Flower-Specific Overproduction of Cytokinins Altered Flower Development and Sex Expression in the Perennial Woody Plant Jatropha curcas L.
Abstract
:1. Introduction
2. Results
2.1. Characterization of the JcTM6 Promoter in J. curcas
2.2. Cytokinin Contents in JcTM6:AtIPT4 Transgenic J. curcas
2.3. The Flower Number was Increased in JcTM6:AtIPT4 Transgenic J. curcas
2.4. JcTM6:AtIPT4 transgenic J. curcas Produced Bisexual Flowers
2.5. Flowers of JcTM6:AtIPT4 Transgenic J. curcas Developed Abnormally
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Plasmid Construction
4.3. Plant Transformation
4.4. Histochemical and Fluorometric GUS Assay
4.5. Quantification of CKs
4.6. qRT-PCR Analysis
4.7. Tissue Preparation for Confocal Analysis
4.8. Confocal Microscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AG | AGAMOUS |
| AGL1 | AGAMOUS-like 1 |
| AHK | ARABIDOPSIS HISTIDINE KINASE |
| AP1 | APETALA1 |
| AP3 | APETALA3 |
| ARR | ARABIDOPSIS RESPONSE REGULATOR |
| 6-BA | 6-benzylaminopurine |
| CKX | cytokinin oxidases/dehydrogenase |
| cZ | cis-zeatin |
| cZR | cis-zeatin riboside |
| DAP | days after pollination |
| GUS | β-glucuronidase |
| iP | isopentenyladenine |
| iPR | isopentenyladenosine |
| IPT | isopentenyltransferase |
| LOG | LONELY GUY |
| STM | SHOOT MERISTEMLESS |
| SyGI | Shy Girl |
| TDZ | thidiazuron |
| TM6 | TOMATO MADS BOX GENE 6 |
| tZ | trans-zeatin |
| tZR | trans-zeatin riboside |
References
- Makkar, H.P.S.; Becker, K. Jatropha curcas, a promising crop for the generation of biodiesel and value-added coproducts. Eur. J. Lipid Sci. Technol. 2009, 111, 773–787. [Google Scholar] [CrossRef]
- Pramanik, K. Properties and use of jatropha curcas oil and diesel fuel blends in compression ignition engine. Renew. Energy 2003, 28, 239–248. [Google Scholar] [CrossRef]
- Mohibbeazam, M.; Waris, A.; Nahar, N. Prospects and potential of fatty acid methyl esters of some non-traditional seed oils for use as biodiesel in India. Biomass Bioenerg 2005, 29, 293–302. [Google Scholar] [CrossRef]
- Raju, A.J.S.; Ezradanam, V. Pollination ecology and fruiting behaviour in a monoecious species, Jatropha curcas L. (Euphorbiaceae). Curr. Sci. 2002, 83, 1395–1398. [Google Scholar]
- Ha, J.; Shim, S.; Lee, T.; Kang, Y.J.; Hwang, W.J.; Jeong, H.; Laosatit, K.; Lee, J.; Kim, S.K.; Satyawan, D.; et al. Genome sequence of Jatropha curcas L., a non-edible biodiesel plant, provides a resource to improve seed-related traits. Plant Biotechnol. J. 2019, 17, 517–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurcher, E.; Muller, B. Cytokinin Synthesis, Signaling, and Function—Advances and New Insights. Int. Rev. Cell Mol. Biol. 2016, 324, 1–38. [Google Scholar]
- Wybouw, B.; De Rybel, B. Cytokinin—A Developing Story. Trends Plant Sci. 2019, 24, 177–185. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [Green Version]
- Werner, T.; Schmulling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmulling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golovko, A.; Sitbon, F.; Tillberg, E.; Nicander, B. Identification of a tRNA isopentenyltransferase gene from Arabidopsis thaliana. Plant Mol. Biol. 2002, 49, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Sakakibara, H.; Sugiyama, T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 26405–26410. [Google Scholar] [CrossRef] [Green Version]
- Kakimoto, T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001, 42, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [Green Version]
- Sakakibara, H.; Kasahara, H.; Ueda, N.; Kojima, M.; Takei, K.; Hishiyama, S.; Asami, T.; Okada, K.; Kamiya, Y.; Yamaya, T.; et al. Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc. Natl. Acad. Sci. USA 2005, 102, 9972–9977. [Google Scholar] [CrossRef] [Green Version]
- Li, X.G.; Su, Y.H.; Zhao, X.Y.; Li, W.; Gao, X.Q.; Zhang, X.S. Cytokinin overproduction-caused alteration of flower development is partially mediated by CUC2 and CUC3 in Arabidopsis. Gene 2010, 450, 109–120. [Google Scholar] [CrossRef]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmulling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhao, B.; Yuan, D.; Duan, M.; Qian, Q.; Tang, L.; Wang, B.; Liu, X.; Zhang, J.; Wang, J.; et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. USA 2013, 110, 3167–3172. [Google Scholar] [CrossRef] [Green Version]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denay, G.; Chahtane, H.; Tichtinsky, G.; Parcy, F. A flower is born: An update on Arabidopsis floral meristem formation. Curr. Opin. Plant Biol. 2017, 35, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.F.; Sang, Y.L.; Wang, L.; Meng, W.J.; Zou, C.H.; Dong, Y.X.; Bie, X.M.; Cheng, Z.J.; Zhang, X.S. Type-B ARRs Control Carpel Regeneration Through Mediating AGAMOUS Expression in Arabidopsis. Plant Cell Physiol. 2018, 59, 756–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.; Hedden, P.; Tsiantis, M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005, 15, 1560–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scofield, S.; Dewitte, W.; Murray, J.A. The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J. 2007, 50, 767–781. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita-Tsujimura, K.; Kakimoto, T. Cytokinin receptors in sporophytes are essential for male and female functions in Arabidopsis thaliana. Plant Signal. Behav. 2011, 6, 66–71. [Google Scholar] [CrossRef] [Green Version]
- Riefler, M.; Novak, O.; Strnad, M.; Schmulling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Olalde, J.I.; Zuniga-Mayo, V.M.; Serwatowska, J.; Chavez Montes, R.A.; Lozano-Sotomayor, P.; Herrera-Ubaldo, H.; Gonzalez-Aguilera, K.L.; Ballester, P.; Ripoll, J.J.; Ezquer, I.; et al. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet. 2017, 13, e1006726. [Google Scholar] [CrossRef]
- Hui, W.; Wang, Y.; Yan, S.; Shi, J.; Huang, W.; Zayed, M.Z.; Peng, C.; Chen, X.; Wu, G. Simultaneous analysis of endogenous plant growth substances during floral sex differentiation in Jatropha curcas L. using HPLC–ESI–MS/MS. Sci. Hortic. 2018, 241, 209–217. [Google Scholar] [CrossRef]
- Hui, W.; Yang, Y.; Wu, G.; Peng, C.; Chen, X.; Zayed, M.Z. Transcriptome profile analysis reveals the regulation mechanism of floral sex differentiation in Jatropha curcas L. Sci. Rep. 2017, 7, 16421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.S.; Pan, B.Z.; Fu, Q.; Tao, Y.B.; Martinez-Herrera, J.; Niu, L.; Ni, J.; Dong, Y.; Zhao, M.L.; Xu, Z.F. Comparative Transcriptome Analysis between Gynoecious and Monoecious Plants Identifies Regulatory Networks Controlling Sex Determination in Jatropha curcas. Front. Plant Sci. 2017, 7, 1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khryanin, V.N. Role of Phytohormones in sex differentiation in plants. Russ. J. Plant Physiol. 2002, 49, 545–551. [Google Scholar] [CrossRef]
- Negi, S.S.; Olmo, H.P. Sex Conversion in a Male Vitis Vinifera L by a Kinin. Science 1966, 152, 1624. [Google Scholar] [CrossRef]
- Chailakhyan, M.K.; Khryanin, V.N. Effect of Growth-Regulators and Role of Roots in Sex Expression in Spinach. Planta 1978, 142, 207–210. [Google Scholar] [CrossRef]
- Louis, J.; Durand, B. Studies with the dioecious angiosperm Mercurialis annua L. (2n = 16): Correlation between genic and cytoplasmic male sterility, sex segregation and feminizing hormones (cytokinins). Mol. Gen. Genet. 1978, 165, 309–322. [Google Scholar] [CrossRef]
- Fu, Q.; Niu, L.; Zhang, Q.; Pan, B.-Z.; He, H.; Xu, Z.-F. Benzyladenine treatment promotes floral feminization and fruiting in a promising oilseed crop Plukenetia volubilis. Ind. Crop. Prod. 2014, 59, 295–298. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.; Shah, F.A.; Liu, W.; Wang, Q.; Wang, D.; Zhao, W.; Lu, W.; Huang, S.; Fu, S.; Wu, L. Comparative transcriptome analysis reveals the regulatory networks of cytokinin in promoting the floral feminization in the oil plant Sapium sebiferum. BMC Plant Biol. 2018, 18, 96. [Google Scholar] [CrossRef] [Green Version]
- Pan, B.-Z.; Xu, Z.-F. Benzyladenine Treatment Significantly Increases the Seed Yield of the Biofuel Plant Jatropha curcas. J. Plant Growth Regul. 2011, 30, 166–174. [Google Scholar] [CrossRef] [Green Version]
- Pan, B.-Z.; Luo, Y.; Song, L.; Chen, M.-S.; Li, J.-L.; Xu, Z.-F. Thidiazuron increases fruit number in the biofuel plant Jatropha curcas by promoting pistil development. Ind. Crop. Prod. 2016, 81, 202–210. [Google Scholar] [CrossRef]
- Kant, S.; Burch, D.; Badenhorst, P.; Palanisamy, R.; Mason, J.; Spangenberg, G. Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.). PLoS ONE 2015, 10, e0116349. [Google Scholar] [CrossRef] [PubMed]
- Pnueli, L.; Abu-Abeid, M.; Zamir, D.; Nacken, W.; Schwarz-Sommer, Z.; Lifschitz, E. The MADS box gene family in tomato: Temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1991, 1, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, Y.; Tang, L.; Zhang, F.; Chen, F. A study on structural features in early flower development of Jatropha curcas L. and the classification of its inflorescences. Afr. J. Agric. Res. 2011, 6, 275–284. [Google Scholar]
- Luo, C.-W.; Li, K.; Chen, Y.; Sun, Y.-Y. Floral display and breeding system of Jatropha curcas L. For. Stud. China 2007, 9, 114–119. [Google Scholar] [CrossRef]
- Ye, J.; Xu, M. Actin bundler PLIM2s are involved in the regulation of pollen development and tube growth in Arabidopsis. J. Plant Physiol. 2012, 169, 516–522. [Google Scholar] [CrossRef]
- Fröschle, M.; Horn, H.; Spring, O. Effects of the cytokinins 6-benzyladenine and forchlorfenuron on fruit-, seed- and yield parameters according to developmental stages of flowers of the biofuel plant Jatropha curcas L. (Euphorbiaceae). Plant Growth Regul. 2016, 81, 293–303. [Google Scholar] [CrossRef]
- Seesangboon, A.; Pokawattana, T.; Eungwanichayapant, P.D.; Tovaranonte, J.; Popluechai, S. Effects of 6-Benzyladenine on Jatropha Gene Expression and Flower Development. Russ. J. Plant Physiol. 2018, 65, 345–356. [Google Scholar] [CrossRef]
- Kiba, T.; Aoki, K.; Sakakibara, H.; Mizuno, T. Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 2004, 45, 1063–1077. [Google Scholar] [CrossRef]
- Akagi, T.; Henry, I.M.; Ohtani, H.; Morimoto, T.; Beppu, K.; Kataoka, I.; Tao, R. A Y-Encoded Suppressor of Feminization Arose via Lineage-Specific Duplication of a Cytokinin Response Regulator in Kiwifruit. Plant Cell 2018, 30, 780–795. [Google Scholar] [CrossRef]
- Zurcher, E.; Tavor-Deslex, D.; Lituiev, D.; Enkerli, K.; Tarr, P.T.; Muller, B. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013, 161, 1066–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartrina, I.; Jensen, H.; Novák, O.; Strnad, M.; Werner, T.; Schmülling, T. Gain-of-function mutants of the cytokinin receptors AHK2 and AHK3 regulate plant organ size, flowering time and plant longevity. Plant Physiol. 2017, 173, 1783–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sa, G.; Mi, M.; He-Chun, Y.; Guo-Feng, L. Anther-specific expression of ipt gene in transgenic tobacco and its effect on plant development. Transgenic Res. 2002, 11, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Sawhney, V.K.; Shukla, A. Male sterility in flowering plants: Are plant growth substances involved? Am. J. Bot. 1994, 81, 1640–1647. [Google Scholar] [CrossRef]
- Huang, S.; Cerny, R.E.; Qi, Y.; Bhat, D.; Aydt, C.M.; Hanson, D.D.; Malloy, K.P.; Ness, L.A. Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol. 2003, 131, 1270–1282. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Ito, T.; Wellmer, F.; Vernoux, T.; Dedieu, A.; Traas, J.; Meyerowitz, E.M. Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development 2009, 136, 1605–1611. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Dinh, T.T.; Li, D.; Shi, B.; Li, Y.; Cao, X.; Guo, L.; Pan, Y.; Jiao, Y.; Chen, X. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 2014, 80, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Prunet, N.; Morel, P.; Thierry, A.M.; Eshed, Y.; Bowman, J.L.; Negrutiu, I.; Trehin, C. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 2008, 20, 901–919. [Google Scholar] [CrossRef] [Green Version]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes directing flower development in Arabidopsis. Plant Cell 1989, 1, 37–52. [Google Scholar]
- Tao, Y.-B.; He, L.-L.; Niu, L.; Xu, Z.-F. Isolation and characterization of the Jatropha curcas APETALA1 (JcAP1) promoter conferring preferential expression in inflorescence buds. Planta 2016, 244, 467–478. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Li, C.; Tang, M.; Tao, Y.B.; Pan, B.Z.; Zhang, L.; Niu, L.; He, H.; Wang, X.; Xu, Z.F. An efficient protocol for Agrobacterium-mediated transformation of the biofuel plant Jatropha curcas by optimizing kanamycin concentration and duration of delayed selection. Plant Biotechnol. Rep. 2015, 9, 405–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Liu, Z.; Wei, F.; Feng, Y.-Q. Determination of cytokinins in plant samples by polymer monolith microextraction coupled with hydrophilic interaction chromatography-tandem mass spectrometry. Anal. Methods 2010, 2, 1676–1685. [Google Scholar] [CrossRef]
- Ding, L.W.; Sun, Q.Y.; Wang, Z.Y.; Sun, Y.B.; Xu, Z.F. Using silica particles to isolate total RNA from plant tissues recalcitrant to extraction in guanidine thiocyanate. Anal. Biochem. 2008, 374, 426–428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; He, L.L.; Fu, Q.T.; Xu, Z.F. Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using real-time quantitative PCR. Int. J. Mol. Sci. 2013, 14, 24338–24354. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, J.; Shi, B.; Yu, T.; Qi, J.; Meyerowitz, E.M.; Jiao, Y. The Stem Cell Niche in Leaf Axils Is Established by Auxin and Cytokinin in Arabidopsis. Plant Cell 2014, 26, 2055–2067. [Google Scholar] [CrossRef] [Green Version]
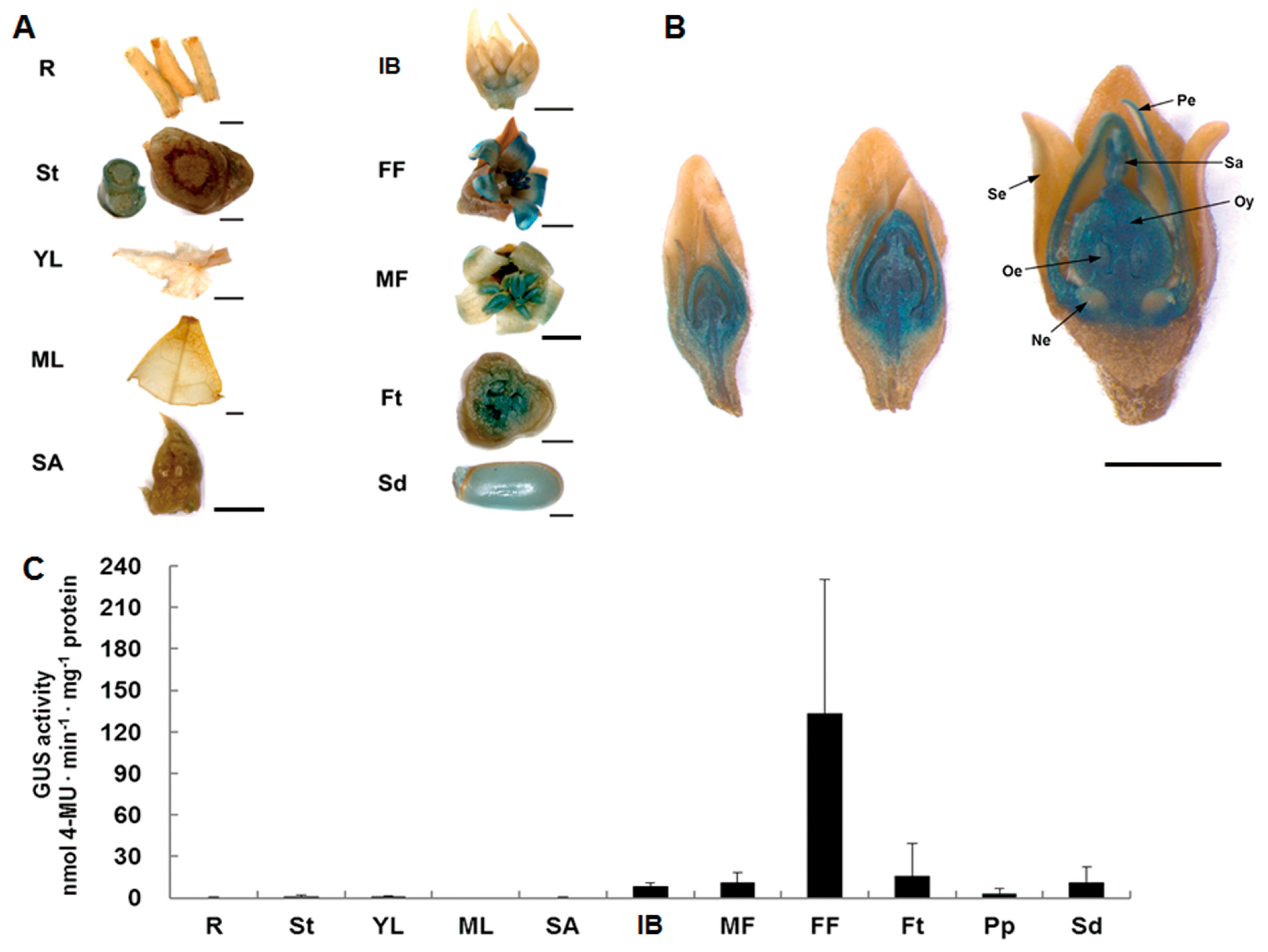

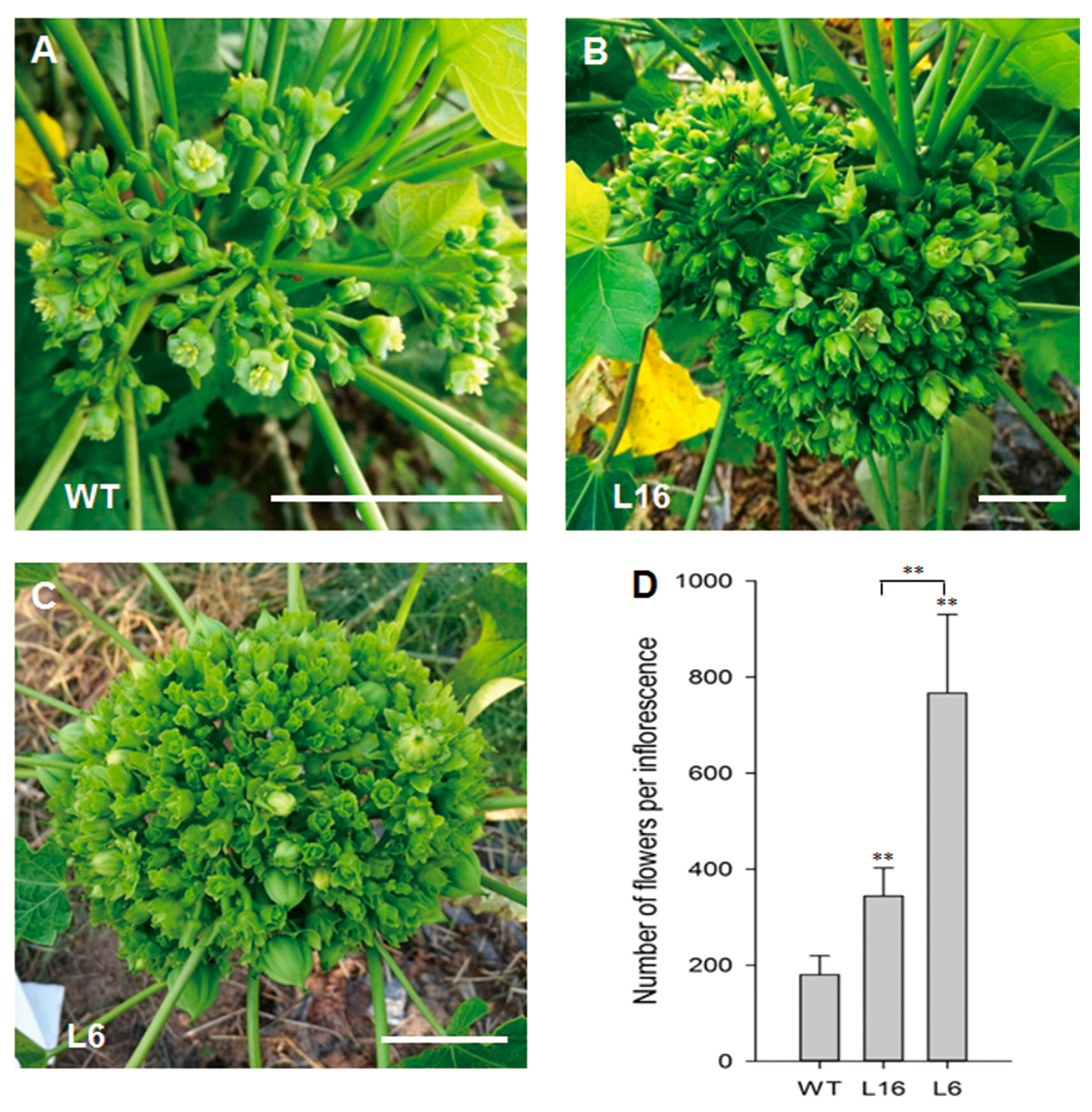


| Total Flowers | Male Flowers | Female Flowers | Bisexual Flowers | |
|---|---|---|---|---|
| WT | 180.25 ± 39.20 | 166.50 ± 35.94 (92.37%) | 13.75 ± 4.68 (7.63%) | 0.00 |
| L16 | 344.00 ± 58.42 | 147.25 ± 37.69 (42.81%) | 0.00 | 196.75 ± 55.06 (57.19%) |
| L6 | 767.00 ± 163.31 | 0.00 | 0.00 | 767.00 ± 163.31 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ming, X.; Tao, Y.-B.; Fu, Q.; Tang, M.; He, H.; Chen, M.-S.; Pan, B.-Z.; Xu, Z.-F. Flower-Specific Overproduction of Cytokinins Altered Flower Development and Sex Expression in the Perennial Woody Plant Jatropha curcas L. Int. J. Mol. Sci. 2020, 21, 640. https://doi.org/10.3390/ijms21020640
Ming X, Tao Y-B, Fu Q, Tang M, He H, Chen M-S, Pan B-Z, Xu Z-F. Flower-Specific Overproduction of Cytokinins Altered Flower Development and Sex Expression in the Perennial Woody Plant Jatropha curcas L. International Journal of Molecular Sciences. 2020; 21(2):640. https://doi.org/10.3390/ijms21020640
Chicago/Turabian StyleMing, Xin, Yan-Bin Tao, Qiantang Fu, Mingyong Tang, Huiying He, Mao-Sheng Chen, Bang-Zhen Pan, and Zeng-Fu Xu. 2020. "Flower-Specific Overproduction of Cytokinins Altered Flower Development and Sex Expression in the Perennial Woody Plant Jatropha curcas L." International Journal of Molecular Sciences 21, no. 2: 640. https://doi.org/10.3390/ijms21020640
APA StyleMing, X., Tao, Y.-B., Fu, Q., Tang, M., He, H., Chen, M.-S., Pan, B.-Z., & Xu, Z.-F. (2020). Flower-Specific Overproduction of Cytokinins Altered Flower Development and Sex Expression in the Perennial Woody Plant Jatropha curcas L. International Journal of Molecular Sciences, 21(2), 640. https://doi.org/10.3390/ijms21020640






