Abstract
Glycoside hydrolase family 5 subfamily 8 (GH5_8) mannanases belong to Firmicutes, Actinomycetia, and Proteobacteria. The presence or absence of carbohydrate-binding modules (CBMs) present a striking difference. While various GH5_8 mannanases need a CBM for binding galactomannans, removal of the CBM did not affect activity of some, whereas it in other cases reduced the catalytic efficiency due to increased KM. Here, monomodular GH5_8 mannanases from Eubacterium siraeum (EsGH5_8) and Xanthomonas citri pv. aurantifolii (XcGH5_8) were produced and characterized to clarify if GH5_8 mannanases from Firmicutes and Proteobacteria without CBM(s) possess distinct properties. EsGH5_8 showed a remarkably high temperature optimum of 55 °C, while XcGH5_8 had an optimum at 30 °C. Both enzymes were highly active on carob galactomannan and konjac glucomannan. Notably, EsGH5_8 was equally active on both substrates, whereas XcGH5_8 preferred galactomannan. The KM values were comparable with those of catalytic domains of truncated GH5_8s, while the turn-over numbers (kcat) were in the higher end. Notably, XcGH5_8 bound to but did not degrade insoluble ivory nut mannan. The findings support the hypothesis that GH5_8 mannanases with CBMs target insoluble mannans found in plant cell walls and seeds, while monomodular GH5_8 members have soluble mannans and mannooligosaccharides as primary substrates.
1. Introduction
Many carbohydrate-active enzymes acting on polymeric substrates, especially insoluble substrates, contain one or more non-catalytic carbohydrate-binding modules (CBMs) together with the catalytic domain. CBMs can improve catalytic function of glycoside hydrolases (GHs) through targeting the enzymes to the substrate and increasing substrate-enzyme proximity as well as disrupting the crystallinity of insoluble substrates or stabilizing the enzyme [1,2,3,4]. Generally, both mono- and multi-modular members are found within carbohydrate-active enzyme families [5]. It is unclear if enzymes composed just of the catalytic domain (CD) have different ways to interact with the substrates. In some enzyme families, so-called substrate binding sites (SBSs) situated on the CD can play the same roles as CBMs [6,7]. Monomodular enzymes represent an advantage since (i) the enzyme is less energy consuming for the organism to produce, and (ii) it is less prone to proteolytic degradation. In particular, the latter is a gain for industrial processes.
A number of endo-β-1,4-mannanases (E.C. 3.2.1.78) have been characterized due to their potential applications in different sectors of industry including detergent, textile, food, animal feed, and bioethanol [8]. They hydrolyze internal backbone β-1,4-linkages in mannans including in substituted mannans such as galactomannans from carob/locust bean (low and high viscosity; CGM-lv and CGM-hv) and guar bean (guar gum; GG), as well as in the linear konjac glucomannan (KGM) containing β-1,4-linked mannose and glucose units.
Endo-β-1,4-mannanases occur in GH families 5, 26, 45, 113, and 134 according to the Carbohydrate Active Enzymes database (CAZy; http://www.cazy.org, accessed on 8 March 2022) [5] with GH5 and GH26 mannanases being the best characterized. GH family 5 contains a wide range of enzymes acting on β-linked oligo-, polysaccharides, and glycoconjugates from a large spectrum of organisms and endo-β-1,4-mannanase activity is recognized in GH5 subfamilies 7, 8, 10, 17, 25, and 36 [9]. According to the CAZy database, subfamilies GH5_7 and GH5_8 are the largest. Only a subset of GH5_7 members have one or more CBMs (of families 2, 3, 23, 27, 35, and 65), while GH5_8 members largely contain at least one CBM (of families 2, 3, 10, 13, 32, 35, 37, 59, 64, 65) with the cellulose-binding CBM2 being the most common. The Xanthomonas enzymes, constituting 30% of the GH5_8 members, are very similar and clear exceptions, as they never have a CBM. GH26 includes several different specificities and among the 61 characterised GH26 mannanases, 26 have CBMs (mainly CBM35). Interestingly, CBM occurs very rarely with the two β-mannanase families GH113 and GH134 [5].
The influence of CBMs on the activity of GH5_8 mannanases has been addressed by truncation analysis. Only one monomodular GH5_8 enzyme from Xanthomonas campestris was studied previously [10]; however, no specific activity or kinetics data are available. Six GH5_8 members, all occurring with a CBM, have been structure determined [11,12,13,14,15,16], but only one structure, of the Bacillus sp. JAMB-602 mannanase (PDB entry 1WKY), included the CBM (a CBM59) [11]. Complex structures with mannooligosaccharides have been reported for three of the enzymes and none have an indication of carbohydrate binding at SBSs outside the active sites. A GH113 mannanase found without CBMs has a recognized SBSs [17].
The CD of a GH5_8 mannanase from Saccharophagus degradans (SdGH5_8-CBM10x3) containing three CBM10s did not bind galactomannan, which was bound strongly by the full-length enzyme [18]. The same was observed for a Bifidobacterium animalis subsp. lactis Bl-0 mannanase (BaGH5_8-CBM10) with one CBM10 [19]. Notably, removal of the CBM10 from BaGH5_8 did not significantly affect kinetics parameters towards CGM-lv. By contrast, the kinetic parameters of SdGH5_8-CBM10x3 changed upon CBM removal. However, the scenario with its three CBM10s was more complicated, as they showed distinct binding specificity and affinity [18]. A GH134 β-1,4-mannanase with a CBM10 from Streptomyces sp. NRRL B-24484 has been shown to bind microcrystalline cellulose, β-mannan, and chitin regardless of the presence or absence of the CBM10, which was, however, shown to be important for protein stability [20].
Here, two monomodular GH5_8 mannanases from the gut bacterium Eubacterium siraeum (EsGH5_8) and from the plant pathogen Xanthomonas citri pv. aurantifolii (XcGH5_8) are recombinantly produced and biochemically characterized to investigate if monomodular GH5_8 mannanases display other properties than multimodular GH5_8 mannanases with regard to activity and polysaccharide binding. The two enzymes represent two phyla.
2. Results
2.1. Bioinformatics Analysis
The phylogenetic tree generated based on the GH5_8 CDs alone shows a clear grouping mainly based on the origin of the proteins, hence it seems like the evolution of the GH5_8 CDs reflected the taxonomy (Figure 1). Furthermore, the presence and type of CBM(s) are not necessarily following a specific pattern, though the majority of Actinomycetia GH5_8 members have one CBM, in most cases a CBM2, but with a subfraction having CBM10(s). In general, very few of the enzymes have a module N-terminally to the GH5_8 CD. Interestingly, a Firmicutes group appears among the Actinomycetia members. Out of the 298 protein sequences included in the tree, only 29 are predicted by dbCAN2 to not have a signal peptide. These intracellular proteins are distributed all over the tree and seven of them lack CBMs (data not shown).
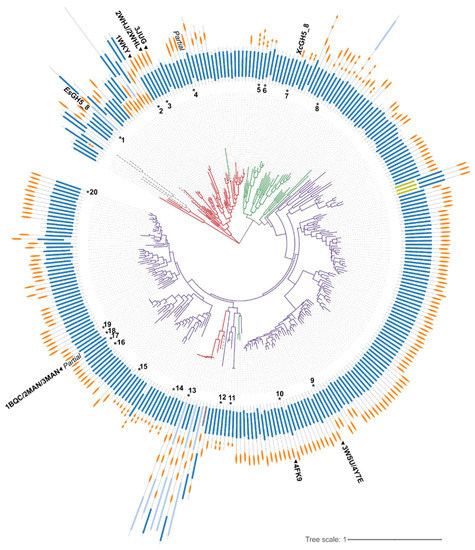
Figure 1.
Phylogenetic tree generated based on a structure-guided multiple alignment (see Figure S1, Supplementary Materials) of the GH5_8 CDs alone. The colouring of the branches follows the origin of the protein sequence (red, Firmicutes; green, Proteobacteria; purple, Actinomycetia; grey dashed lines, other organisms including uncultured organisms). The domain architectures of the full-length proteins are show in the outer ring (dark blue, GH5_8 CD; light blue, CDs from other GH families; light green, carbohydrate esterase family 3 CD; grey, auxiliary activities family 9 CD; orange, CBMs). The numbers refer to GH5_8 enzymes with specific activity and/or kinetic analysis included here (see Table 1 and Table 2 for details). Structure determined GH5_8 members are indicated by a triangle and their PDB entries. “Partial” refers to only partial sequence being available. See Figure S2 for the phylogenetic tree with accession numbers and information about domain families.
EsGH5_8 is found in a small subgroup of other gut Firmicutes (Ruminococcus species) GH5_8 enzymes, which have remarkably different modular architecture (Figure 1). Otherwise, in EsGH5_8, only one more Eubacterium GH5_8 sequence is found in CAZy and it also lacks a CBM [5]. XcGH5_8 is found in a subgroup together with monomodular GH5_8 proteins from other Xanthomonas species. The Xanthomonas protein sequences account for 230 out of the 785 members of GH5_8 in the CAZy database [4] and none of them contain a CBM.
The models of XcGH5_8 and EsGH5_8 were predicted using AlphaFold 2 and, as expected, were overall very similar to known GH5_8 structures (Figure 2A). The root mean square deviation (RMSD) between the models of EsGH5_8 and XcGH5_8 was 0.652 Å2, while it was 0.432–0.596 Å2 between the models and the structure determined GH5_8 mannanases included in Figure 2. A comparison of subsite residues of the Streptomyces thermolilacinus GH5_8 structure (PDB entry 4Y7E) [15], which has ligands covering both plus (aglycone) and minus (glycone) subsites, with the two models showing that most residues are conserved; although, residues around subsites +2 and +3 differ/are absent (Figure 2B). Furthermore, the residues proposed by Kumagai et al. to be involved in interaction with galactose branches in the substrate differ (residues 308–310) [15]. EsGH5_8 is closely related to the GH5_8 structures of other Firmicutes, while for XcGH5_8 it is important to note, that there is no available structure of a GH5_8 from the Actinomycetia phylum. When the electrostatic surface of the model of XcGH5_8 was compared with one of the closest related structures from S. thermolilacinus (PDB entry 4Y7E), it was clearly different. XcGH5_8 has large positively charged patches (Figure 2C), also in agreement with the high pI predicted to 9.0. Moreover, it had more of a closed active site cleft as compared to the structure of S. thermolilacinus (PDB entry 4Y7E). EsGH5_8 resembled the other Firmicutes GH5_8 structure (PDB entry 2WHL) with a relatively long and open active site cleft (Figure 2C).
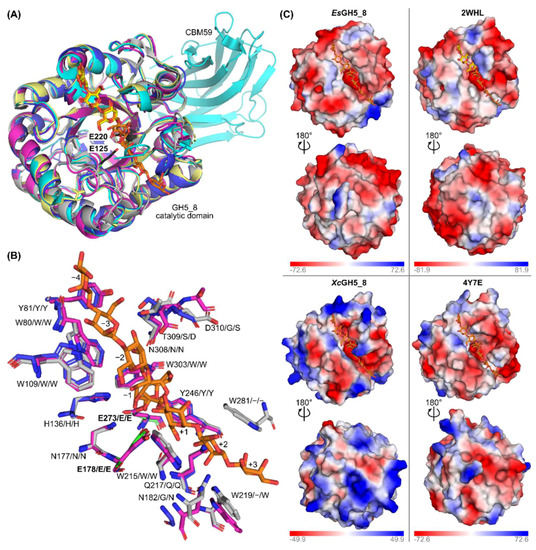
Figure 2.
Comparison of the AlphaFold models of EsGH5_8 and XcGH5_8 and the structures of GH5_8 from Bacillus agaradhaerens (PDB entry 2WHL), Bacillus sp. JAMB-602 (PDB entry 1WKY), and Streptomyces thermolilacinus (PDB entry 4Y7E). (A) Superimposition of the two models (EsGH5_8, dark blue; XcGH5_8, pink) and three structures (1WKY, cyan; 2WHL, pale yellow; 4Y7E, grey). The ligands from 2WHL (yellow) and 4Y7E (orange) is shown as sticks, while the catalytic residues of 2WHL are shown as black sticks. (B) Comparison of subsite residues of S. thermolilacinus (green sticks, catalytic residues; grey sticks, other subsite residues; orange, mannooligosaccharide ligands) (Kumagai 2015) with EsGH5_8 (blue sticks) and XcGH5_8 (pink sticks). Residue numbers refer to PDB entry 4Y7E, while following residue letters refer to EsGH5_8 and XcGH5_8, respectively. (C) Comparison of the electrostatic surface of the two models and the two related structures. The mannooligosaccharide ligands of 4Y7E is superimposed on all models and structures (orange sticks), while the ligand (mannotriose) of 2WHL is shown as yellow sticks.
2.2. Enzymatic Activity of EsGH5_8 and XcGH5_8
EsGH5_8 and XcGH5_8 were produced and purified to homogeneity in yields of 14.3 and 2.6 mg/g cells, respectively. The pH optimum of both EsGH5_8 and XcGH5_8 was 7, and the optimal activity was around 30 °C of XcGH5_8 and 55 °C of EsGH5_8 (Figure 3). Furthermore, melting temperatures (Tm) of XcGH5_8 and EsGH5_8 were determined by differential scanning fluorimentry to 37.9 °C and 57.8 °C, respectively (Figure S3). While the specific activity and kinetic parameters of XcGH5_8 were determined at its optimal temperature, EsGH5_8 was analysed at 37 °C, since most mannanases despite their activity temperature optimum are assayed at either 37 °C or 40 °C, which was also a biologically relevant temperature as EsGH5_8 is a gut bacterium. EsGH5_8 showed equally good activity towards CGMs and KGM, whereas XcGH5_8 had a clear preference for CGMs (Table 1). Both enzymes showed low activity on the more densely branched guar gum, as compared with other GH5_8 mannanases (Table 1). XcGH5_8 did not degrade ivory nut mannan (INM), and EsGH5_8 had low activity on INM, as compared to the few other GH5_8 mannanases characterized on this substrate (Table 1). None of the enzymes had activity on xanthan from X. campestris.
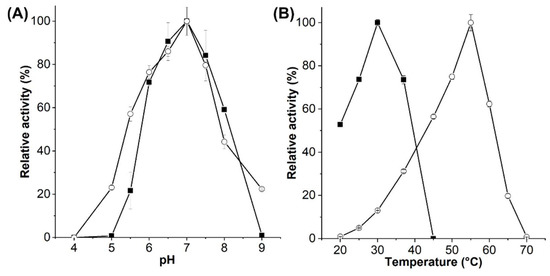
Figure 3.
pH (A) and temperature (B) optimum of EsGH5_8 (■) and XcGH5_8 (○).

Table 1.
Specific activity of GH5_8 mannanases. Enzymes are listed according to the groups on the phylogenetic tree (Figure 1).
Table 1.
Specific activity of GH5_8 mannanases. Enzymes are listed according to the groups on the phylogenetic tree (Figure 1).
| Origin (GenBank Accession; No. in Phylogenetic Tree) | Modular Structure of Charac. Protein | CGM-lv U/mg; 1/s (Relative 1, %) | CGM-hv U/mg; 1/s (Relative 1, %) | KGM U/mg 1/s (Relative 1, %) | GG U/mg; 1/s (Relative 1, %) | INM U/mg; 1/s (Relative 1, %) | Ref. |
|---|---|---|---|---|---|---|---|
| Eubacterium siraeum (CBK96294; #1) | GH5_8 | 625 ± 28.8; 397 ± 18.3 (97) | 578 ± 13.0; 367 ± 8.3 (90) | 644 ± 11.4; 409 ± 7.2 (100) | 3.6 ± 0.2; 2.3 ± 0.1 (0.6) | 7.5 ± 0.1; 4.8 ± 0.1 (1.2) | Present study |
| Saccharophagus degradans (ABD79918; #5) | GH5_8-CBM10x3 | 1972 ± 80; 1729 ± 70 (56) | 2212 ± 78; 1939 ± 68 (62) | 3544 ± 110; 3107 ± 96 (100) | 40 ± 8; 35 ± 7 (1.1) | 9 ± 1; 8 ± 1 (0.3) | [18] |
| GH5_8-ΔCBM10x3 | 2906 ± 53; 1695 ± 31 (64) | 3151 ± 304; 1838 ± 177 (69) | 4556 ± 108; 2658 ± 63 (100) | 108 ± 25; 63 ± 15 (2.4) | 81 ± 7; 47 ± 4 (1.8) | [18] | |
| Xanthomonas citri (AMU98328; #7) | GH5_8 | 713 ± 12.0; 414 ± 7.0 (100) | 709 ± 23.0; 411 ± 13,3 (99) | 247 ± 19.3; 143 ± 11.2 (35) | 1.1 ± 0.1; 0.6 ± 0.1 (0.2) | N.D. 2 | Present study |
| Bifidobacterium animalis (ACS46797; #11) | GH5_8-CBM10 | 1380; 872 (55) | 1920; 1213 (76) | 2520; 1592 (100) | N.D. 2 | 120; 76 (5) | [19] |
| Cellulosimicrobium sp. strain HY-13 (AEE43708; #12) | GH5_8-CBM10x2 | 8498 ± 105; 6232 ± 77 (58) | - | - | 967 ± 18; 709 ± 13 (6.6) | 14,711 ± 183; 10,788 ± 134 (100) | [21] |
| Streptomyces sp. S27 (ADK91085; #14) | GH5_8-CBM10 | 2107 ± 182; 1510 ± 130 (100) | - | 1312 ± 110; 940 ± 79 (62) | 74 ± 12; 53 ± 8.6 (3.5) | - | [22] |
| Streptomyces lividans (AAA26710; #15) | GH5_8-CBM10 | 141 ± 1.7 3,4 (100) | - | 55 ± 0.7 4 (39) | 21 ± 1.7 4 (15) | 18.8 ± 1.2 4 (13) | [23] |
| GH5_8-ΔCBM10 | 97 ± 1.4 3,4 (100) | - | 61 ± 0.45 4 (63) | 23 ± 3.1 4 (23) | 19 ± 0.7 4 (20) | [23] | |
| Streptomyces thermoluteus (BAM62868; #20) | GH5_8-CBM2 | 51 ± 1.6 3,4 (78) | - | 66 ± 1.5 4 (100) | 27 ± 1.4 4 (41) | 20.5 ± 0.4 4 (31) | [23] |
| StGH5_8-ΔCBM2 | 39 ± 0.6 3,4 (86) | - | 45 ± 0.5 4 (100) | 20 ± 3.7 4 (44) | 14 ± 1.3 4 (32) | [23] |
1 Relative to the best substrate of a given enzyme. 2 No activity detected. 3 It is not clear if assay was performed with low- or high-viscosity CGM. 4 1/s.
EsGH5_8 and XcGH5_8 showed different hydrolysis patterns of linear mannooligosaccharides (Figure 4): None of the enzymes hydrolysed mannobiose or mannotriose, but EsGH5_8 hydrolysed mannotetraose into mannose and mannotriose, while XcGH5_8 showed no activity on this oligosaccharide. Furthermore, both enzymes hydrolysed mannopentaose and mannohexaose, but the product profiles differed: EsGH5_8 released solely mannobiose and mannotriose from mannopentaose, while XcGH5_8 released mannose to mannotetraose. EsGH5_8 hydrolysed mannohexaose hydrolysed to mannose, mannobiose, and mannotriose, while XcGH5_8 produced mannobiose, mannotriose, and mannotetraose. This profile of XcGH5_8 on mannohexaose is in line with its inability to hydrolyse mannotetraose. The hydrolysis profile of CGM-lv is similar for the two enzymes (Figure 4).
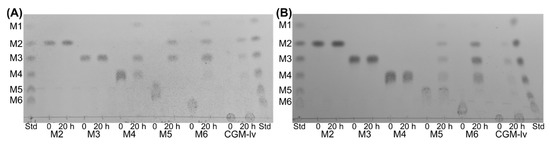
Figure 4.
Thin layer chromatography (TLC) of the hydrolytic products generated by EsGH5_8 (A) and XcGH5_8 (B) on mannooligosaccharides (mannobiose, M2, to mannohexaose, M6) and CGM-lv. The standard (Std) contained a mix of linear mannooligosaccharides (mannose to mannohexaose; M1–M6).
Kinetic analysis of the two enzymes on CGM-lv gave a Michaelis–Menten constant (KM) for EsGH5_8 of 4.6 mg/mL and 2.6 mg/mL for XcGH5_8 (Table 2). EsGH5_8 had a slightly higher highest turn-over number (kcat), but its relatively higher KM resulted in the lower catalytic efficiency (kcat/KM) of these two monomodular GH5_8 enzymes.

Table 2.
Kinetic parameters for GH5_8 enzymes on CGM-lv. The enzymes listed according to the groups of the phylogenetic tree (Figure 1).
Table 2.
Kinetic parameters for GH5_8 enzymes on CGM-lv. The enzymes listed according to the groups of the phylogenetic tree (Figure 1).
| Origin (Genbank Accession; No. in Phylogenetic Tree) | Modular Structure of Charac. Protein | KM (mg/mL) | kcat (1/s) | kcat/KM (mg/(mL s) | Binding CGM? | Ref. |
|---|---|---|---|---|---|---|
| Eubacterium siraeum (CBK96294; #1) | GH5_8 | 4.6 ± 0.5 | 850 ± 47 | 185 ± 23 | Yes, weakly | Present study |
| Bacillus sp. JAMB-602 (BAD99527; #2) | GH5_8-CBM59 | 3.1 | 135 1 | ND 2 | [24] | |
| Bacillus agaradhaerens (AAN27517; #3) | GH5_8-CBM59 | 1.8 | 633 | 250 | ND 2 | [13] |
| Bacillus nealsonii PN-11 (AGU71466; #4) | GH5_8-CBM59 | 7.2 ± 0.3 | 750 ± 55 3 | 104 ± 2 3 | ND 2 | [25] |
| Saccharophagus degradans (ABD79918; #5) | GH5_8-CBM10x3 | 2.1 ± 0.1 | 2333 ± 55 | 1096 ± 71 | Yes, Kd < 0.125 mg/mL | [18] |
| SdGH5_8-ΔCBM10x3 | 2.4 ± 0.1 | 3440 ± 75 | 1413 ± 82 | No | [18] | |
| Cellvibrio japonicus (ACE84673/AAO31759; #6) | GH5_8-ΔCBM10x2 | 8.5 ± 1.5 | 2381 ± 66 | 246.0 | No | [26] |
| Xanthomonas citri (AMU98328; #7) | GH5_8 | 2.6 ± 0.3 | 732 ± 36 | 282 ± 35 | ND 2 | Present study |
| Cellvibrio japonicus (AAO31760; #8) | GH5_8-ΔCBM10-CBM2 | 2.2 ± 0.3 | 1075 ± 27 | 446 | No | [26] |
| Streptomyces thermolilacinus (BAK26781; #9) | GH5_8-ΔCBM2 | 4.9 ± 1.0 | 21 ± 2 | 4 ± 1 | ND 2 | [15,27] |
| Streptomyces sp. SirexAA-E (AEN10237; #10) | GH5_8-Fn3-CBM2 | 2 ± 0.2 | 41 ± 2 | 21 ± 10 | Yes, weakly | [14] |
| GH5_8-ΔFn3-CBM2 | 2 ± 0.4 | 41 ± 3 | 21 ± 8 | No | [14] | |
| Bifidobacterium animalis (ACS46797; #11) | GH5_8-CBM10 | 1.6 ± 0.2 | 1828 ± 87 | 1157 ± 177 | Yes, Kd = 0.31 mg/ml | [19] |
| GH5_8-ΔCBM10 | 1.8 ± 0.5 | 2005 ± 179 | 1146 ± 324 | No | [19] | |
| Caldicellulosiruptor bescii (ACM60953; #13) | GH9-CBM3x3-GH5_8 | 0.6 ± 0.3 | 1420 ± 158 | 2290 | ND 2 | [28] |
| CBM3x3-GH5_8 | 1.8 ± 0.5 | 3446 ± 367 | 1893 | ND 2 | [28] | |
| Streptomyces sp. S27 (ADK91085; #14) | GH5_8-CBM10 | 0.16 | 3739 3 | 23369 3 | ND 2 | [22] |
| Streptomyces lividans (AAA26710; #15) | GH5_8-CBM10 | 3.5 ± 0.5 | 197 ± 11 | 60 ± 7 | ND 2 | [23] |
| GH5_8-ΔCBM10 | 4.3 ± 0.7 | 139 ± 9 | 33 ± 4 | ND 2 | [23] | |
| Thermobifida halotolerans (AHB89704; #16) | GH5_8-CBM2 | 1.3 ± 0.3 | 78 ± 9 | 60 | ND 2 | [29] |
| Thermobifida cellulosilytica (AHB89703; #17) | GH5_8-CBM2 | 0.8 ± 0.2 | 89 ± 5 | 106 | ND 2 | [29] |
| Thermobifida fusca (AAZ54938; #18) | GH5_8-ΔCBM2 | 10.4 ± 2.6 | 96 ± 14 | 9 ± 3 | ND 2 | [15] |
| Thermobifida fusca TM51 (AHB89702; #19) | GH5_8-CBM2 | 1.7 ± 0.4 | 122 ± 11 | 74 | ND 2 | [29] |
| Streptomyces thermoluteus (BAM62868; #20) | GH5_8-CBM2 | 5.5 ± 1.6 | 101 ± 17 | 18 ± 3 | ND 2 | [23] |
| GH5_8-ΔCBM2 | 5.5 ± 1.5 | 75 ± 12 | 14 ± 2 | ND 2 | [23] |
1Vmax (mg mannose/min/mg protein). 2 Not determined. 3 Vmax (µmol/mL/min) and Vmax/Km (µmol/min/mg).
2.3. Polysaccharide Interaction
Binding between EsGH5_8 to soluble CGM-lv was analysed using affinity gel electrophoresis (AGE). The migration of EsGH5_8 was slightly retarded in a CGM-lv concentration dependent manner (Figure 5A), but a binding constant could not be determined due to a combination of too low affinity and too high viscosity of CGM-lv at higher concentrations (highest in-gel concentration tested was 5 mg/mL). The high pI (9.0) of XcGH5_8 prevented use of the conventional AGE setup [30] and various alternative attempts were not successful. However, using a pull-down assay it was shown that XcGH5_8 bound to INM, whereas both EsGH5_8 and the control bovine serum albumin (BSA) showed very weak binding (Figure 5B). None of the proteins bound to microcrystalline cellulose (Avicel) or starch granules (data not shown).
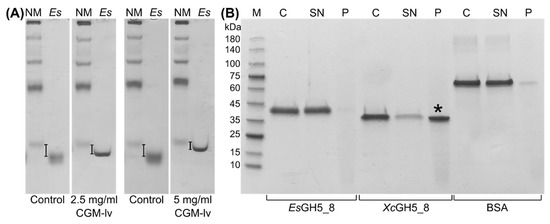
Figure 5.
Analysis of binding of EsGH5_8 and XcGH5_8 to polysaccharides. (A) Affinity gel-electrophoresis of EsGH5_8 (Es) at 2.5 and 5.0 mg/mL CGM-lv in the gel (NM, native marker functioning as reference). Control gels without CGM-lv are at the left. (B) Qualitative pull-down assay with INM (M, marker; C, the protein stock used for assay acting as a control; SN, supernatant; P, pellet). Asterisk indicates the significant binding of XcGH5_8 to INM. The original gels can be found in Figure S4.
3. Discussion
The EsGH5_8 and XcGH5_8 represent the first well-characterized monomodular GH5_8 mannanases. Though, temperature and pH optima were reported of a close XcGH5_8 homologue from X. campestris studied due to its function as virulence factor [10]. It had a temperature optimum around 37 °C, thus slightly higher than XcGH5_8 (Figure 3B), while both enzymes had a pH optimum of 7. EsGH5_8 has a remarkably high temperature optimum and Tm. The S. lividans GH5_8 with one CBM10 had a comparable Tm (60 °C) in the presence of calcium ions, but this decreased by approximately 15 °C after treatment with EDTA. Furthermore, it was concluded that the CBM10 was not affecting the thermal stability [23]. GH5_8 mannanases from the thermotolerant Thermobifida bacteria have temperature optima at 70–75 °C [29]. However, their turn-over numbers were in the range of 80–120 1/s at 50 °C, hence 7–10-fold lower than that of EsGH5_8 at 37 °C (Table 2), thus far below its optimal temperature.
Notably, EsGH5_8 degrades CGM and KGM equally good (Table 1). This is in contrast to the other characterized GH5_8 mannanases, which do prefer either CGM or KGM (Table 1). This seems to be reflected in the kinetics of CGM hydrolysis, as EsGH5_8 has a relatively high KM, but still displays better catalytic efficiency than the Thermobifida mannanases and the majority of the Streptomyces mannanases (Table 2). Some of the Streptomyces mannanases even show clear preferences for CGM (Table 1). Hence, EsGH5_8 is an interesting candidate for industrial applications due to its remarkable temperature stability and its high activity on both CGM and KGM.
The GH5_8 endo-β-1,4-mannanase from X. campestris was shown to be required for full virulence of the bacterium to plants. It was suggested that the role of the enzyme during disease is to promote transitions from an aggregated or biofilm lifestyle to a planktonic lifestyle [10,31]. However, the enzyme cannot degrade the mannose-containing exopolysaccharide xanthan produced by X. campestris [31]. In the present study, neither XcGH5_8 nor EsGH5_8 could degrade xanthan (data not shown).
Interestingly, the product profile of mannooligosaccharide hydrolysis by EsGH5_8 and XcGH5_8 was different indicating differences in subsite availability. The complete lack of activity of XcGH5_8 on mannotetraose is uncommon among the characterized GH5_8 mannanases. Notably, the observation is in agreement with mannotetraose being a hydrolysis product from mannohexaose. In the case of EsGH5_8, no mannotetraose is observed, most likely because it is efficiently hydrolysed. Usually, some activity of GH5_8 mannanases is detected on mannotetraose, though the activity generally increases significantly when increasing the length to 5 and 6 mannose units [13,15,19,26].
The kinetics of the Thermobifida enzymes clearly show that their CBM2 is important for activity. Thus, removal of CBM2 by truncation induced a six-fold increase of KM, and hence a crucial loss of efficiency (Table 2). The CBM is also important for activity of other characterized Actinomycetia GH5_8, but without similar strong impact on KM and the effect being seen on the turn-over number (Table 2). Interestingly, the removal of the CBM10 influenced the substrate preference of mannanase from S. lividans; the activity being reduced for the favored CGM and increased on KGM (Table 1).
EsGH5_8 is found in a subgroup of the phylogenetic tree with proteins having a very diverse modular architecture (Figure 1), but all originating from gut bacteria. This suggests that the catalytic domain evolved first and that the additional modules were incorporated later in evolution.
Interestingly, XcGH5_8 did not degrade INM, but could bind to INM, unlike EsGH5_8. The active site of both enzymes are predicted to have the same cleft-like shape (Figure 2B), hence the differences in activity on INM cannot be easily explained. Importantly, the interaction between XcGH5_8 and INM seemed to be specific, as no interaction was detected with microcrystalline cellulose (β-1,4-linked glucose units) or granular starch (α-1,4-linked glucose units). Notably, the activity of EsGH5_8 on INM was in the same range as of the full-length enzyme from S. degradans, the CD of which showed about a ten-fold better activity (Table 1). There seems to be no real pattern with regard to the effect of the presence of CBM on the activity on insoluble INM. Full-length GH5_8-CBM10x2 from C. japonicus bound to crystalline mannan, while the CD alone did not. The same was seen for the other C. japonicus mannanase (GH5_8-CBM10-CBM2), here CBM10 was important for mannan binding. In addition, none of the CBMs from the two C. japonicus mannanases was able to bind soluble CGM or KGM [26]. Unfortunately, the catalytic constants were only determined for the CDs alone (Table 2), which did not act on INM [26]. Interestingly, a mannanase (GH5_8-CBM10x2) from Cellulosimicrobium sp. HY-13 was also shown to bind INM as well as having the best specific activity towards INM (1.7-fold better than on CGM) (Table 1) [21].
The CBM2 and CBM10 of GH5_8 mannanases are known to facilitate binding to cellulose, despite the enzymes not being active on cellulose [14,18,21,26]. It has been suggested that the function of these CBMs is to target the enzymes to the mannan-containing plant cell-wall through interaction with the simple crystalline cellulose surface [18]. Along these lines, it is suggested that the GH5_8 mannanases with CBMs target insoluble mannans in plant cell-walls and seeds, while the monomodular GH5_8 members have soluble mannans and mannooligosaccharides as their primary substrates [13]. The latter also goes for GH5_8 members having CBMs not binding to insoluble polysaccharides, i.e., the mannanase from B. animalis, which has a CBM10 only capable of binding CGM but not insoluble cellulose or INM [19]. The monomodular group and the group of GH5_8 with CBMs not binding to insoluble polysaccharides might act together with other enzymes capable of degrading the insoluble mannans.
4. Materials and Methods
4.1. Carbohydrates
CGM-lv, INM, KGM, and linear β-1,4-mannooligosaccharides were from Megazyme (Wicklow, Ireland); CGM-hv (locust bean gum), GG, xanthan from Xanthomonas campestris, Avicel (microcrystalline cellulose), and unmodified wheat starch were from Sigma-Aldrich (Darmstadt, Germany).
4.2. Bioinformatics Analysis of GH5_8
Protein sequences for all GH5_8 proteins in the CAZy database [1] (778 sequences) were retrieved from NCBI and redundancy was reduced using CD-HIT [32] with a 90% identity cut-off. It was ensured that characterized protein sequences were maintained in the dataset. The resulting dataset contained 298 sequences. A structure-based alignment was generated using PROMALS3D [33] including only the GH5_8 catalytic domain (as predicted by dbCAN2 [34]). Representative structures of the six structures determined GH5_8 enzymes guided the alignment. Phylogenetic analysis was performed using the maximum likelihood method and the bootstrapping procedure with 500 bootstrap trials from the MEGA11 software suite [35]. The tree was displayed with the Interactive Tree Of Life (iTOL) online tool (https://itol.embl.de/ accessed on 8 March 2022) [36].
Models of EsGH5_8 and XcGH5_8 were predicted using AlphaFold run in the Colab notebook version [37].
4.3. Recombinant Protein Production
The genes encoding EsGH5_8 (GenBank accession no.: CBK96294) and XcGH5_8 (GenBank accession no.: AMU98328) without predicted signal peptides (amino acids 1–27 and 1–26, respectively) were purchased (GenScript, Leiden, The Netherlands) sub-cloned into pET28a using the NheI and XhoI restriction sites resulting in a cleavable N-terminal His-tag. The plasmids were transformed into E. coli BL21. Starter cultures (10 mL) were made by inoculating LB medium including 50 µg/mL kanamycin with a single colony and incubated at 37 °C overnight. LB medium (750 mL) containing 10 mM glucose and 50 µg/mL kanamycin in shake flask was inoculated with the overnight culture and propagated (37 °C, 160 rpm) to an absorbance at 600 nm of 0.6. The temperature was decreased to 16 °C and expression was induced by a final concentration of 0.1 mM IPTG. Cells were harvested (4000× g, 20 min, 4 °C) after approximately 20 h and stored at −20 °C until protein purification.
4.4. Protein Purification
The proteins were purified in two steps. Cells were resuspended in HisTrap equilibration buffer (10 mM Hepes, pH 7.4, 10 mM imidazole, 0.5 M NaCl, and 10% glycerol), lysed using a high-pressure homogenizer at 1 bar, added 3 μL of Benzonase Nuclease (Sigma-Aldrich), and centrifuged (40,000× g, 4 °C, 30 min). The supernatant was loaded onto a 5 mL HisTrap HP column (Cytiva, Marlborough, MA, USA) pre-equilibrated with HisTrap equilibration buffer and eluted using HisTrap elution buffer (10 mM Hepes, pH 7.4, 320 mM imidazole, 0.5 M NaCl, and 10% glycerol). The eluate was further purified by gel filtration (Superdex 16/60 75). EsGH5_8 was purified using 10 mM Hepes pH 7.4, 150 mM NaCl, while XcGH5_8 was purified using 10 mM Hepes at a pH of 7.0, 150 mM NaCl, and 10% glycerol. Fractions containing pure recombinant protein of correct size as based on SDS-PAGE were pooled and concentrated to 2–3 mg/mL (10 kDa Amicon Ultra; Merck-Millipore, Darmstadt, Germany)). XcGH5_8 was stored in 10 mM Hepes pH 7.0, 150 mM NaCl, and 30% glycerol, while EsGH5_8 was stored in the gel filtration buffer. The absorbance at 280 nm was measured, and protein concentration was determined using theoretical extinction coefficients (EsGH5_8, 78,380 M−1 cm−1 and molecular weight 38.1 kDa; XcGH5_8, 68,410 M−1 cm−1 and molecular weight 34.8 kDa).
4.5. Determination of Melting Temperature
The melting temperatures (Tm) of EsGH5_8 and XcGH5_8 were determined by differential scanning fluorimetry using a Prometheus Panta instrument (NanoTemper Technologies, München, Germany). Fluorescence at 330 nm and 350 nm upon excitation at 280 nm were measured using Prometheus NT.48 High Sensitivity capillaries (NanoTemper Technologies) with a protein concentration of 2 µM. The excitation power was determined from a pre-scan of the samples and set to 61%. The unfolding was followed by ramping the temperature from 20 to 95 °C with a temperature increment of 1 °C/min.
4.6. Enzymatic Assays
Enzymatic activity was determined using a standard reducing end 3,5-dinitrosalicylic acid (DNS) assay essentially as previously described [18,19,38]: 360 µL of substrate in assay buffer was preincubated at a given temperature for 5 min before the reaction was initiated by addition of 40 µL of enzyme solution in assay buffer. The reaction was terminated by addition of 600 µL of DNS reagent followed by heat treatment at 95 °C for 15 min. After 15 min in ice water, the samples were centrifuged at 20,000× g for 10 min. Finally, the absorbance was measured at 540 nm. Mannose was used as standard. One unit of mannanase activity was defined as the amount of enzyme that liberates reducing sugars equivalent to one µmol mannose per minute.
pH optimum was determined using the standard assay in triplicate using a final concentration of 2.5 mg/mL CGM-lv with enzyme (EsGH5_8, 7.4 nM; XcGH5_8, 15.3 nM) in a universal buffer (10 mM Hepes, 10 mM MES, 10 mM sodium acetate, and 75 mM NaCl) [39] in the range from pH 4 to 9.
The temperature optimum was determined using the standard assay with 2.5 mg/mL of CGM-lv at different temperatures (XcGH5_8, 20–45 °C; EsGH5_8, 20–65 °C) in 50 mM sodium phosphate-citrate pH 7.0 including 0.005% Triton X-100, as a buffer.
The specific activity was determined using the standard assay with 50 mM sodium phosphate-citrate pH 7.0, 0.005% Triton X-100, as buffer at 37 °C (EsGH5_8) and 30 °C (XcGH5_8). EsGH5_8 (3.5–28 nM) and XcGH5_8 (5.1–51.1 nM) were incubated with low- and high-viscosity CGMs (2.5 mg/mL, 10 min), KGM (2.5 mg/mL, 10 min), GG (2.5 mg/mL, 2.5 h), xanthan (2.5 mg/mL, 2.5 h), or INM (5 mg/mL, 45 min) in triplicate.
The kinetics on CGM-lv were determined using the same buffer and temperatures as for specific activity determination with a range of substrate concentrations (0.45–9 mg/mL CGM-lv) and enzyme (EsGH5_8, 5.0 nM; XcGH5_8, 5.4 nM) in 2000 μL and withdrawing 400 μL aliquots at 3, 6, 9, and 12 min. The kinetic assays were done in triplicate. The Michaelis–Menten model was fitted to the initial velocity data using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA).
4.7. Thin Layer Chromatography (TLC)
The product profile from hydrolysis of mannooligosaccharides was analysed using TLC: 50 µL of the reaction mixture, containing 2 mM of mannooligosaccharide (mannobiose to mannohexaose) or 2.5 mg/mL of CGM-lv and enzyme (EsGH5_8, 7 nM; XcGH5_8, 10 nM) in 50 mM sodium phosphate-citrate pH 7.0, 0.005% Triton X-100, was incubated at 25 °C (XcGH5_8) or 37 °C (EsGH5_8) for 20 h followed by 10 min at 95 °C to inactivate the enzyme. A sample (2 µL) was applied twice on a silica gel 60 F454 plate (Merck Millipore) and 2 µL of a mixture of 2 mM mannooligosaccharides (mannose to mannohexaose) was applied as standard. The separation was carried out in isopropanol:n-butanol:water (12:4:5) (v/v) as the mobile phase and run twice. Sugars were visualized with a diphenylamine alanine solution (12 mM diphenylamine, 2% aniline, and 8.5% phosphoric acid in methanol) and heat treatment.
4.8. Affinity Gel-Electrophoresis (AGE)
Qualitative screening of binding of EsGH5_8 was performed by AGE with 2.5 mg/mL and 5 mg/mL of CGM-lv in 10% acrylamide gels without stacking gels at pH 7.4 (43 mM imidazole and 35 mM Hepes) [30]. Protein (5 μL 0.4 mg/mL) in loading buffer was applied to polysaccharide-containing and control (without polysaccharide) gels, respectively. NativeMark unstained protein standard (Invitrogen) was included on all gels for normalization. The gels were run (4 h at 4 °C and 100 V) and stained using InstantBlue (Expedeon, Cambridge, UK).
4.9. Pull-Down Assay
A qualitative screening of the binding of EsGH5_8 and XcGH5_8 to insoluble crystalline INM, microcrystalline cellulose (Avicel), and wheat starch, was performed by a pull-down assay, where the samples were analysed by SDS-PAGE: 10 mg insoluble polysaccharide (prewashed with buffer three times) was mixed with 200 μL of 0.5 mg/mL protein in an assay buffer (50 mM sodium phosphate-citrate buffer pH 7.0, 0.005% Triton X-100). Bovine serum albumin (BSA; Sigma-Aldrich) was used as a control. The samples were incubated at 4 °C for 1 h with gentle agitation and then centrifuged (20,000× g, 10 min, 4 °C). Supernatants were transferred to fresh tubes and centrifuged again, before 4 μL of a supernatant was heat treated in the presence of SDS-loading buffer and subjected to SDS-PAGE. The pellets from the pull-down assay were washed with 250 μL of assay buffer and pelleted as aforementioned, resuspended in 200 μL of assay buffer, added 50 μL of SDS-loading buffer, boiled for 10 min, and applied (5 μL) on the SDS-PAGE. For each protein, a sample of the 0.5 mg/mL protein stock used for the pull-down assay was treated as the supernatant sample and included on the gel.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/molecules27061915/s1, Figure S1: Structure-guided multiple sequence alignment of GH5_8 catalytic domains; Figure S2: Phylogenetic tree with accession and domain family numbers; Figure S3: Differential scanning fluorimetry (DSF) results for EsGH5_8 and XcGH5_8. Figure S4: Original AGE gels related to Figure 5.
Funding
This research was funded by Independent Research Fund Denmark|Natural Science (DFF-6108-00476B) by a grant to the project “Roles and structural determinants of low-affinity carbohydrate-protein interactions”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All available data is included in the article.
Acknowledgments
I thank Birte Svensson (Technical University of Denmark) for great discussions and constructive advice. Karina Jansen (Technical University of Denmark) is thanked for technical assistance.
Conflicts of Interest
The author declares no conflict of interest.
Sample Availability
All compounds used in the study is commercially available.
References
- Gilbert, H.J.; Knox, J.P.; Boraston, A.B. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 2013, 23, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Sidar, A.; Albuquerque, E.D.; Voshol, G.P.; Ram, A.F.J.; Vijgenboom, E.; Punt, P.J. Carbohydrate binding modules: Diversity of domain architecture in amylases and cellulases from filamentous microorganisms. Front. Bioeng. Biotechnol. 2020, 8, 871. [Google Scholar] [CrossRef] [PubMed]
- Guillén, D.; Sánchez, S.; Rodríguez-Sanoja, R. Carbohydrate-binding domains: Multiplicity of biological roles. Appl. Microbiol. Biotechnol. 2010, 85, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Cockburn, D.; Svensson, B. Surface binding sites in carbohydrate active enzymes: An emerging picture of structural and functional diversity. In Carbohydrate Chemistry; Rauter, A.P., Lindhorst, T., Eds.; Royal Society of Chemistry: Cambridge, UK, 2013; Volume 39, pp. 204–221. ISBN 9781849735872. [Google Scholar]
- Cuyvers, S.; Dornez, E.; Delcour, J.A.; Courtin, C.M. Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit. Rev. Biotechnol. 2012, 32, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Dawood, A.; Ma, K. Applications of microbial β-mannanases. Front. Bioeng. Biotechnol. 2020, 8, 598630. [Google Scholar] [CrossRef]
- Aspeborg, H.; Coutinho, P.M.; Wang, Y.; Brumer, H.; Henrissat, B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 2012, 12, 186. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, Y.M.; Liu, Y.F.; Fang, M.C.; Tseng, Y.H. Transcriptional regulation and molecular characterization of the manA gene encoding the biofilm dispersing enzyme mannan endo-1,4-β-mannosidase in Xanthomonas campestris. J. Agric. Food Chem. 2010, 58, 1653–1663. [Google Scholar] [CrossRef]
- Akita, M.; Takeda, N.; Hirasawa, K.; Sakai, H.; Kawamoto, M.; Yamamoto, M.; Grant, W.D.; Hatada, Y.; Ito, S.; Horikoshi, K. Crystallization and preliminary X-ray study of alkaline mannanase from an alkaliphilic Bacillus isolate. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, D60, 1490–1492. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhang, Y.; Cao, Y.; Qi, J.; Mao, L.; Xue, Y.; Gao, F.; Peng, H.; Wang, X.; Gao, G.F.; et al. Structural analysis of alkaline β-Mannanase from alkaliphilic Bacillus sp. N16-5: Implications for adaptation to alkaline conditions. PLoS ONE 2011, 6, e14608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tailford, L.E.; Ducros, V.M.A.; Flint, J.E.; Roberts, S.M.; Morland, C.; Zechel, D.L.; Smith, N.; Bjørnvad, M.E.; Borchert, T.V.; Wilson, K.S.; et al. Understanding how diverse β-mannanases recognize heterogeneous substrates. Biochemistry 2009, 48, 7009–7018. [Google Scholar] [CrossRef] [PubMed]
- Takasuka, T.E.; Acheson, J.F.; Bianchetti, C.M.; Prom, B.M.; Bergeman, L.F.; Book, A.J.; Currie, C.R.; Fox, B.G. Biochemical properties and atomic resolution structure of a proteolytically processed β-mannanase from cellulolytic Streptomyces sp. SirexAA-E. PLoS ONE 2014, 9, e94166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumagai, Y.; Yamashita, K.; Tagami, T.; Uraji, M.; Wan, K.; Okuyama, M.; Yao, M.; Kimura, A.; Hatanaka, T. The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition. FEBS J. 2015, 282, 4001–4014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilge, M.; Gloor, S.M.; Rypniewski, W.; Sauer, O.; Heightman, T.D.; Zimmermann, W.; Winterhalter, K.; Piontek, K. High-resolution native and complex structures of thermostable β-mannanase from Thermomonospora fusca-substrate specificity in glycosyl hydrolase family 5. Structure 1998, 6, 1433–1444. [Google Scholar] [CrossRef]
- You, X.; Qin, Z.; Yan, Q.; Yang, S.; Li, Y.; Jiang, Z. Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 β-1,4-mannanase from Amphibacillus xylanus. J. Biol. Chem. 2018, 293, 11746–11757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Møller, M.S.; El Bouaballati, S.; Henrissat, B.; Svensson, B. Functional diversity of three tandem C-terminal carbohydrate-binding modules of a β-mannanase. J. Biol. Chem. 2021, 296, 100638. [Google Scholar] [CrossRef] [PubMed]
- Morrill, J.; Kulcinskaja, E.; Sulewska, A.M.; Lahtinen, S.; Stålbrand, H.; Svensson, B.; Abou Hachem, M. The GH5 1,4-β-mannanase from Bifidobacterium animalis subsp. lactis Bl-04 possesses a low-affinity mannan-binding module and highlights the diversity of mannanolytic enzymes. BMC Biochem. 2015, 16, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakai, K.; Kimoto, S.; Shinzawa, Y.; Minezawa, M.; Suzuki, K.; Jindou, S.; Kato, M.; Shimizu, M. Characterization of pH-tolerant and thermostable GH 134 β-1,4-mannanase SsGH134 possessing carbohydrate binding module 10 from Streptomyces sp. NRRL B-24484. J. Biosci. Bioeng. 2018, 125, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Ham, S.J.; Lee, H.J.; Cho, H.Y.; Kim, J.H.; Kim, Y.J.; Shin, D.H.; Rhee, Y.H.; Son, K.H.; Park, H.Y. Cloning and characterization of a modular GH5 β-1,4-mannanase with high specific activity from the fibrolytic bacterium Cellulosimicrobium sp. strain HY-13. Bioresour. Technol. 2011, 102, 9185–9192. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Yuan, T.; Zhao, J.; Huang, H.; Luo, H.; Meng, K.; Wang, Y.; Yao, B. Genetic and biochemical characterization of a protease-resistant mesophilic β-mannanase from Streptomyces sp. S27. J. Ind. Microbiol. Biotechnol. 2011, 38, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Kawakami, K.; Uraji, M.; Hatanaka, T. Binding of bivalent ions to actinomycete mannanase is accompanied by conformational change and is a key factor in its thermal stability. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Takeda, N.; Hirasawa, K.; Uchimura, K.; Nogi, Y.; Hatada, Y.; Akita, M.; Usami, R.; Yoshida, Y.; Grant, W.D.; Ito, S.; et al. Alkaline mannanase from a novel species of alkaliphilic Bacillus. J. Appl. Glycosci. 2004, 51, 229–236. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, P.S.; Sharma, P.; Puri, N.; Gupta, N. Purification and characterization of an alkali-thermostable β-mannanase from Bacillus nealsonii PN-11 and its application in mannooligosaccharides preparation having prebiotic potential. Eur. Food Res. Technol. 2014, 238, 927–936. [Google Scholar] [CrossRef]
- Hogg, D.; Pell, G.; Dupree, P.; Goubet, F.; Martín-Orúe, S.M.; Armand, S.; Gilbert, H.J. The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem. J. 2003, 371, 1027–1043. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Uraji, M.; Wan, K.; Okuyama, M.; Kimura, A.; Hatanaka, T. Molecular insights into the mechanism of thermal stability of actinomycete mannanase. FEBS Lett. 2016, 590, 2862–2869. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Mackie, R.I.; Cann, I.K.O. Biochemical and mutational analyses of a multidomain cellulase/mannanase from Caldicellulosiruptor bescii. Appl. Environ. Microbiol. 2012, 78, 2230–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tóth, Á.; Barna, T.; Szabó, E.; Elek, R.; Hubert, Á.; Nagy, I.; Nagy, I.; Kriszt, B.; Táncsics, A.; Kukolya, J. Cloning, expression and biochemical characterization of endomannanases from Thermobifida species isolated from different niches. PLoS ONE 2016, 11, e155769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cockburn, D.; Wilkens, C.; Svensson, B. Chapter 9: Affinity electrophoresis for analysis of catalytic module-carbohydrate interactions. In Protein-Carbohydrate Interactions: Methods and Protocols; Abbott, D.W., Lammerts van Bueren, A., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1588, pp. 119–127. ISBN 978-1-4939-6898-5. [Google Scholar]
- Dow, J.M.; Crossman, L.; Findlay, K.; He, Y.Q.; Feng, J.X.; Tang, J.L. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 2003, 100, 10995–11000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Pei, J.; Tang, M.; Grishin, N. V PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, W30–W34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Zhang, H.; Wu, P.; Entwistle, S.; Li, X.; Yohe, T.; Yi, H.; Yang, Z.; Yin, Y. dbCAN-seq: A database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 2018, 46, D516–D521. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Dilokpimol, A.; Nakai, H.; Gotfredsen, C.H.; Baumann, M.J.; Nakai, N.; Abou Hachem, M.; Svensson, B. Recombinant production and characterisation of two related GH5 endo-β-1,4-mannanases from Aspergillus nidulans FGSC A4 showing distinctly different transglycosylation capacity. Biochim. Biophys. Acta Proteins Proteom. 2011, 1814, 1720–1729. [Google Scholar] [CrossRef] [PubMed]
- Brooke, D.; Movahed, N.; Bothner, B. Universal buffers for use in biochemistry and biophysical experiments. AIMS Biophys. 2015, 2, 336–342. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).