Bioassay-Guided Isolation and Identification of Antibacterial Compounds from Invasive Tree of Heaven Stem and Trunk Bark
Abstract
1. Introduction
2. Results and Discussion
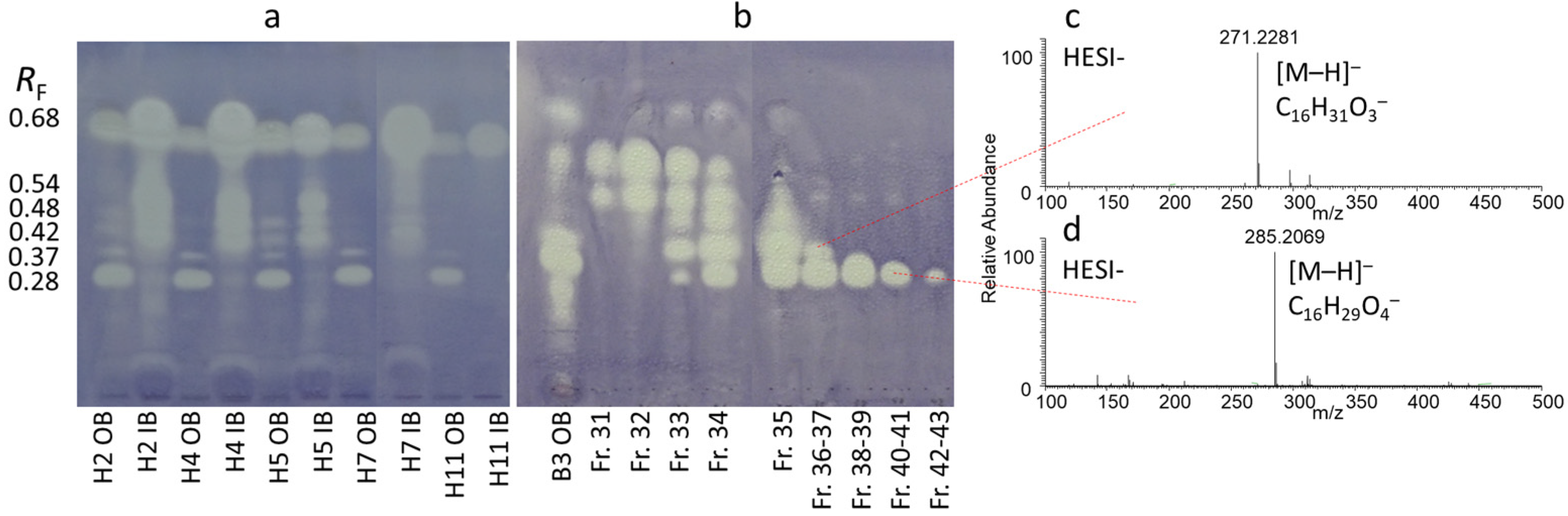
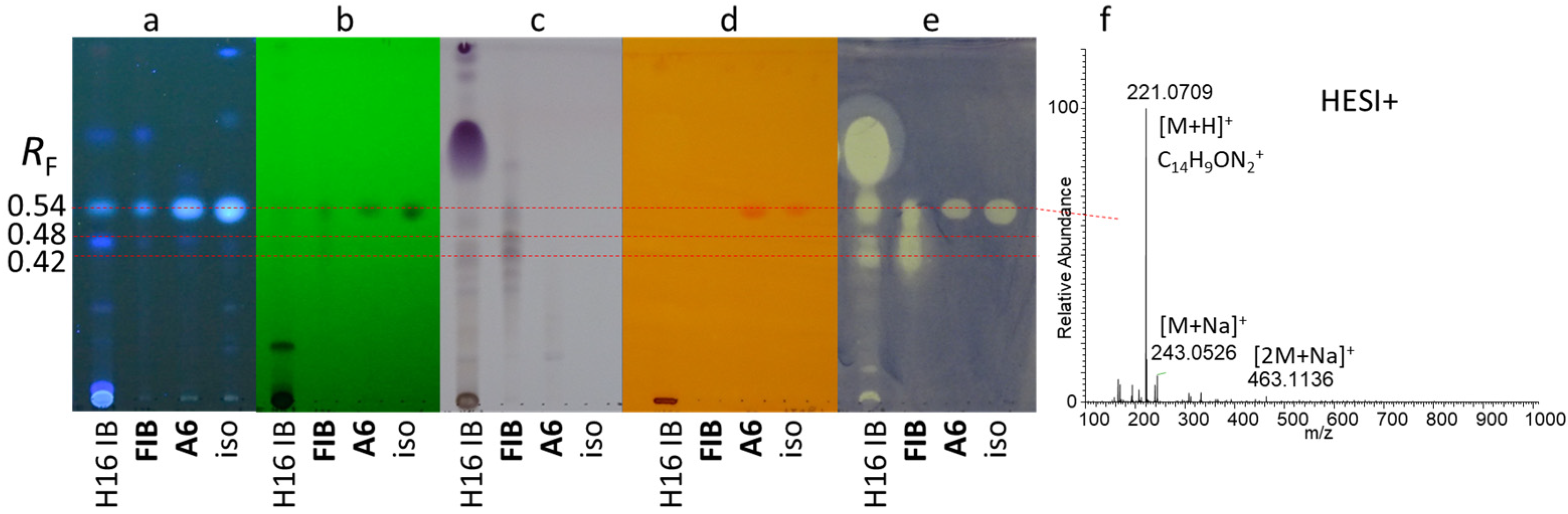
2.1. Detection and Isolation of Antibacterial Compounds
2.2. NMR Results
2.3. Structure Elucidation of the Isolates
2.4. Confirmation of the Antibacterial Activity of Isolated Compounds
3. Materials and Methods
3.1. Materials
3.2. Sample Origin and Preparation
3.3. Thin-Layer Chromatography
3.4. TLC–DB
3.5. TLC–HRMS/MS and FIA–HRMS/MS
3.6. Fractionation and Isolation of Bioactive Compounds
3.6.1. Fractionation and Isolation of Bioactive Compounds from the Stem Bark of Young Branches
3.6.2. Fractionation and Isolation of Bioactive Compounds from Outer Trunk Bark
3.6.3. Inner Trunk Bark
3.7. NMR Spectroscopy
3.8. Bacillus subtilis Microplate Assay of the Isolated Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.J.; Chipeta, M.G.; Haines-Woodhouse, G.; Kumaran, E.P.A.; Hamadani, B.H.K.; Zaraa, S.; Henry, N.J.; Deshpande, A.; Reiner, R.C.; Day, N.P.J.; et al. Global Antibiotic Consumption and Usage in Humans, 2000–2018: A Spatial Modelling Study. Lancet Planet Health 2021, 5, 893–904. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic Resistance: A Rundown of a Global Crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Huemer, M.; Mairpady Shambat, S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic Resistance and Persistence—Implications for Human Health and Treatment Perspectives. EMBO Rep. 2020, 21, 51034. [Google Scholar] [CrossRef] [PubMed]
- Chopra, B.; Dhingra, A.K. Natural Products: A Lead for Drug Discovery and Development. Phytother. Res. 2021, 35, 4660–4702. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and Resupply of Pharmacologically Active Plant-Derived Natural Products: A Review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed]
- Porras, G.; Chassagne, F.; Lyles, J.T.; Marquez, L.; Dettweiler, M.; Salam, A.M.; Samarakoon, T.; Shabih, S.; Farrokhi, D.R.; Quave, C.L. Ethnobotany and the Role of Plant Natural Products in Antibiotic Drug Discovery. Chem. Rev. 2021, 121, 3495–3560. [Google Scholar] [CrossRef]
- Kowarik, I.; Säumel, I. Biological Flora of Central Europe: Ailanthus altissima (Mill.) Swingle. Perspect. Plant Ecol. Evol. Syst. 2007, 8, 207–237. [Google Scholar] [CrossRef]
- Soler, J.; Izquierdo, J. The Invasive Ailanthus altissima: A Biology, Ecology, and Control Review. Plants 2024, 13, 931. [Google Scholar] [CrossRef]
- Sladonja, B.; Sušek, M.; Guillermic, J. Review on Invasive Tree of Heaven (Ailanthus altissima (Mill.) Swingle) Conflicting Values: Assessment of Its Ecosystem Services and Potential Biological Threat. Environ. Manag. 2015, 56, 1009–1034. [Google Scholar] [CrossRef] [PubMed]
- Casella, F.; Vurro, M. Ailanthus altissima (Tree of Heaven): Spread and Harmfulness in a Case-Study Urban Area. Arboric. J. 2013, 35, 172–181. [Google Scholar] [CrossRef]
- Wickert, K.L.; O’Neal, E.S.; Davis, D.D.; Kasson, M.T. Seed Production, Viability, and Reproductive Limits of the Invasive Ailanthus altissima (Tree-of-Heaven) within Invaded Environments. Forests 2017, 8, 226. [Google Scholar] [CrossRef]
- Motard, E.; Muratet, A.; Clair-Maczulajtys, D.; MacHon, N. Does the Invasive Species Ailanthus altissima Threaten Floristic Diversity of Temperate Peri-Urban Forests? Comptes Rendus Biol. 2011, 334, 872–879. [Google Scholar] [CrossRef]
- Terzi, M.; Fontaneto, D.; Casella, F. Effects of Ailanthus altissima Invasion and Removal on High-Biodiversity Mediterranean Grasslands. Environ. Manag. 2021, 68, 914–927. [Google Scholar] [CrossRef]
- Martí-Garrido, J.; Corominas, M.; Castillo-Fernández, M.; Belmonte, J.; Pineda, F.; Lleonart, R. Allergy to Ailanthus altissima Pollen: A Local Allergen to Consider. J. Investig. Allergol. Clin. Immunol. 2020, 30, 452–454. [Google Scholar] [CrossRef]
- Bennett, W.O.; Paget, J.T.; MacKenzie, D. Surgery for a Tree Surgeon? Acute Presentation of Contact Dermatitis Due to Ailanthus altissima. J. Plast. Reconstr. Aesthetic Surg. 2013, 66, 79–80. [Google Scholar] [CrossRef]
- Prenzel, F.; Treudler, R.; Lipek, T.; Vom Hove, M.; Kage, P.; Kuhs, S.; Kaiser, T.; Bastl, M.; Bumberger, J.; Genuneit, J.; et al. Invasive Growth of Ailanthus altissima Trees Is Associated with a High Rate of Sensitization in Atopic Patients. J. Asthma Allergy 2022, 15, 1217–1226. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Ma, S.; Zhao, Q.; Wu, J.; Duan, L.; Xie, Y.; Wang, S. Traditional Uses, Phytochemistry, and Pharmacology of Ailanthus altissima (Mill.) Swingle Bark: A Comprehensive Review. J. Ethnopharmacol. 2021, 275, 114121. [Google Scholar] [CrossRef]
- Fang, C.; Wu, W.; Ni, Z.; Liu, Y.; Luo, J.; Zhou, Y.; Gong, C.; Hu, D.; Yao, C.; Chen, X.; et al. Ailanthone Inhibits Non-Small Cell Lung Cancer Growth and Metastasis through Targeting UPF1/GAS5/ULK1 Signaling Pathway. Phytomedicine 2024, 128, 155333. [Google Scholar] [CrossRef]
- Kim, H.M.; Kim, S.J.; Kim, H.Y.; Ryu, B.; Kwak, H.; Hur, J.; Choi, J.H.; Jang, D.S. Constituents of the Stem Barks of Ailanthus altissima and Their Potential to Inhibit LPS-Induced Nitric Oxide Production. Bioorg. Med. Chem. Lett. 2015, 25, 1017–1020. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.M.; Lee, J.S.; Sezirahiga, J.; Kwon, J.; Jeong, M.; Lee, D.; Choi, J.H.; Jang, D.S. A New Canthinone-Type Alkaloid Isolated from Ailanthus altissima Swingle. Molecules 2016, 21, 642. [Google Scholar] [CrossRef] [PubMed]
- Vieira Torquato, H.F.; Ribeiro-Filho, A.C.; Buri, M.V.; Araújo Júnior, R.T.; Pimenta, R.; de Oliveira, J.S.R.; Filho, V.C.; Macho, A.; Paredes-Gamero, E.J.; de Oliveira Martins, D.T. Canthin-6-One Induces Cell Death, Cell Cycle Arrest and Differentiation in Human Myeloid Leukemia Cells. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 958–967. [Google Scholar] [CrossRef]
- Yang, J.G.; Dang, Y.G.; Li, G.Y.; Guo, L.J.; Wang, W.T.; Tan, Q.W.; Lin, Q.Y.; Wu, Z.J.; Xie, L.H. Note: Anti-Viral Activity of Ailanthus altissima Crude Extract on Rice stripe Virus in Rice Suspension Cells. Phytoparasitica 2008, 36, 405–408. [Google Scholar] [CrossRef]
- Pedersini, C.; Bergamin, M.; Aroulmoji, V.; Baldini, S.; Picchio, R.; Gutierrez Pesce, P.; Ballarin, L.; Murano, E. Herbicide Activity of Extracts from Ailanthus altissima (Simaroubaceae). Nat. Prod. Commun. 2011, 6, 593–596. [Google Scholar] [CrossRef]
- Tanasković, S.; Gvozdenac, S.; Kolarov, R.; Bursić, V.; Konstantinović, B.; Prvulović, D. Antifeeding and Insecticidal Activity of Ailanthus altissima and Morus alba Extracts against Gipsy Moth (Lymantria dispar (L.), Lepidoptera, Lymantridae) Larvae under Laboratory Conditions. J. Entomol. Res. Soc. 2021, 23, 197–212. [Google Scholar] [CrossRef]
- Morlock, G.E. High-Performance Thin-Layer Chromatography Combined with Effect-Directed Assays and High-Resolution Mass Spectrometry as an Emerging Hyphenated Technology: A Tutorial Review. Anal. Chim. Acta 2021, 1180, 338644. [Google Scholar] [CrossRef]
- Móricz, Á.M.; Ott, P.G. Effects-Directed Detection: Cell-Based Assays. In Instrumental Thin-Layer Chromatography, 2nd ed.; Poole, C.F., Ed.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 259–296. [Google Scholar] [CrossRef]
- Móricz, Á.M.; Ott, P.G.; Yüce, I.; Darcsi, A.; Béni, S.; Morlock, G.E. Effect-Directed Analysis via Hyphenated High-Performance Thin-Layer Chromatography for Bioanalytical Profiling of Sunflower Leaves. J. Chromatogr. A 2018, 1533, 213–220. [Google Scholar] [CrossRef]
- Azadniya, E.; Goldoni, L.; Bandiera, T.; Morlock, G.E. Same Analytical Method for Both (Bio)Assay and Zone Isolation to Identify/Quantify Bioactive Compounds by Quantitative Nuclear Magnetic Resonance Spectroscopy. J. Chromatogr. A 2020, 1628, 461434. [Google Scholar] [CrossRef]
- Móricz, Á.M.; Jamshidi-Aidji, M.; Krüzselyi, D.; Darcsi, A.; Böszörményi, A.; Csontos, P.; Béni, S.; Ott, P.G.; Morlock, G.E. Distinction and Valorization of 30 Root Extracts of Five Goldenrod (Solidago) Species. J. Chromatogr. A 2020, 1611, 460602. [Google Scholar] [CrossRef]
- Móricz, Á.M.; Baglyas, M.; Darcsi, A.; Balla, J.; Morlock, G.E. New Antioxidant Caffeate Esters of Fatty Alcohols Identified in Robinia pseudoacacia. Molecules 2024, 29, 5673. [Google Scholar] [CrossRef]
- Baglyas, M.; Ott, P.G.; Garádi, Z.; Glavnik, V.; Béni, S.; Vovk, I.; Móricz, Á.M. High-Performance Thin-Layer Chromatography—Antibacterial Assay First Reveals Bioactive Clerodane Diterpenes in Giant Goldenrod (Solidago gigantea Ait.). J. Chromatogr. A 2022, 1677, 463308. [Google Scholar] [CrossRef]
- Baglyas, M.; Ott, P.G.; Schwarczinger, I.; Nagy, J.K.; Darcsi, A.; Bakonyi, J.; Móricz, Á.M. Antimicrobial Diterpenes from Rough Goldenrod (Solidago rugosa Mill.). Molecules 2023, 28, 3790. [Google Scholar] [CrossRef]
- Jork, H.; Funk, W.; Fischer, W.; Wimmer, H.; Burns, D.T. Thin-Layer Chromatography: Reagents and Detection Methods; VCH: Weinheim, Germany, 1990. [Google Scholar]
- Móricz, Á.M.; Krüzselyi, D.; Alberti, Á.; Darcsi, A.; Horváth, G.; Csontos, P.; Béni, S.; Ott, P.G. Layer Chromatography-Bioassays Directed Screening and Identification of Antibacterial Compounds from Scotch Thistle. J. Chromatogr. A 2017, 1524, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Casillas-Vargas, G.; Ocasio-Malavé, C.; Medina, S.; Morales-Guzmán, C.; Del Valle, R.G.; Carballeira, N.M.; Sanabria-Ríos, D.J. Antibacterial Fatty Acids: An Update of Possible Mechanisms of Action and Implications in the Development of the next-Generation of Antibacterial Agents. Prog. Lipid Res. 2021, 82, 101093. [Google Scholar] [CrossRef]
- Koch, E.; Löwen, A.; Kampschulte, N.; Plitzko, K.; Wiebel, M.; Rund, K.M.; Willenberg, I.; Schebb, N.H. Beyond Autoxidation and Lipoxygenases: Fatty Acid Oxidation Products in Plant Oils. J. Agric. Food Chem. 2023, 71, 13092–13106. [Google Scholar] [CrossRef] [PubMed]
- Trapp, M.A.; Kai, M.; Mithöfer, A.; Rodrigues-Filho, E. Antibiotic Oxylipins from Alternanthera brasiliana and Its Endophytic Bacteria. Phytochemistry 2015, 110, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-Y.; Jiang, G.-H.; Sun, Q.H.; Luo, Q.; Liu, B.; Zhan, R.; Aisa, H.A.; Chen, Y.-G. Compounds from the Leaves and Stems of Machilus salicina. Chem. Nat. Compd. 2023, 59, 765–767. [Google Scholar] [CrossRef]
- Kim, C.S.; Subedi, L.; Oh, J.; Kim, S.Y.; Choi, S.U.; Lee, K.R. A New Phenolic Compound from Salix glandulosa. Heterocycles 2018, 96, 931–942. [Google Scholar] [CrossRef]
- Ganßer, D.; Spiteller, G. Aromatase Inhibitors from Urtica dioica Roots. Planta Med. 1995, 61, 138–140. [Google Scholar] [CrossRef]
- Kerboua, M.; Monia, A.A.; Samba, N.; Silva, L.; Raposo, C.; Díez, D.; Rodilla, J.M. Phytochemical Composition of Lichen Parmotrema hypoleucinum (J. Steiner) Hale from Algeria. Molecules 2022, 27, 5229. [Google Scholar] [CrossRef]
- Johansen, K.T.; Wubshet, S.G.; Nyberg, N.T.; Jaroszewski, J.W. From Retrospective Assessment to Prospective Decisions in Natural Product Isolation: HPLC-SPE-NMR Analysis of Carthamus oxyacantha. J. Nat. Prod. 2011, 74, 2454–2461. [Google Scholar] [CrossRef]
- Mojarrab, M.; Delazar, A.; Moghadam, S.B.; Nazemiyeh, H.; Nahar, L.; Kumarasamy, Y.; Asnaashari, S.; Hadjiakhoondi, A.; Sarker, S.D. Armenin and Isoarmenin–Two Prenylated Coumarins from the Aerial Parts of Artemisia armeniaca. Chem. Biodivers. 2011, 8, 2097–2103. [Google Scholar] [CrossRef] [PubMed]
- Wube, A.A.; Wenzig, E.M.; Gibbons, S.; Asres, K.; Bauer, R.; Bucar, F. Constituents of the Stem Bark of Discopodium penninervium and Their LTB4 and COX-1 and -2 Inhibitory Activities. Phytochemistry 2008, 69, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.P.T.; Roullier, C.; Genta-Jouve, G.; Boumard, M.C.; Robiou du Pont, T.; Nazih, H.; Pouchus, Y.F.; Grovel, O. C25 Steroids from the Marine Mussel-Derived Fungus Penicillium ubiquetum MMS330. Phytochem. Lett. 2019, 34, 18–24. [Google Scholar] [CrossRef]
- Qiu, S.; Khan, S.I.; Wang, M.; Zhao, J.; Ren, S.; Khan, I.A.; Steffek, A.; Pfund, W.P.; Li, X.C. Chemometrics-Assisted Identification of Anti-Inflammatory Compounds from the Green Alga Klebsormidium flaccidum Var. Zivo. Mol. 2020, 25, 1048. [Google Scholar] [CrossRef]
- Franich, R.A.; Volkman, J.K. Constituent acids of Pinus radiata stem cutin. Phytochemistry 1982, 21, 2687–2689. [Google Scholar] [CrossRef]
- Yadav, L.; Rani, G.; Kalidhar, S.B. Chemical Examination of Citrus pectinifera Juice. J. Indian Chem. Soc. 2009, 86, 1352–1353. [Google Scholar]
- Millan, A.F.S.; Gamir, J.; Farran, I.; Larraya, L.; Veramendi, J. Identification of New Antifungal Metabolites Produced by the Yeast Metschnikowia pulcherrima Involved in the Biocontrol of Postharvest Plant Pathogenic Fungi. Postharvest Biol. Technol. 2022, 192, 111995. [Google Scholar] [CrossRef]
- Wilson, S.; Panagabko, C.; Laleye, T.; Robinson, M.; Jagas, S.; Bowman, D.; Atkinson, J. Synthesis of a Photocleavable Bola-Phosphatidylcholine. Bioorg. Med. Chem. 2023, 93, 117465. [Google Scholar] [CrossRef]
- Ray, A.K.; Lin, Y.Y.; Gerard, H.C.; Chen, Z.-J.; Osman, S.F.; Fett, W.F.; Moreau, R.A.; Stark, R.E. Separation and identification of lime cutin monomers by high performance liquid chromatography and mass spectrometry. Phytochemistry 1995, 38, 1361–1369. [Google Scholar] [CrossRef]
- Brieskorn, C.H.; Kabelitz, L. Hydroxyfettsäuren aus dem Cutin des Blattes von Rosmarinus officinalis. Phytochemistry 1971, 10, 3195–3204. [Google Scholar] [CrossRef]
- Bian, X.; Yao, L.; Si, E.; Meng, Y.; Li, B.; Ma, X.; Yang, K.; Lai, Y.; Shang, X.; Li, C.; et al. Characterization of Glossy Spike Mutants and Identification of Candidate Genes Regulating Cuticular Wax Synthesis in Barley (Hordeum vulgare L.). Int. J. Mol. Sci. 2022, 23, 13025. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.J.; Kao, C.L.; Yeh, H.C.; Li, H.T.; Cheng, M.J.; Wu, M.D.; Chen, C.Y. A New Phenylalkanoid of Alpinia galanga. Chem. Nat. Compd. 2023, 59, 852–855. [Google Scholar] [CrossRef]
- Ohmoto, T.; Yamaguchi, K. Constituents of Pollen. XV.: Constituents of Biota orientalis (L.) ENDL. (1). Chem. Pharm. Bull. 1988, 36, 807–809. [Google Scholar] [CrossRef][Green Version]
- Omosa, L.K.; Nchiozem-Ngnitedem, V.A.; Mukavi, J.; Atieno Okoko, B.; Ombui Nyaboke, H.; Hashim, I.; Obegi Matundura, J.; Efferth, T.; Spiteller, M. Cytotoxic Alkaloids from the Root of Zanthoxylum paracanthum (Mildbr) Kokwaro. Nat. Prod. Res. 2022, 36, 2518–2525. [Google Scholar] [CrossRef]
- Aono, H.; Koike, K.; Kaneko, J.; Ohmoto, T. Alkaloids and Quassinoids from Ailanthus malabarica. Phytochemistry 1994, 37, 579–584. [Google Scholar] [CrossRef]
- Shao, Q.; Li, T.-Y.; Quan, W.; Wang, H.; Yan, K.-Y.; Qiu, P.-C.; Tang, H.-F.; Lu, Y.-Y. The Alkaloids with Neuroprotective Effect from the Root Bark of Ailanthus altissima. Nat. Prod. Res. 2023, 37, 3751–3757. [Google Scholar] [CrossRef]
- Bucar, F.; Roberts, M.F.; El-Seedi, H.R. Sterones and Indole Alkaloid from Ailanthus altissima Callus Cultures. Chem. Nat. Compd. 2007, 43, 234–236. [Google Scholar] [CrossRef]
- Mundt, S.; Kreitlow, S.; Jansen, R. Fatty Acids with Antibacterial Activity from the Cyanobacterium Oscillatoria Redekei HUB 051. J. Appl. Phycol. 2003, 15, 263–267. [Google Scholar] [CrossRef]
- Li, Z.; Tran, V.H.; Duke, R.K.; Ng, M.C.H.; Yang, D.; Duke, C.C. Synthesis and Biological Activity of Hydroxylated Derivatives of Linoleic Acid and Conjugated Linoleic Acids. Chem. Phys. Lipids 2009, 158, 39–45. [Google Scholar] [CrossRef]
- Prema; Wong, C.P.; Nugroho, A.E.; Awouafack, M.D.; Win, Y.Y.; Win, N.N.; Ngwe, H.; Morita, H.; Morita, H. Two New Quassinoids and Other Constituents from Picrasma javanica Wood, and Their Biological Activities. J. Nat. Med. 2019, 73, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.K.; Dan, W.J.; Li, N.; Du, H.T.; Zhang, J.W.; Wang, J.R. Synthesis, in Vitro Antibacterial Activities of a Series of 3-N-Substituted Canthin-6-Ones. Bioorg. Med. Chem. Lett. 2016, 26, 580–583. [Google Scholar] [CrossRef]
- Dan, W.; Gao, J.; Zhang, J.; Cao, Y.; Liu, J.; Sun, Y.; Wang, J.; Dai, J. Antibacterial Activity and Mechanism of Action of Canthin-6-One against Staphylococcus aureus and Its Application on Beef Preservation. Food Control 2023, 147, 109604. [Google Scholar] [CrossRef]
- O’donnell, G.; Gibbons, S. Antibacterial Activity of Two Canthin-6-One Alkaloids from Allium neapolitanum. Phytother. Res. 2007, 21, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Gray, A.I.; Khondkar, P.; Islam, M.A. Antimicrobial Activities of Alkaloids and Lignans from Zanthoxylum budrunga. Nat. Prod. Commun. 2008, 3, 45–47. [Google Scholar] [CrossRef]
- Geubelle, P.; Gilissen, J.; Dilly, S.; Poma, L.; Dupuis, N.; Laschet, C.; Abboud, D.; Inoue, A.; Jouret, F.; Pirotte, B.; et al. Identification and Pharmacological Characterization of Succinate Receptor Agonists. Br. J. Pharmacol. 2017, 174, 796–808. [Google Scholar] [CrossRef]
- Fang, H.L.; Liu, M.L.; Li, S.Y.; Song, W.Q.; Ouyang, H.; Xiao, Z.P.; Zhu, H.L. Identification, Potency Evaluation, and Mechanism Clarification of α-Glucosidase Inhibitors from Tender Leaves of Lithocarpus polystachyus Rehd. Food Chem. 2022, 371, 131128. [Google Scholar] [CrossRef]
- Móricz, Á.M.; Häbe, T.T.; Böszörményi, A.; Ott, P.G.; Morlock, G.E. Tracking and Identification of Antibacterial Components in the Essential Oil of Tanacetum vulgare L. by the Combination of High-Performance Thin-Layer Chromatography with Direct Bioautography and Mass Spectrometry. J. Chromatogr. A 2015, 1422, 310–317. [Google Scholar] [CrossRef]


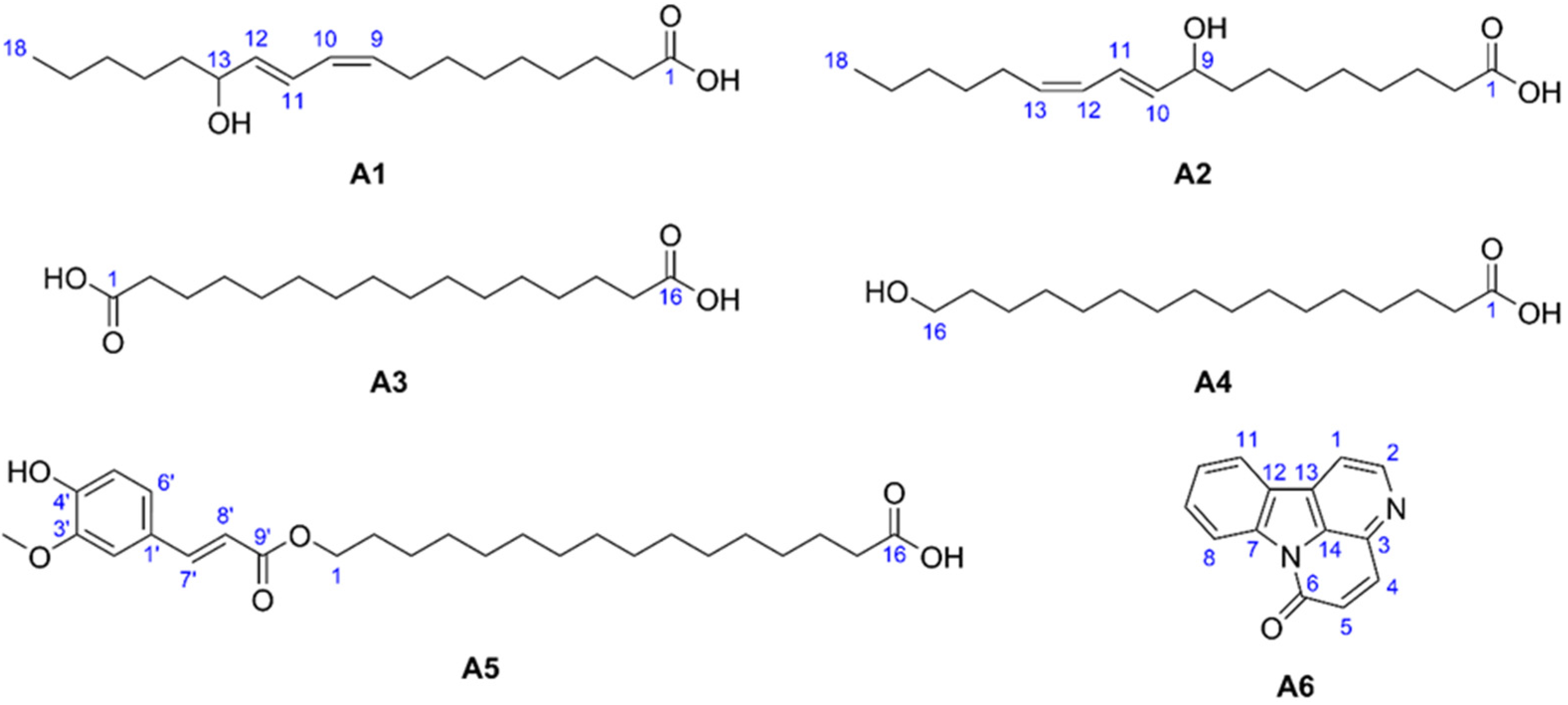
| Isolate | Name | MIC (µg/mL) |
|---|---|---|
| A1 | (9Z,11E)-13-hydroxy-9,11-octadecadienoic acid (13-HODE) | 66.7 |
| A2 | (10E,12Z)-9-hydroxy-10,12-octadecadienoic acid (9-HODE) | 66.7 |
| A3 | hexadecanedioic acid (thapsic acid) | >133.3 * |
| A4 | 16-hydroxyhexadecanoic acid (juniperic acid) | 66.7 |
| A5 | alpinagalanate | >133.3 |
| A6 | canthin-6-one | 8.3 |
| gentamicin | 0.8 |
| Sample | Collection Time | Collection Area | Collected Tissue(s) | Voucher Code |
|---|---|---|---|---|
| H1 | 2 May 2022 | Harta | stem bark | Aa.H1.8 |
| H2 | 16 May 2022 | Harta | stem bark outer trunk bark inner trunk bark | Aa.H2.8 Aa.H2.9OB Aa.H2.9IB |
| H3 | 30 May 2022 | Harta | stem bark | Aa.H3.8 |
| H4 | 3 July 2022 | Harta | stem bark outer trunk bark inner trunk bark | Aa.H4.8 Aa.H4.9OB Aa.H4.9IB |
| H5 | 13 August 2022 | Harta | outer trunk bark inner trunk bark | Aa.H5.9OB Aa.H5.9IB |
| H7 | 24 October 2022 | Harta | outer trunk bark inner trunk bark | Aa.H7.9OB Aa.H7.9IB |
| H11 | 10 April 2023 | Harta | outer trunk bark inner trunk bark | Aa.H11.9OB Aa.H11.9IB |
| H16 | 20 May 2023 | Harta | stem bark inner trunk bark | Aa.H16.8 Aa.H16.9IB |
| H19 | 31 July 2023 | Harta | inner trunk bark | Aa.H19.9IB |
| B3 | 26 July 2022 | Balatongyörök | outer trunk bark | Aa.B3.9OB |
| L1 | 21 May 2022 | Leányfalu | stem bark | Aa.L1.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cselőtey, A.; Baglyas, M.; Király, N.; Ott, P.G.; Glavnik, V.; Vovk, I.; Móricz, Á.M. Bioassay-Guided Isolation and Identification of Antibacterial Compounds from Invasive Tree of Heaven Stem and Trunk Bark. Molecules 2024, 29, 5846. https://doi.org/10.3390/molecules29245846
Cselőtey A, Baglyas M, Király N, Ott PG, Glavnik V, Vovk I, Móricz ÁM. Bioassay-Guided Isolation and Identification of Antibacterial Compounds from Invasive Tree of Heaven Stem and Trunk Bark. Molecules. 2024; 29(24):5846. https://doi.org/10.3390/molecules29245846
Chicago/Turabian StyleCselőtey, Anna, Márton Baglyas, Nóra Király, Péter G. Ott, Vesna Glavnik, Irena Vovk, and Ágnes M. Móricz. 2024. "Bioassay-Guided Isolation and Identification of Antibacterial Compounds from Invasive Tree of Heaven Stem and Trunk Bark" Molecules 29, no. 24: 5846. https://doi.org/10.3390/molecules29245846
APA StyleCselőtey, A., Baglyas, M., Király, N., Ott, P. G., Glavnik, V., Vovk, I., & Móricz, Á. M. (2024). Bioassay-Guided Isolation and Identification of Antibacterial Compounds from Invasive Tree of Heaven Stem and Trunk Bark. Molecules, 29(24), 5846. https://doi.org/10.3390/molecules29245846








