]
- Call, D. Challenges and opportunities for pathogen detection using DNA microarrays. Crit. Rev. Microbiol. 2005, 31, 91–99. [Google Scholar] [CrossRef]
- Qi, Z.; Bharate, P.; Lai, C.-H.; Ziem, B.; Böttcher, C.; Schulz, A.; Beckert, F.; Hatting, B.; Mülhaupt, R.; Seeberger, P.H.; et al. Multivalency at interfaces: Supramolecular carbohydrate-functionalized graphene derivatives for bacterial capture, release, and disinfection. Nano Lett. 2015, 15, 6051–6057. [Google Scholar] [CrossRef] [Green Version]
- El-Boubbou, K.; Gruden, C.; Huang, X. Magnetic glyco-nanoparticles: A unique tool for rapid pathogen detection, decontamination, and strain differentiation. J. Am. Chem. Soc. 2007, 129, 13392–13393. [Google Scholar] [CrossRef]
- Kim, D.; Kwon, S.-J.; Wu, X.; Sauve, J.; Lee, I.; Nam, J.; Kim, J.; Dordick, J.S. Selective killing of pathogenic bacteria by antimicrobial silver nanoparticle-cell wall binding domain conjugate. ACS Appl. Mater. Interfaces 2018, 10, 13317–13324. [Google Scholar] [CrossRef]
- Kasai, Y.; Kobayashi, H.; Tsuchido, Y.; Hashimoto, T.; Kanzawa, N.; Hayashita, T. Staphylococcus aureus detection by fluorescent silica nanoparticles modified with metal dipicolylamine complexes. Chem. Lett. 2016, 45, 749–751. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-J.; Jeong, K.J.; Hashimoto, M.; Kwon, A.H.; Rwei, A.; Shankarappa, S.A.; Tsui, J.H.; Kohane, D.S. Synthetic ligand-coated magnetic nanoparticles for microfluidic bacterial separation from blood. Nano Lett. 2014, 14, 1–5. [Google Scholar] [CrossRef]
- Leevy, W.M.; Lambert, T.N.; Johnson, J.R.; Morris, J.; Smith, B.D. Quantum dot probes for bacteria distinguish Escherichia coli mutants and permit in vivo imaging. Chem. Commun. 2008, 2331–2333. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Swamy, K.M.K.; Lee, K.M.; Jagdale, A.R.; Kim, Y.; Kim, S.-J.; Yoo, K.H.; Yoon, J. Pyrophosphate selective fluorescent chemosensors based on coumarin–DPA–Cu(II) complexes. Chem. Commun. 2009, 46, 7215–7217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngo, H.T.; Liu, X.; Jolliffe, K.A. Anion recognition and sensing with Zn(II)-dipicolylamine complexes. Chem. Soc. Rev. 2012, 41, 4928–4965. [Google Scholar] [CrossRef]
- Cabral, A.D.; Rafiei, N.; De Araujo, E.D.; Radu, T.B.; Toutah, K.; Nino, D.; Murcar-Evans, B.I.; Milstein, J.N.; Kraskouskaya, D.; Gunning, P.T. Sensitive detection of broad-Spectrum bacteria with small- molecule fluorescent excimer chemosensors. ACS Sens. 2020, 5, 2753–2762. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xu, K.; Wang, H.; Tan, P.K.J.; Fan, W.; Venkatraman, S.S.; Li, L.; Yang, Y.-Y. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat. Nanotechnol. 2009, 4, 457–463. [Google Scholar] [CrossRef]
- Hayden, S.C.; Zhao, G.; Saha, K.; Phillips, R.L.; Li, X.; Miranda, O.R.; Rotello, V.M.; El-Sayed, M.A.; Schmidt-Krey, I.; Bunz, U.H.F. Aggregation and interaction of cationic nanoparticles on bacterial surfaces. J. Am. Chem. Soc. 2012, 134, 6920–6923. [Google Scholar] [CrossRef]
- Dumitriu, S. (Ed.) Polysaccharides: Structural Diversity and Functional Versatility, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2014; Chapter 1. [Google Scholar]
- Bull, S.D.; Davidson, M.G.; van den Elsen, J.M.H.; Fossey, J.S.; Jenkins, A.T.A.; Jiang, Y.-B.; Kubo, Y.; Marken, F.; Sakurai, K.; Zhao, J.; et al. Exploiting the reversible covalent bonding of boronic acids: Recognition, sensing, and assembly. Acc. Chem. Res. 2013, 46, 312–326. [Google Scholar] [CrossRef]
- Wu, X.; Li, Z.; Chen, X.-X.; Fossey, J.S.; James, T.D.; Jiang, Y.-B. Selective sensing of saccharides using simple boronic acids and their aggregates. Chem. Soc. Rev. 2013, 42, 8032–8048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchido, Y.; Horiuchi, R.; Hashimoto, T.; Ishihara, K.; Kanzawa, N.; Hayashita, T. Rapid and selective discrimination of Gram-positive and Gram-negative bacteria by boronic acid-modified poly(amidoamine) dendrimer. Anal. Chem. 2019, 91, 3929–3935. [Google Scholar] [CrossRef]
- Mikagi, A.; Tsurufusa, R.; Tsuchido, Y.; Hashimoto, T.; Hayashita, T. Fast and sensitive bacteria detection by boronic acid modified fluorescent dendrimer. Sensors 2021, 21, 3115. [Google Scholar] [CrossRef]
- Hua, X.; Bao, Y.; Wang, H.; Chen, Z.; Wu, F. Bacteria-derived fluorescent carbon dots for microbial live/dead differentiation. Nanoscale 2017, 9, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Quan, K.; Jiang, G.; Liu, J.; Zhang, Z.; Ren, Y.; Busscher, H.J.; Van der Mei, H.C.; Peterson, B.W. Influence of interaction between surface-modified magnetic nanoparticles with infectious biofilm components in artificial channel digging and biofilm eradication by antibiotics in vitro and in vivo. Nanoscale 2021, 13, 4644–4653. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, J.; Yu, Q.; Zhang, J.; Niu, X.; Hao, L.; Yang, L.; Zhao, Y. Effects of salts on the self-assembly behavior and antibacterial activity of a surfactant-like peptide. Soft Matter 2018, 16, 9758–9768. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Yuan, X.; Song, Z.; Xu, S.; Yang, Y.; Yang, X. Gram-negative Escherichia coli promotes deposition of olymer-capped ilver nanoparticles in saturated porous media. Environ. Sci. Nano 2018, 5, 1495–1505. [Google Scholar] [CrossRef]
- Medvedová, A.; Valík, L. Staphylococcus aureus: Characterisation and quantitative growth description in milk and artisanal raw milk cheese production. In Structure and Function of Food Engineering; Eissa, A.A., Ed.; IntechOpen Limited: London, UK, 2012; Chapter 3. [Google Scholar]
- Wilks, J.C.; Slonczewski, J.L. pH of the Cytoplasm and periplasm of Escherichia coli: Rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 2007, 189, 5601–5607. [Google Scholar] [CrossRef] [Green Version]
- De Jonge, R.; Takumi, K.; Ritmeester, W.S.; Van Leusden, F.M. The adaptive response of Escherichia coli O157 in an environment with changing pH. J. Appl. Microbiol. 2003, 94, 555–560. [Google Scholar] [CrossRef]
- Sadekar, S.; Thiagarajan, G.; Bartlett, K.; Hubbard, D.; Ray, A.; McGill, L.D.; Ghandehari, H. Poly(amidoamine) dendrimers as absorption enhancers for oral delivery of camptothecin. Int. J. Pharm. 2013, 456, 175–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 31 December 2021).


| Probe | PAMAM Dendrimer | Phenylboronic Acid | DMT-MM | Number of Modifications | Yield |
|---|---|---|---|---|---|
| B4-PAMAM | 1 mL (1 eq, 5.73 μmol) | 5.72 mg (6 eq, 34.5 μmol) | 48.0 mg (30 eq, 174 μmol) | 4 | 73.6 mg |
| B7-PAMAM | 2 mL (1 eq, 11.46 μmol) | 15.18 mg (8.2 eq, 93.3 μmol) | 227.7 mg (72 eq, 823 μmol) | 7 | 101 mg |
| B8-PAMAM | 2 mL (1 eq, 11.46 μmol) | 18.97 mg (10 eq, 114 μmol) | 158 mg (50 eq, 572 μmol) | 8 | 126 mg |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikagi, A.; Manita, K.; Yoyasu, A.; Tsuchido, Y.; Kanzawa, N.; Hashimoto, T.; Hayashita, T. Rapid Bacterial Recognition over a Wide pH Range by Boronic Acid-Based Ditopic Dendrimer Probes for Gram-Positive Bacteria. Molecules 2022, 27, 256. https://doi.org/10.3390/molecules27010256
Mikagi A, Manita K, Yoyasu A, Tsuchido Y, Kanzawa N, Hashimoto T, Hayashita T. Rapid Bacterial Recognition over a Wide pH Range by Boronic Acid-Based Ditopic Dendrimer Probes for Gram-Positive Bacteria. Molecules. 2022; 27(1):256. https://doi.org/10.3390/molecules27010256
Chicago/Turabian StyleMikagi, Ayame, Koichi Manita, Asuka Yoyasu, Yuji Tsuchido, Nobuyuki Kanzawa, Takeshi Hashimoto, and Takashi Hayashita. 2022. "Rapid Bacterial Recognition over a Wide pH Range by Boronic Acid-Based Ditopic Dendrimer Probes for Gram-Positive Bacteria" Molecules 27, no. 1: 256. https://doi.org/10.3390/molecules27010256
APA StyleMikagi, A., Manita, K., Yoyasu, A., Tsuchido, Y., Kanzawa, N., Hashimoto, T., & Hayashita, T. (2022). Rapid Bacterial Recognition over a Wide pH Range by Boronic Acid-Based Ditopic Dendrimer Probes for Gram-Positive Bacteria. Molecules, 27(1), 256. https://doi.org/10.3390/molecules27010256







