Molecular Analysis of UV-C Induced Resveratrol Accumulation in Polygonum cuspidatum Leaves
Abstract
1. Introduction
2. Results
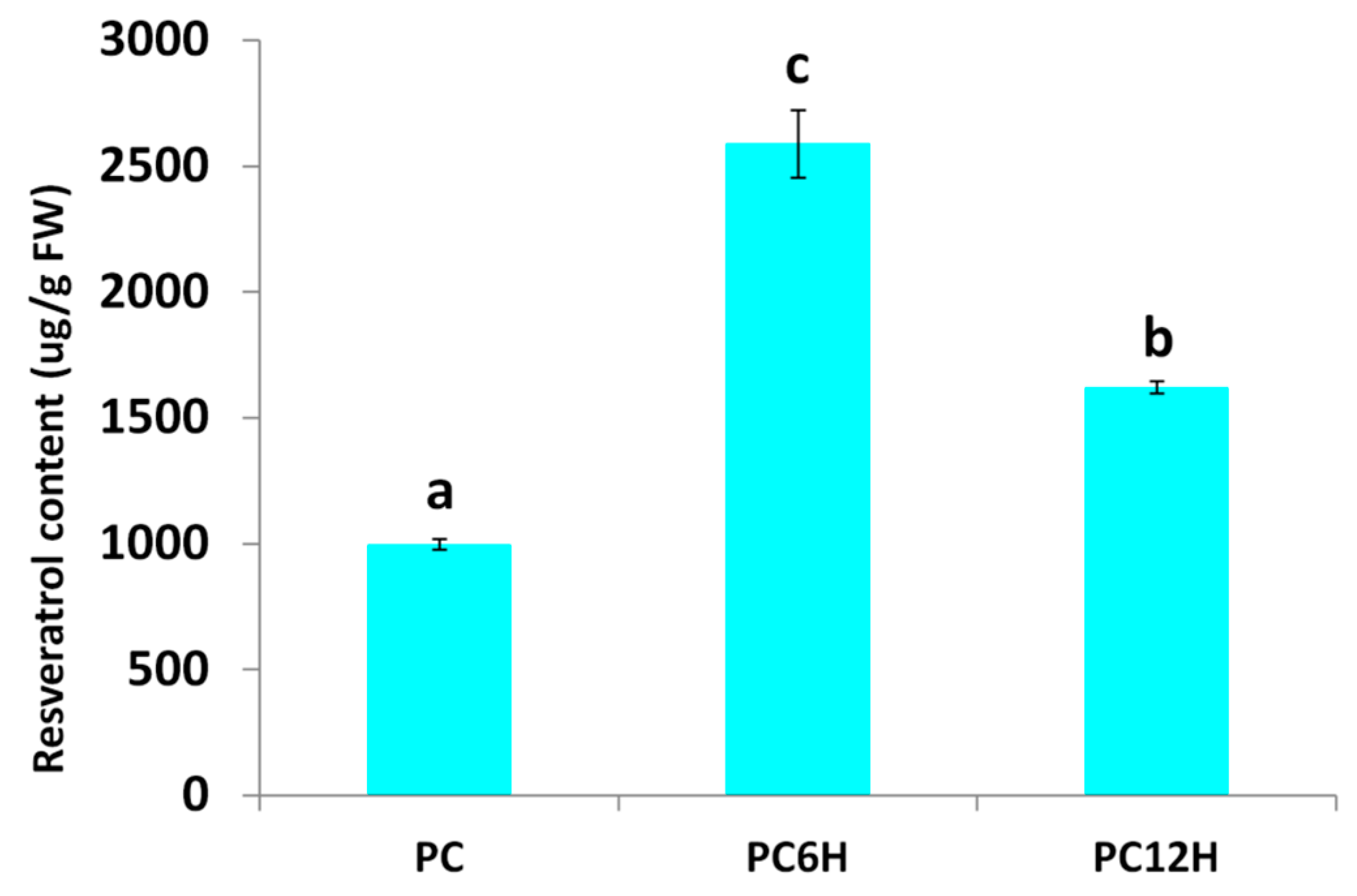
2.1. Effect of UV-C Irradiation on Leaf Resveratrol Content in Polygonum cuspidatum
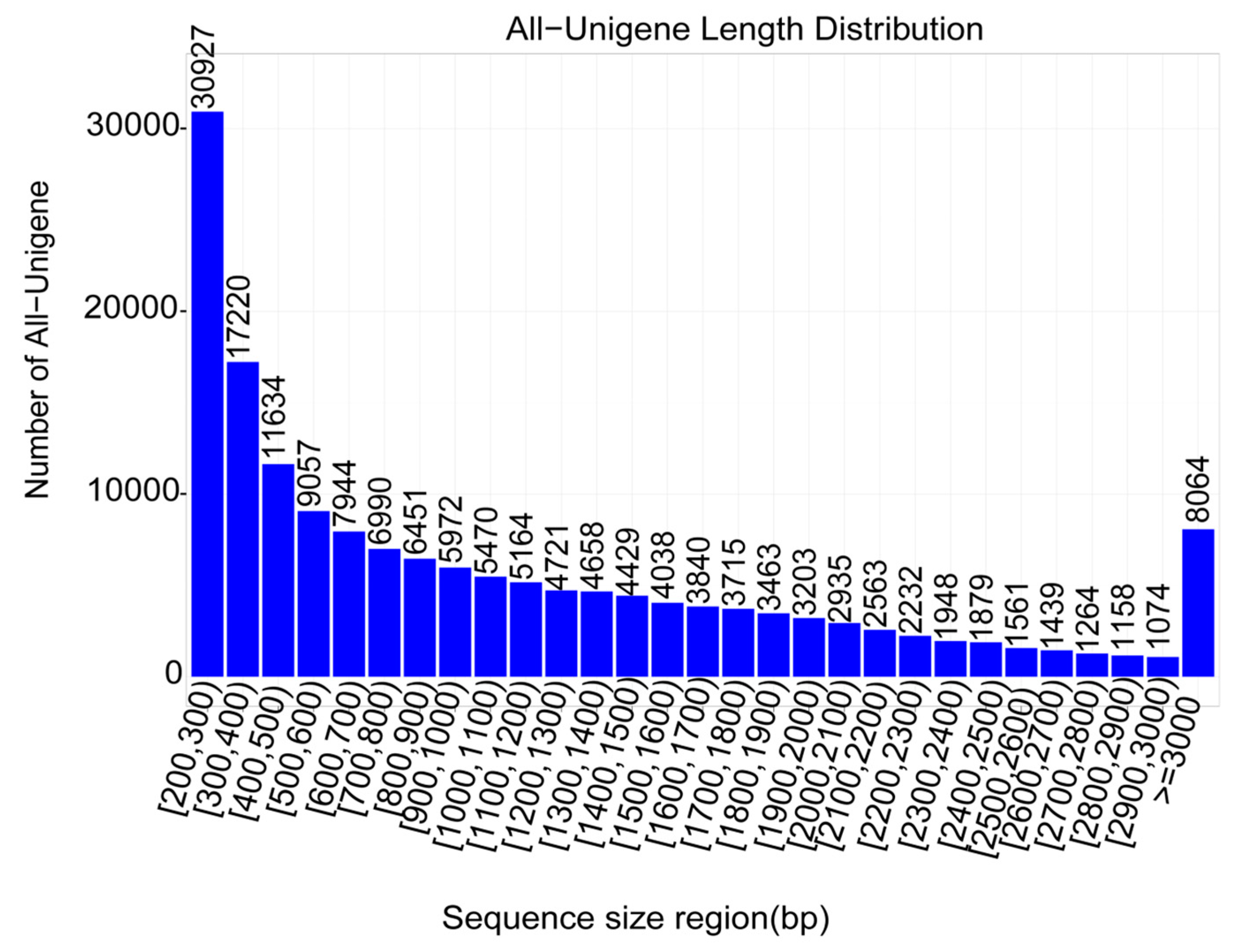
2.2. De Novo Transcriptome Sequencing and Unigene Assembly
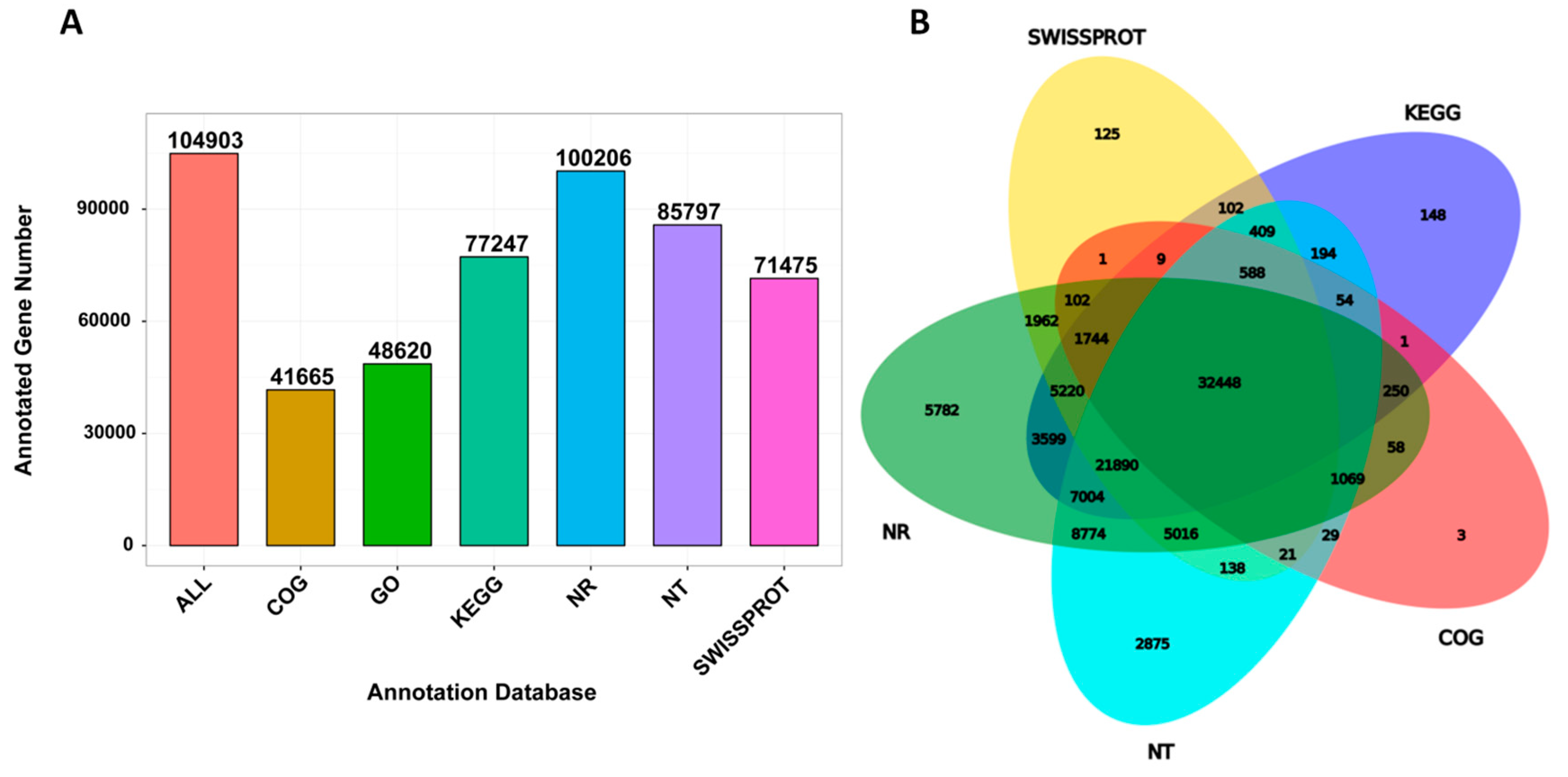
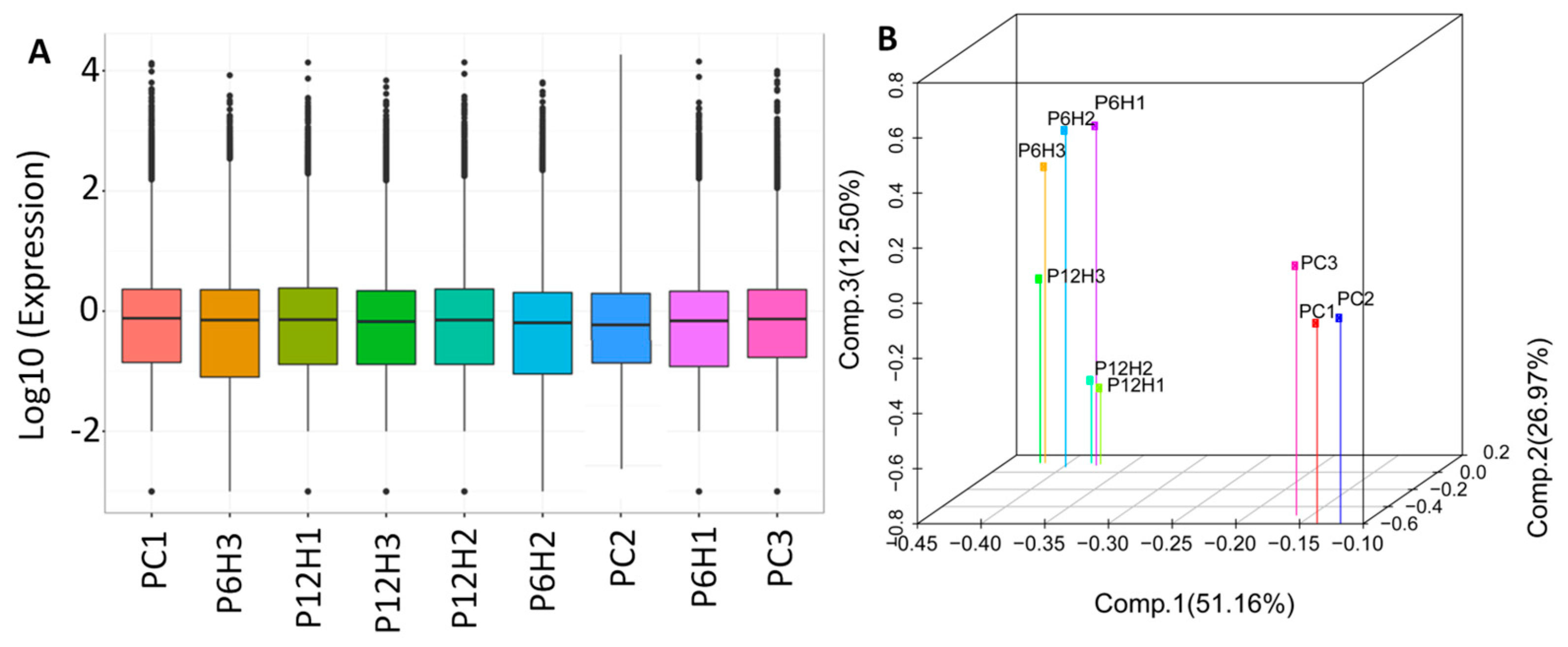
2.3. Differential Expressed Genes (DEG) between Control and UV-C Treated Leaf Samples
2.4. Mapping of DEGs Related to the Resveratrol Biosynthetic Pathway
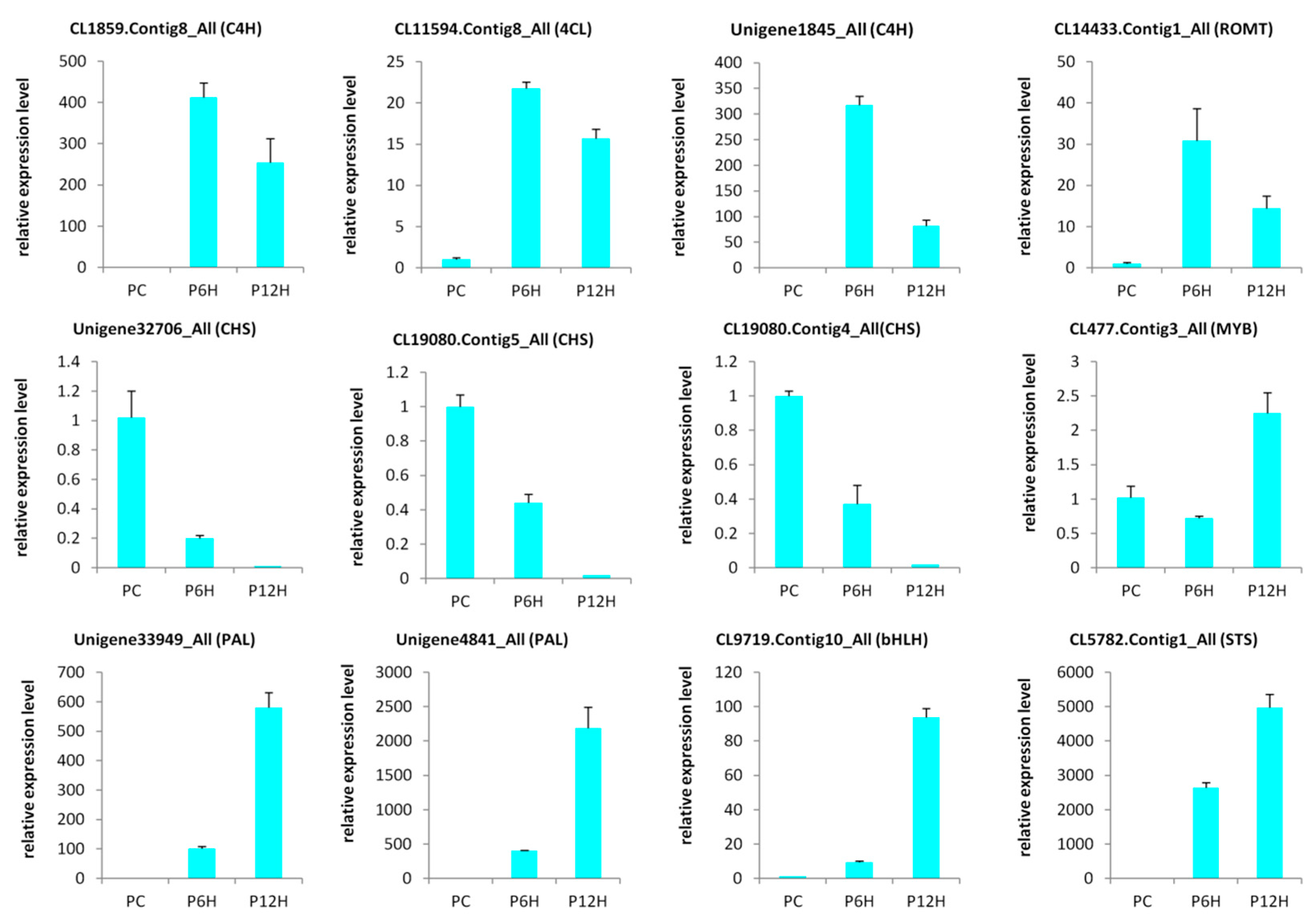
2.5. qRT-PCR Validation of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Quantification of Resveratrol in Polygonum cuspidatum Leaves
4.3. RNA Extraction, cDNA Library Construction and Transcriptome Sequencing
4.4. Quality Check, Cleaning and de novo Assembly
4.5. Functional Annotation of the Unigenes
4.6. Analysis of Transcription Factors
4.7. Gene Expression and Differential Expression Analysis
4.8. Gene Expression Using Quantitative Real Time-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Seiger, L. Mechanical control of Japanese knotweed (Fallopia japonica [Houtt.] Ronse Decraene): Effects of cutting regime on rhizomatous reserves. Nat. Area. J. 1997, 17, 341–345. [Google Scholar]
- Bailey, J. Chapter 14. The rise and fall of Japanese knotweed? In Invasive and Introduced Plants and Animals Human Perceptions, Attitudes and Approaches to Management; Rotherham, I., Lambert, R., Eds.; Routledge: New York, NY, USA, 2013; pp. 221–232. [Google Scholar]
- Jones, D.; Bruce, G.; Fowler, M.S.; Law-Cooper, R.; Graham, I.; Abel, A.; Street-Perrott, F.A.; Eastwood, D. Optimising physiochemical control of invasive Japanese knotweed. Biol. Invasions 2018, 20, 2091–2105. [Google Scholar] [CrossRef]
- Yan, S.; Li, L.; Yu, S.; Xiao, P.G. Effect of Tabellae Polygonum Cuspidatum on Blood Lipids and Rheological Property in Rats. China J. Chin. Mat. Med. 1993, 18, 617–619. [Google Scholar]
- Xiao, K.; Xuan, L.; Xu, Y.; Bai, D.; Zhong, D. Constituents from Polygonum cuspidatum. Chem. Pharm. Bull. (Tokyo) 2002, 50, 605–608. [Google Scholar] [CrossRef]
- Peng, W.; Qin, R.; Li, X.; Zhou, H. Botany, phytochemistry, pharmacology, and potential application of Polygonum cuspidatum Sieb.et Zucc.: A review. J. Ethnopharmacol. 2013, 148, 729–745. [Google Scholar] [CrossRef]
- Dong, J.; Wang, H.; Wan, L.; Hashi, Y.; Chen, S. Identification and determination of major constituents in Polygonum cuspidatum Sieb. et Zucc. by high performance liquid chromatography/electrospray ionization-ion traptime-of-flight mass spectrometry. Se Pu 2009, 27, 425–430. [Google Scholar]
- Lachowicz, S.; Oszmiański, J. Profile of Bioactive Compounds in the Morphological Parts of Wild Fallopia japonica (Houtt) and Fallopia sachalinensis (F. Schmidt) and Their Antioxidative Activity. Molecules 2019, 24, 1436. [Google Scholar] [CrossRef]
- Baur, J.A.; Sinclair, D.A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov. 2006, 5, 493–506. [Google Scholar] [CrossRef]
- Truong, V.L.; Jun, M.; Jeong, W.S. Role of resveratrol in regulation of cellular defense systems against oxidative stress. BioFactors. 2018, 44, 36–49. [Google Scholar] [CrossRef]
- Renaud, S.; de Lorgeril, M. Wine, alcohol, platelets, and the French paradox for coronary heart disease. Lancet. 1992, 339, 1523–1526. [Google Scholar] [CrossRef]
- Novelle, M.G..; Wahl, D.; Dieguez, C.; Bernier, M.; de Cabo, R. Resveratrol supplementation, where are we now and where should we go? Ageing Res. Rev. 2015, 21, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nonomura, S.; Kanagawa, H.; Makimoto, A. Chemical constituents of polygonaceous plants. I. Studies on the components of Ko-jo-kon. Yakugaku Zasshi. 1963, 83, 988–990. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Ma, P.; Mu, J.; Chen, S.; Xiao, P.; Peng, Y.; Sun, C. De novo characterization of the root transcriptome of a traditional Chinese medicinal plant Polygonum cuspidatum. Sci. China Life Sci. 2012, 55, 452–466. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Bae, H. An Overview of Stress-Induced Resveratrol Synthesis in Grapes: Perspectives for Resveratrol-Enriched Grape Products. Molecules. 2017, 22, 294. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, U463–U465. [Google Scholar]
- Halls, C.; Yu, O. Potential for metabolic engineering of resveratrol biosynthesis. Trends Biotechnol. 2008, 26, 77–81. [Google Scholar] [CrossRef]
- Lanz, T.; Tropf, S.; Marner, F.J.; Schroder, J.; Schroder, G. The role of cysteines in polyketide synthases. Site-directed mutagenesis of resveratrol and chalcone synthases, two key enzymes in different plant-specific pathways. J. Biol. Chem. 1991, 266, 9971–9976. [Google Scholar]
- Langcake, P.; Pryce, R.J. The production of resveratrol by Vitis vinifera and other members of the Vitaceae as a response to infection or injury. Physiol. Plant Pathol. 1976, 9, 77–86. [Google Scholar] [CrossRef]
- Höll, J.; Vannozzi, A.; Czemmel, S.; D’Onofrio, C.; Walker, A.R.; Rausch, T.; Lucchin, M.; Boss, P.K.; Dry, I.B.; Bogs, J. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 2013, 25, 4135–4149. [Google Scholar] [CrossRef]
- Jiang, J.; Xi, H.; Dai, Z.; Lecourieux, F.; Yuan, L.; Liu, X.; Patra, B.; Wei, Y.; Li, S.; Wang, L. VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J. Exp. Bot. 2019, 70, 715–729. [Google Scholar] [CrossRef]
- Langcake, P. Disease resistance of Vitis spp. and the production of the stress metabolites resveratrol, ”-viniferin, -viniferin, and pterostilbene. Physiol. Plant Pathol. 1981, 18, 213–226. [Google Scholar] [CrossRef]
- Dercks, W.; Creasy, L.L. The significance of stilbene phytoalexins in the Plasmopara viticola-grapevine interaction. Physiol. Mol. Plant Pathol. 1989, 34, 189–202. [Google Scholar] [CrossRef]
- Tassoni, A.; Fornale, S.; Franceschetti, M.; Musiani, F.; Michael, A.J.; Perry, B. Jasmonates and Na-orthovanadate promote resveratrol production in Vitis vinifera cv. Barbera cell cultures. New Phytol. 2005, 166, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.F.; Chen, J.Y.; Kong, W.F.; Pan, Q.H.; Wan, S.B.; Huang, W.D. Salicylic acid induced the expression of phenylalanine ammonia-lyase gene in grape berry. Plant Sci. 2005, 169, 928–934. [Google Scholar] [CrossRef]
- González-Barrio, R.; Beltrán, D.; Cantos, E.; Gil, M.I.; Espín, J.C.; Tomás-Barberán, F.A. Comparison of ozone and UV-C treatments on the postharvest stilbenoid monomer, dimer, and trimer induction in var. ‘Superior’ white table grapes. J. Agric. Food Chem. 2006, 54, 4222–4228. [Google Scholar] [CrossRef] [PubMed]
- Schmidlin, L.; Poutaraud, A.; Claudel, P.; Mestre, P.; Prado, E.; Santos-Rosa, M. A stress-inducible resveratrol O-methyltransferase involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol. 2008, 148, 1630–1639. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Ma, L.; Xi, H.F.; Duan, W.; Wang, J.F.; Li, S.H. Individual and combined effects of CaCl2 and UV-C on the biosynthesis of resveratrols in grape leaves and berry skins. J. Agric. Food Chem. 2013, 61, 7135–7141. [Google Scholar] [CrossRef]
- Banerjee, N.; Zhang, M.Q. Identifying cooperativity among transcription factors controlling the cell cycle in yeast. Nucleic Acids Res. 2003, 31, 7024–7031. [Google Scholar] [CrossRef]
- Hollósy, F. Effects of ultraviolet radiation on plant cells. Micron 2002, 33, 179–197. [Google Scholar] [CrossRef]
- Kasim, M.U.; Kasim, R.; Erkal, S. UV-C treatments on fresh-cut green onions enhanced antioxidant activity, maintained green color and controlled ‘telescoping’. J. Food Agric. Environ. 2008, 6, 63–67. [Google Scholar]
- Rivera-Pastrana, D.M.; Gardea, A.A.; Yahia, E.M.; Martínez-Téllez, M.A.; González-Aguilar, G.A. Effect of UV-C irradiation and low temperature storage on bioactive compounds, antioxidant enzymes and radical scavenging activity of papaya fruit. J. Food Sci. Technol. 2014, 51, 3821–3829. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Crupi, P.; Pichierri, A.; Basile, T.; Antonacci, D. Postharvest stilbenes and flavonoids enrichment of table grape cv Redglobe (Vitis vinifera L.) as affected by interactive UV-C exposure and storage conditions. Food Chem. 2013, 141, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tang, K.; Yang, H.R.; Wen, P.F.; Zhang, P.; Wang, H.L.; Huang, W.D. Distribution of resveratrol and stilbene synthase in young grape plants (Vitis vinifera L. cv. Cabernet Sauvignon) and the effect of UV-C on its accumulation. Plant Physiol. Biochem. 2010, 48, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Ma, L.; Xi, H.F.; Wang, L.J.; Li, S.H. Resveratrol synthesis under natural conditions and after UV-C irradiation in berry skin is associated with berry development stages in ‘Beihong’ (V. vinifera × V. amurensis). Food Chem. 2015, 168, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.; Liu, C.; Chang, E.; Ji, J.; Yao, X.; Yue, J.; Bartish, I.V.; Chen, L.; Jiang, Z.; Shi, S. High temperature and UV-C treatments affect stilbenoid accumulation and related gene expression levels in Gnetum parvifolium. Electron. J. Biotechnol. 2017, 25, 43–49. [Google Scholar] [CrossRef]
- Cantos, E.; Espin, J.C.; Fernandez, M.J.; Oliva, J.; Tomas-Barberan, F.A. Postharvest UV-C-irradiated grapes as a potential source for producing stilbene-enriched red wines. J. Agric. Food Chem. 2003, 51, 1208–1214. [Google Scholar] [CrossRef]
- Guerrero, R.F.; Puertas, B.; Fernández, M.I.; Palma, M.; Cantos-Villar, E. Induction of stilbenes in grapes by UV-C: Comparison of different subspecies of Vitis. Innov. Food Sci. Emerg. Technol. 2010, 11, 231–238. [Google Scholar] [CrossRef]
- Cantos, E.; Espin, J.C.; Tomas-Barberan, F.A. Postharvest stilbene-enrichment of red and white table grape varieties using UV-C irradiation pulses. J. Agric. Food Chem. 2002, 50, 6322–6329. [Google Scholar] [CrossRef]
- Yin, X.; Singer, S.D.; Qiao, H.; Liu, Y.; Jiao, C.; Wang, H.; Li, Z.; Fei, Z.; Wang, Y.; Fan, C. Insights into the Mechanisms Underlying Ultraviolet-C Induced Resveratrol Metabolism in Grapevine (V. amurensis Rupr.) cv. “Tonghua-3”. Front Plant Sci. 2016, 7, 503. [Google Scholar] [CrossRef]
- Xi, H.; Ma, L.; Liu, G.; Wang, N.; Wang, J.; Wang, L. Transcriptomic analysis of grape (Vitis vinifera L.) leaves after exposure to ultraviolet C irradiation. PLoS One 2014, 9, e113772. [Google Scholar] [CrossRef]
- Suzuki, M.; Nakabayashi, R.; Ogata, Y.; Sakurai, N.; Tokimatsu, T.; Goto, S. Multi omics in grape berry skin revealed specific induction of stilbene synthetic pathway by UV-C irradiation. Plant Physiol. 2015, 168, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Liu, P.; Chen, J.; Du, G.; Li, H.; Zhou, J. Characterization of mutants of a tyrosine ammonia-lyase from Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 2016, 100, 10443–10452. [Google Scholar] [CrossRef] [PubMed]
- Walle, T.F.; Hsieh, M.H.; DeLegge, J.E.; Oatis, U.K., Jr. Walle High absorption but very low bioavailability of oral resveratrol in humans Drug. Metab. Dispos. 2004, 32, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.J.; An, C.H.; Woo, S.G.; Jeong, H.J.; Kim, Y.M.; Park, S.J.; Yoon, B.D.; Kim, C.Y. Production of pinostilbene compounds by the expression of resveratrol O-methyltransferase genes in Escherichia coli. Enzyme Microb. Technol. 2014, 54, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Márquez, A.; Morante-Carriel, J.A.; Ramírez-Estrada, K.; Cusidó, R.M.; Palazon, J.; Bru-Martínez, R. Production of highly bioactive resveratrol analogues pterostilbene and piceatannol in metabolically engineered grapevine cell cultures. Plant Biotechnol. J. 2016, 14, 1813–1825. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Fang, L.; Hou, Y.; Wang, L.; Xin, H.; Wang, N.; Li, S. Myb14, a direct activator of STS, is associated with resveratrol content variation in berry skin in two grape cultivars. Plant Cell Rep. 2014, 33, 1629–1640. [Google Scholar] [CrossRef]
- Childs, K.L.; Davidson, R.M.; Buell, C.R. Gene coexpression network analysis as a source of functional annotation for rice genes. PLoS ONE 2011, 6, e22196. [Google Scholar] [CrossRef]
- Weston, D.J.; Karve, A.A.; Gunter, L.E.; Jawdy, S.S.; Yang, X.; Allen, S.M.; Wullschleger, S.D. Comparative physiology and transcriptional networks underlying the heat shock response in Populus trichocarpa, Arabidopsis thaliana and Glycine max. Plant Cell Environ. 2011, 34, 1488–1506. [Google Scholar]
- Downs, G.S.; Bi, Y.M.; Colasanti, J.; Wu, W.; Chen, X.; Zhu, T.; Rothstein, S.J.; Lukens, L.N. A developmental transcriptional network for Zea mays defines coexpression modules. Plant Physiol. 2013, 161, 1830–1843. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ding, Z.; Li, P. Maize network analysis revealed gene modules involved in development, nutrients utilization, metabolism, and stress response. BMC Plant Biol. 2017, 17, 131. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Hour, A.; Liu, L.D. Microarray meta-analysis to explore abiotic stress-specific gene expression patterns in Arabidopsis. Botanical Studies 2017, 58, 22. [Google Scholar] [CrossRef] [PubMed]
- Shahan, R.; Zawora, C.; Wight, H.; Sittmann, J.; Wang, W.; Mount, S.M.; Liu, Z. Consensus coexpression network analysis identifies key regulators of flower and fruit development in wild strawberry. Plant Physiol. 2018, 178, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Mmadi, M.A.; Zhou, R.; Zhang, T.; Su, R.; Zhang, Y.; Wang, L.; You, J.; Zhang, X. Depicting the core transcriptome modulating multiple abiotic stresses responses in sesame (Sesamum indicum L.). Int. J. Mol. Sci. 2019, 20, 3930. [Google Scholar] [CrossRef] [PubMed]
- Corso, M.; Vannozzi, A.; Maza, E.; Vitulo, N.; Meggio, F.; Pitacco, A.; Telatin, A.; D’Angelo, M.; Feltrin, E.; Negri, A.S.; et al. Comprehensive transcript profiling of two grapevine rootstock genotypes contrasting in drought susceptibility links the phenylpropanoid pathway to enhanced tolerance. J. Exp. Bot. 2015, 66, 5739–5752. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.C.J.; Schlechter, R.; Vannozzi, A.; Höll, J.; Hmmam, I.; Bogs, J.; Tornielli, G.B.; Castellarin, S.D.; Matus, J.T. A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res. 2016, 23, 451–466. [Google Scholar] [CrossRef]
- Vannozzi, A.; Wong, D.C.J.; Höll, J.; Hmmam, I.; Matus, J.T.; Bogs, J.; Ziegler, T.; Dry, I.; Barcaccia, G.; Lucchin, M. Combinatorial Regulation of Stilbene Synthase Genes by WRKY and MYB Transcription Factors in Grapevine (Vitis vinifera L.). Plant Cell Physiol. 2018, 59, 1043–1059. [Google Scholar] [CrossRef]
- Xi, H.F.; Ma, L.; Wang, L.N.; Li, S.H.; Wang, L.J. Differential response of the biosynthesis of resveratrols and flavonoids to UV-C irradiation in grape leaves. New Zealand J. Crop Hortic. Sci. 2015, 43, 163–172. [Google Scholar] [CrossRef]
- Zhang, S.; Ying, H.; Pingcuo, G.; Wang, S.; Zhao, F.; Cui, Y.; Shi, J.; Zeng, H.; Zeng, X. Identification of Potential Metabolites Mediating Bird’s Selective Feeding on Prunus mira Flowers. Biomed Res. Int. 2019, 2019, 8. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full length transcriptome assembly from RNA Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G. , Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; Tsai, J.; Quackenbush, J. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 2004. 32, D277–D280. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A. The COG database: A tool for genome scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004. 32, D115–D119. [CrossRef]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.P.; Chen, Y.W.; He, F.C. Integrated nr Database in Protein Annotation System and Its Localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 1997. 25, 3389–3402. [Google Scholar] [CrossRef]
- Iseli, C.; Jongeneel, C.V.; Bucher, P. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999, 138–148. [Google Scholar]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, T.; Liu, C.J.; Song, S.; Zhang, X.; Liu, W.; Jia, H.; Xue, Y.; Guo, A.Y. AnimalTFDB 2.0: A resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 2015, 43, D76–D81. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, J.; Jang, S.; Kim, S.; Kong, S.; Choi, J.; Ahn, K.; Kim, J.; Lee, S.; Kim, S. FTFD: An informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics. 2008, 24, 1024–1025. [Google Scholar] [CrossRef] [PubMed]
- Kummerfeld, S.K.; Teichmann, S.A. DBD: A transcription factor prediction database. Nucleic Acids Res. 2006, 34, D74–D81. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Colin, N.D. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Dossa, K.D.; Li, D.J.; Yu, J.L.; Wang, Y.; Zhang, J.; You, R.; Zhou, M.A.; Mmadi, A.; Li, D.; Fonceka, D.; et al. The genetic basis of drought tolerance in the high oil crop Sesamum indicum. Plant Biotechnol. J. 2019, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
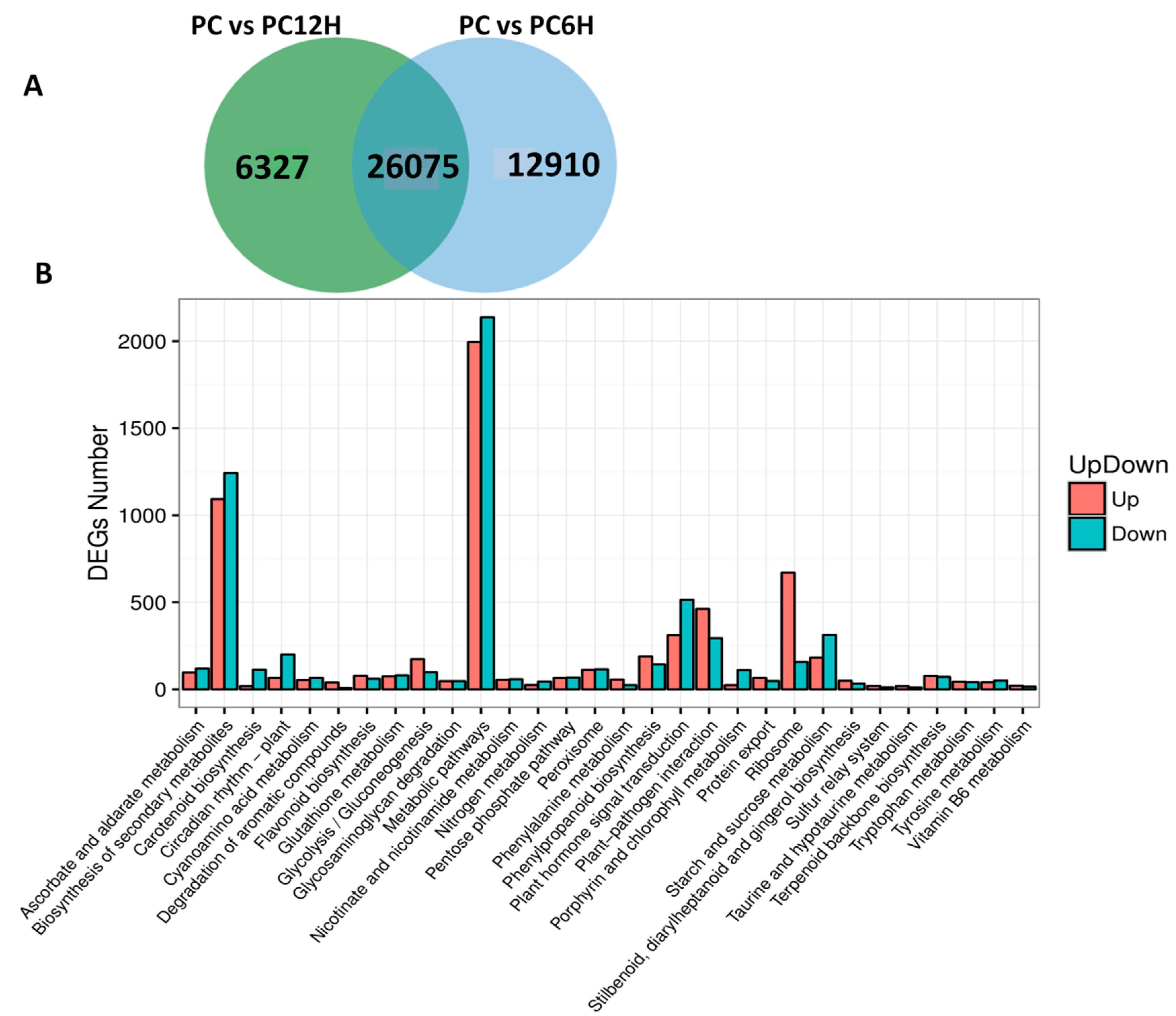

| Type | Read Length | Total Clean Reads | Clean Reads Q20 (%) | Clean Reads Q30 (%) | Detected Genes |
|---|---|---|---|---|---|
| P12H1 | 150 | 55,184,328 | 97.81 | 93.76 | 122,534 |
| P12H2 | 150 | 57,685,708 | 97.47 | 93.21 | 123,131 |
| P12H3 | 150 | 60,727,642 | 97.83 | 93.85 | 124,005 |
| P6H1 | 150 | 51,083,608 | 97.66 | 93.41 | 122,200 |
| P6H2 | 150 | 53,076,416 | 97.64 | 93.36 | 120,570 |
| P6H3 | 150 | 48,587,712 | 97.74 | 93.56 | 119,464 |
| PC1 | 150 | 53,966,128 | 97.52 | 93.14 | 123,390 |
| PC2 | 150 | 51,267,598 | 97.5 | 93.19 | 117,489 |
| PC3 | 150 | 52,633,030 | 97.77 | 93.69 | 125,163 |
| Sample | Total_Number | Min_Length | Max_length | Mean_Length | N50 | N90 | GC |
|---|---|---|---|---|---|---|---|
| All | 165,013 | 200 | 17,269 | 1,102.73 | 1,744 | 489 | 0.4216 |
| TF | Count | TF | Count | TF | Count |
|---|---|---|---|---|---|
| LFY | 3 | M-type | 5 | HB-PHD | 17 |
| SAP | 23 | GATA | 105 | E2F/DP | 23 |
| WRKY | 298 | DBB | 44 | Trihelix | 126 |
| RAV | 13 | ZF-HD | 35 | ARF | 137 |
| CPP | 35 | LBD | 50 | MIKC | 98 |
| HRT-like | 3 | MYB | 402 | SRS | 15 |
| HB-other | 45 | LSD | 14 | NF-X1 | 8 |
| FAR1 | 67 | WOX | 15 | Nin-like | 43 |
| ERF | 341 | C2H2 | 204 | MYB_related | 272 |
| Whirly | 11 | CAMTA | 23 | VOZ | 18 |
| ARR-B | 57 | GRAS | 109 | G2-like | 157 |
| HSF | 90 | GRF | 32 | NF-YA | 56 |
| NZZ/SPL | 8 | EIL | 36 | Dof | 119 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Xu, J.; Wu, X.; Wang, Y.; Lin, Y.; Wu, D.; Zhang, H.; Qin, J. Molecular Analysis of UV-C Induced Resveratrol Accumulation in Polygonum cuspidatum Leaves. Int. J. Mol. Sci. 2019, 20, 6185. https://doi.org/10.3390/ijms20246185
Liu Z, Xu J, Wu X, Wang Y, Lin Y, Wu D, Zhang H, Qin J. Molecular Analysis of UV-C Induced Resveratrol Accumulation in Polygonum cuspidatum Leaves. International Journal of Molecular Sciences. 2019; 20(24):6185. https://doi.org/10.3390/ijms20246185
Chicago/Turabian StyleLiu, Zhongyu, Junxiong Xu, Xiang Wu, Yanyan Wang, Yanli Lin, Duanyang Wu, Hongjie Zhang, and Jianbing Qin. 2019. "Molecular Analysis of UV-C Induced Resveratrol Accumulation in Polygonum cuspidatum Leaves" International Journal of Molecular Sciences 20, no. 24: 6185. https://doi.org/10.3390/ijms20246185
APA StyleLiu, Z., Xu, J., Wu, X., Wang, Y., Lin, Y., Wu, D., Zhang, H., & Qin, J. (2019). Molecular Analysis of UV-C Induced Resveratrol Accumulation in Polygonum cuspidatum Leaves. International Journal of Molecular Sciences, 20(24), 6185. https://doi.org/10.3390/ijms20246185





