Mining the Microbiome and Microbiota-Derived Molecules in Inflammatory Bowel Disease
Abstract
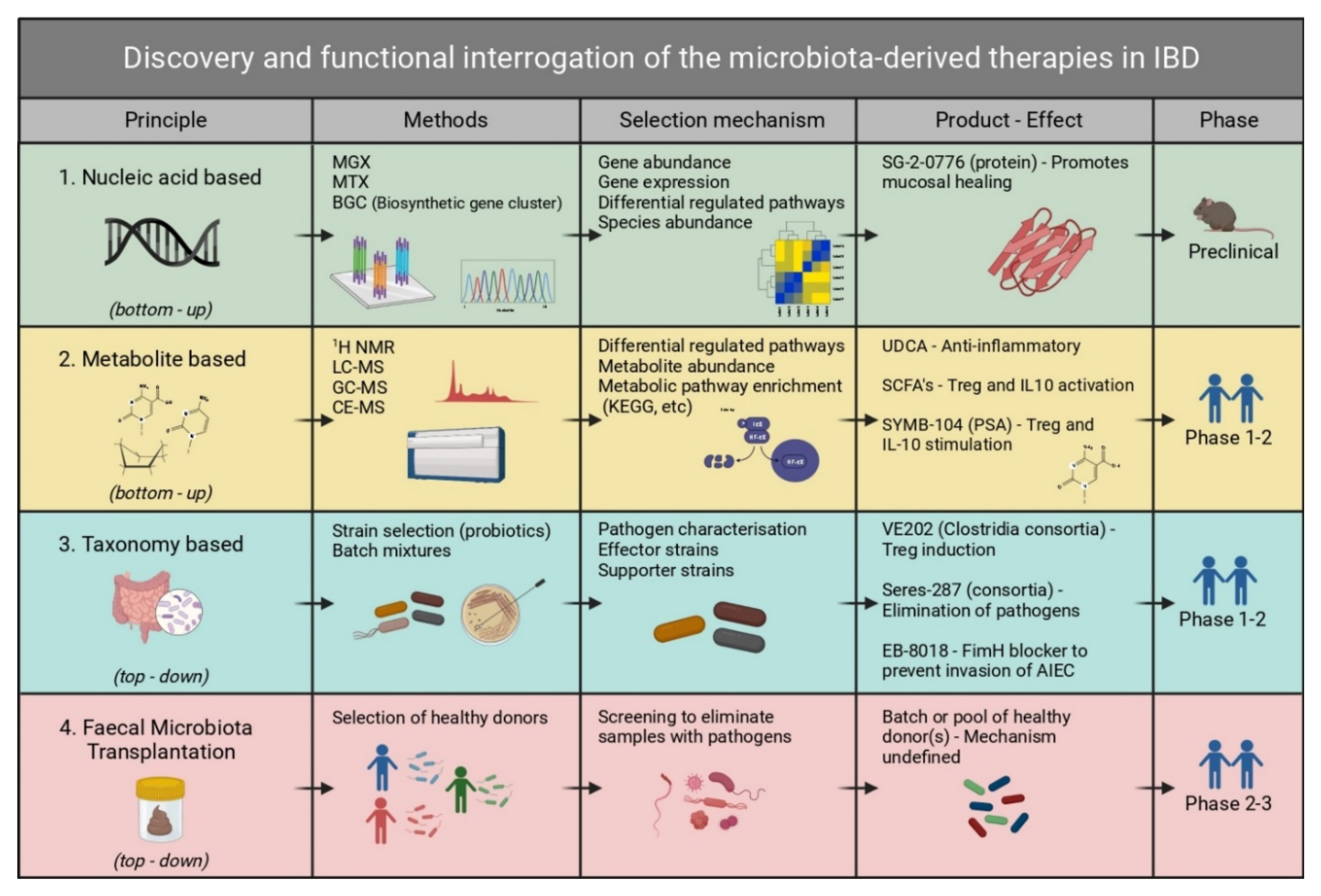
1. Introduction
2. Taxonomic Characterization of the Gut Microbiome in IBD
3. Fecal Microbiota Transplantation
4. Live Biotherapeutic Products
5. Metabolomics of the Gut Microbiome in IBD
6. Sequencing-Based Approaches
6.1. 16S rRNA Sequencing
6.2. Whole Metagenome Shotgun Sequencing (Metagenomics) and Analysis
6.3. Metatranscriptomics
7. Fractionation and Elimination Approach
8. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Ungaro, R.; Mehandru, S.; Allen, P.B.; Peyrin-Biroulet, L.; Colombel, J.F. Ulcerative colitis. Lancet 2017, 389, 1756–1770. [Google Scholar] [CrossRef]
- Torres, J.; Mehandru, S.; Colombel, J.F.; Peyrin-Biroulet, L. Crohn’s disease. Lancet 2017, 389, 1741–1755. [Google Scholar] [CrossRef]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef]
- Ott, S.J.; Musfeldt, M.; Wenderoth, D.F.; Hampe, J.; Brant, O.; Folsch, U.R.; Timmis, K.N.; Schreiber, S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004, 53, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, M.; Dogan, B.; Rishniw, M.; Weitzman, G.; Bosworth, B.; Yantiss, R.; Orsi, R.H.; Wiedmann, M.; McDonough, P.; Kim, S.G.; et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007, 1, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Schirmer, M.; Franzosa, E.A.; Lloyd-Price, J.; McIver, L.J.; Schwager, R.; Poon, T.W.; Ananthakrishnan, A.N.; Andrews, E.; Barron, G.; Lake, K. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 2018, 3, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.-L.; Barnich, N.; Bringer, M.-A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.-F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef]
- Reinisch, W.; Hébuterne, X.; Buisson, A.; Schreiber, S.; Desreumaux, P.; Paillarse, J.; Bonny, C. P568 An open-label, multicenter, phase ib, pharmacokinetic (pk) and safety study of a fimh blocker, Sibofimloc (TAK-018/EB8018), in patients with Crohn’s disease (CD). J. Crohns Colitis 2020, 14, S479–S480. [Google Scholar] [CrossRef]
- Youngster, I.; Russell, G.H.; Pindar, C.; Ziv-Baran, T.; Sauk, J.; Hohmann, E.L. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA 2014, 312, 1772–1778. [Google Scholar] [CrossRef]
- Bakken, J.S.; Borody, T.; Brandt, L.J.; Brill, J.V.; Demarco, D.C.; Franzos, M.A.; Kelly, C.; Khoruts, A.; Louie, T.; Martinelli, L.P. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011, 9, 1044–1049. [Google Scholar] [CrossRef]
- DeFilipp, Z.; Bloom, P.P.; Torres Soto, M.; Mansour, M.K.; Sater, M.R.; Huntley, M.H.; Turbett, S.; Chung, R.T.; Chen, Y.-B.; Hohmann, E.L. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 2019, 381, 2043–2050. [Google Scholar] [CrossRef]
- Bennet, J.; Brinkman, M. Treatment of ulcerative colitis by implantation of normal colonic flora. Lancet 1989, 1, 164. [Google Scholar] [CrossRef]
- Paramsothy, S.; Kamm, M.A.; Kaakoush, N.O.; Walsh, A.J.; van den Bogaerde, J.; Samuel, D.; Leong, R.W.; Connor, S.; Ng, W.; Paramsothy, R. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: A randomised placebo-controlled trial. Lancet 2017, 389, 1218–1228. [Google Scholar] [CrossRef]
- Costello, S.P.; Hughes, P.A.; Waters, O.; Bryant, R.V.; Vincent, A.D.; Blatchford, P.; Katsikeros, R.; Makanyanga, J.; Campaniello, M.A.; Mavrangelos, C. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: A randomized clinical trial. JAMA 2019, 321, 156–164. [Google Scholar] [CrossRef]
- Rossen, N.G.; Fuentes, S.; van der Spek, M.J.; Tijssen, J.G.; Hartman, J.H.; Duflou, A.; Löwenberg, M.; van den Brink, G.R.; Mathus-Vliegen, E.M.; de Vos, W.M. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 2015, 149, 110–118.e4. [Google Scholar] [CrossRef] [PubMed]
- Moayyedi, P.; Surette, M.G.; Kim, P.T.; Libertucci, J.; Wolfe, M.; Onischi, C.; Armstrong, D.; Marshall, J.K.; Kassam, Z.; Reinisch, W. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 2015, 149, 102–109.e6. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Chuai, S.; Nessel, L.; Lichtenstein, G.R.; Aberra, F.N.; Ellenberg, J.H. Use of the noninvasive components of the Mayo score to assess clinical response in ulcerative colitis. Inflamm. Bowel Dis. 2008, 14, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Imdad, A.; Nicholson, M.R.; Tanner-Smith, E.E.; Zackular, J.P.; Gomez-Duarte, O.G.; Beaulieu, D.B.; Acra, S. Fecal transplantation for treatment of inflammatory bowel disease. Cochrane Database Syst Rev. 2018, 11, CD012774. [Google Scholar] [CrossRef]
- Kaiko, G.E.; Stappenbeck, T.S. Host–microbe interactions shaping the gastrointestinal environment. Trends Immunol. 2014, 35, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Ruff, W.E.; Greiling, T.M.; Kriegel, M.A. Host–microbiota interactions in immune-mediated diseases. Nat. Rev. Microbiol. 2020, 18, 521–538. [Google Scholar] [CrossRef]
- Guo, C.-J.; Allen, B.M.; Hiam, K.J.; Dodd, D.; Van Treuren, W.; Higginbottom, S.; Nagashima, K.; Fischer, C.R.; Sonnenburg, J.L.; Spitzer, M.H.; et al. Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science 2019, 366, 6471. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Ando, M.; Kamada, N.; Nagano, Y.; Narushima, S.; Suda, W.; Imaoka, A.; Setoyama, H.; Nagamori, T. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015, 163, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Viladomiu, M.; Kivolowitz, C.; Abdulhamid, A.; Dogan, B.; Victorio, D.; Castellanos, J.G.; Woo, V.; Teng, F.; Tran, N.L.; Sczesnak, A.; et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci. Transl. Med. 2017, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Sartor, R.B. Manipulating resident microbiota to enhance regulatory immune function to treat inflammatory bowel diseases. J. Gastroenterol. 2020, 55, 4–14. [Google Scholar] [CrossRef]
- Charbonneau, M.R.; Isabella, V.M.; Li, N.; Kurtz, C.B. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Ross, J.J.; Boucher, P.E.; Bhattacharyya, S.P.; Kopecko, D.J.; Sutkowski, E.M.; Rohan, P.J.; Chandler, D.K.; Vaillancourt, J. Considerations in the development of live biotherapeutic products for clinical use. Curr. Issues Mol. Biol. 2008, 10, 13–16. [Google Scholar]
- Claesen, J.; Fischbach, M.A. Synthetic microbes as drug delivery systems. ACS Synth. Biol. 2015, 4, 358–364. [Google Scholar] [CrossRef]
- Schultz, M. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm. Bowel Dis. 2008, 14, 1012–1018. [Google Scholar] [CrossRef]
- Dignass, A.; Lindsay, J.O.; Sturm, A.; Windsor, A.; Colombel, J.-F.; Allez, M.; D’Haens, G.; D’Hoore, A.; Mantzaris, G.; Novacek, G. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis part 2: Current management. J. Crohns Colitis 2012, 6, 991–1030. [Google Scholar] [CrossRef] [PubMed]
- Losurdo, G.; Iannone, A.; Contaldo, A.; Ierardi, E.; Di Leo, A.; Principi, M. Escherichia coli Nissle 1917 in ulcerative colitis treatment: Systematic review and meta-analysis. J. Gastrointestin. Liver Dis. 2015, 24, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Praveschotinunt, P.; Duraj-Thatte, A.M.; Gelfat, I.; Bahl, F.; Chou, D.B.; Joshi, N.S. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 2019, 10, 5580. [Google Scholar] [CrossRef] [PubMed]
- Prantera, C.; Scribano, M.; Falasco, G.; Andreoli, A.; Luzi, C. Ineffectiveness of probiotics in preventing recurrence after curative resection for Crohn’s disease: A randomised controlled trial with Lactobacillus GG. Gut 2002, 51, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.-H.; Chen, C.-L.; Wang, J.-D.; Yang, Y.-J.; Cun, Y.; Wu, J.-B.; Liu, Y.-H.; Dan, H.-L.; Jian, Y.-T.; Chen, X.-Q. Effects of probiotic on intestinal mucosa of patients with ulcerative colitis. World J. Gastroenterol. 2004, 10, 1521. [Google Scholar] [CrossRef]
- Bjarnason, I.; Sission, G. A randomised, double-blind, placebo-controlled trial of a multi-strain probiotic in patients with asymptomatic ulcerative colitis and Crohn’s disease. Inflammopharmacology 2019, 27, 465–473. [Google Scholar] [CrossRef]
- Sang, L.-X.; Chang, B.; Zhang, W.-L.; Wu, X.-M.; Li, X.-H.; Jiang, M. Remission induction and maintenance effect of probiotics on ulcerative colitis: A meta-analysis. World J. Gastroenterol. WJG 2010, 16, 1908. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K. T reg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Misra, B.; Curran, J.; Herfarth, H.; Jagarlamudi, K.; Oneto, C.; Bhandari, B.; Wiener, G.; Kerman, D.; Moss, A.; Pomerantz, R. P421 SER-287, an investigational microbiome therapeutic, induces remission and endoscopic improvement in a placebo-controlled, double-blind randomised trial in patients with active mild-to-moderate ulcerative colitis. J. Crohns Colitis 2018, 12, S317. [Google Scholar] [CrossRef][Green Version]
- Wrzosek, L.; Miquel, S.; Noordine, M.-L.; Bouet, S.; Chevalier-Curt, M.J.; Robert, V.; Philippe, C.; Bridonneau, C.; Cherbuy, C.; Robbe-Masselot, C.; et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Dreher-Lesnick, S.M.; Stibitz, S.; Carlson, J.J.; Paul, E. US regulatory considerations for development of live biotherapeutic products as drugs. Microbiol. Spectr. 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.J.; Cho, J.H.; Gevers, D.; Chu, H. Genetic factors and the intestinal microbiome guide development of microbe-based therapies for inflammatory bowel diseases. Gastroenterology 2019, 156, 2174–2189. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.S.; Sauk, J.; Gevers, D.; Friedman, J.; Sung, J.; Youngster, I.; Hohmann, E.L.; Staley, C.; Khoruts, A.; Sadowsky, M.J. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 2018, 23, 229–240.e5. [Google Scholar] [CrossRef] [PubMed]
- Cieplak, T.; Soffer, N.; Sulakvelidze, A.; Nielsen, D.S. A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes 2018, 9, 391–399. [Google Scholar] [PubMed]
- Nakamoto, N.; Sasaki, N.; Aoki, R.; Miyamoto, K.; Suda, W.; Teratani, T.; Suzuki, T.; Koda, Y.; Chu, P.-S.; Taniki, N. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol. 2019, 4, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of recent developments in GC-MS approaches to metabolomics-based research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Huda-Faujan, N.; Abdulamir, A.S.; Fatimah, A.B.; Anas, O.M.; Shuhaimi, M.; Yazid, A.M.; Loong, Y.Y. The impact of the level of the intestinal short chain Fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem. J. 2010, 4, 53–58. [Google Scholar] [CrossRef]
- Scheppach, W.; Sommer, H.; Kirchner, T.; Paganelli, G.M.; Bartram, P.; Christl, S.; Richter, F.; Dusel, G.; Kasper, H. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology 1992, 103, 51–56. [Google Scholar] [CrossRef]
- Breuer, R.I.; Soergel, K.H.; Lashner, B.A.; Christ, M.L.; Hanauer, S.B.; Vanagunas, A.; Harig, J.M.; Keshavarzian, A.; Robinson, M.; Sellin, J.H.; et al. Short chain fatty acid rectal irrigation for left-sided ulcerative colitis: A randomised, placebo controlled trial. Gut 1997, 40, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Jamka, M.; Kokot, M.; Kaczmarek, N.; Bermagambetova, S.; Nowak, J.K.; Walkowiak, J. The Effect of Sodium Butyrate Enemas Compared with Placebo on Disease Activity, Endoscopic Scores, and Histological and Inflammatory Parameters in Inflammatory Bowel Diseases: A Systematic Review of Randomised Controlled Trials. Complement. Med. Res. 2021, 28, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.R.; Haileselassie, Y.; Nguyen, L.P.; Tropini, C.; Wang, M.; Becker, L.S.; Sim, D.; Jarr, K.; Spear, E.T.; Singh, G.; et al. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe 2020, 27, 659–670.e5. [Google Scholar] [CrossRef] [PubMed]
- Keely, S.J.; Steer, C.J.; Lajczak-McGinley, N.K. Ursodeoxycholic acid: A promising therapeutic target for inflammatory bowel diseases? Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G872–G881. [Google Scholar] [CrossRef]
- Kolho, K.L.; Pessia, A.; Jaakkola, T.; de Vos, W.M.; Velagapudi, V. Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J. Crohns Colitis 2017, 11, 321–334. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef]
- Fornelos, N.; Franzosa, E.A.; Bishai, J.; Annand, J.W.; Oka, A.; Lloyd-Price, J.; Arthur, T.D.; Garner, A.; Avila-Pacheco, J.; Haiser, H.J.; et al. Growth effects of N-acylethanolamines on gut bacteria reflect altered bacterial abundances in inflammatory bowel disease. Nat. Microbiol. 2020, 5, 486–497. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Ongey, E.L.; Giessmann, R.T.; Fons, M.; Rappsilber, J.; Adrian, L.; Neubauer, P. Heterologous Biosynthesis, Modifications and Structural Characterization of Ruminococcin-A, a Lanthipeptide From the Gut Bacterium Ruminococcus gnavus E1, in Escherichia coli. Front. Microbiol. 2018, 9, 1688. [Google Scholar] [CrossRef]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [PubMed]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37.e6. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Byrd, D.A.; Sinha, R.; Hoffman, K.L.; Chen, J.; Hua, X.; Shi, J.; Chia, N.; Petrosino, J.; Vogtmann, E. Comparison of methods to collect fecal samples for microbiome studies using whole-genome shotgun metagenomic sequencing. mSphere 2020, 5, e00827-19. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; Morgan, X.C.; Segata, N.; Waldron, L.; Reyes, J.; Earl, A.M.; Giannoukos, G.; Boylan, M.R.; Ciulla, D.; Gevers, D. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. USA 2014, 111, E2329–E2338. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, J.D.; van den Bogert, B.; Edwards, J.E.; Boekhorst, J.; Van Gastelen, S.; Saccenti, E.; Plugge, C.M.; Smidt, H. The effect of DNA extraction methods on observed microbial communities from fibrous and liquid rumen fractions of dairy cows. Front. Microbiol. 2018, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Cohen, D.B.; Ravel, J.; Abdo, Z.; Forney, L.J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS ONE 2012, 7, e33865. [Google Scholar] [CrossRef]
- Poretsky, R.; Rodriguez-R, L.M.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9, e93827. [Google Scholar] [CrossRef]
- Young, R.B.; Marcelino, V.R.; Chonwerawong, M.; Gulliver, E.L.; Forster, S.C. Key Technologies for Progressing Discovery of Microbiome-Based Medicines. Front. Microbiol. 2021, 12, 1604. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef] [PubMed]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [PubMed]
- Chehoud, C.; Albenberg, L.G.; Judge, C.; Hoffmann, C.; Grunberg, S.; Bittinger, K.; Baldassano, R.N.; Lewis, J.D.; Bushman, F.D.; Wu, G.D. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2015, 21, 1948–1956. [Google Scholar] [CrossRef]
- Clooney, A.G.; Sutton, T.D.; Shkoporov, A.N.; Holohan, R.K.; Daly, K.M.; O’Regan, O.; Ryan, F.J.; Draper, L.A.; Plevy, S.E.; Ross, R.P.; et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 2019, 26, 764–778.e5. [Google Scholar] [CrossRef]
- Zuo, T.; Lu, X.-J.; Zhang, Y.; Cheung, C.P.; Lam, S.; Zhang, F.; Tang, W.; Ching, J.Y.; Zhao, R.; Chan, P.K.; et al. Gut mucosal virome alterations in ulcerative colitis. Gut 2019, 68, 1169–1179. [Google Scholar] [CrossRef]
- Nelson, M.T.; Pope, C.E.; Marsh, R.L.; Wolter, D.J.; Weiss, E.J.; Hager, K.R.; Vo, A.T.; Brittnacher, M.J.; Radey, M.C.; Hayden, H.S.; et al. Human and extracellular DNA depletion for metagenomic analysis of complex clinical infection samples yields optimized viable microbiome profiles. Cell Rep. 2019, 26, 2227–2240.e5. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef]
- Bharti, R.; Grimm, D.G. Current challenges and best-practice protocols for microbiome analysis. Brief. Bioinform. 2021, 22, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Boisvert, S.; Raymond, F.; Godzaridis, É.; Laviolette, F.; Corbeil, J. Ray Meta: Scalable de novo metagenome assembly and profiling. Genome Biol. 2012, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Leung, H.C.; Yiu, S.-M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 2047-217X. [Google Scholar] [CrossRef] [PubMed]
- Pell, J.; Hintze, A.; Canino-Koning, R.; Howe, A.; Tiedje, J.M.; Brown, C.T. Scaling metagenome sequence assembly with probabilistic de Bruijn graphs. Proc. Natl. Acad. Sci. USA 2012, 109, 13272–13277. [Google Scholar] [CrossRef] [PubMed]
- Sedlar, K.; Kupkova, K.; Provaznik, I. Bioinformatics strategies for taxonomy independent binning and visualization of sequences in shotgun metagenomics. Comput. Struct. Biotechnol. J. 2017, 15, 48–55. [Google Scholar] [CrossRef]
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biol. 2016, 17, 239. [Google Scholar] [CrossRef]
- Albertsen, M.; Hugenholtz, P.; Skarshewski, A.; Nielsen, K.L.; Tyson, G.W.; Nielsen, P.H. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 2013, 31, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Duncan, S.H.; Louis, P.; Flint, H.J. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol. 2014, 22, 267–274. [Google Scholar] [CrossRef]
- Almeida, A.; Mitchell, A.L.; Boland, M.; Forster, S.C.; Gloor, G.B.; Tarkowska, A.; Lawley, T.D.; Finn, R.D. A new genomic blueprint of the human gut microbiota. Nature 2019, 568, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Tørresen, O.K.; Star, B.; Mier, P.; Andrade-Navarro, M.A.; Bateman, A.; Jarnot, P.; Gruca, A.; Grynberg, M.; Kajava, A.V.; Promponas, V.J. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 2019, 47, 10994–11006. [Google Scholar] [CrossRef]
- Liao, Y.-C.; Lin, S.-H.; Lin, H.-H. Completing bacterial genome assemblies: Strategy and performance comparisons. Sci. Rep. 2015, 5, 1–8. [Google Scholar] [CrossRef]
- Woyke, T.; Doud, D.F.; Schulz, F. The trajectory of microbial single-cell sequencing. Nat. Methods 2017, 14, 1045–1054. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Luo, C.; Yajnik, V.; Khalili, H.; Garber, J.J.; Stevens, B.W.; Cleland, T.; Xavier, R.J. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe 2017, 21, 603–610.e603. [Google Scholar] [CrossRef]
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.R.; Griffiths, A.M.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.S.; Hoffmann, C. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015, 18, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R.; et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef]
- Vaughn, B.P.; Vatanen, T.; Allegretti, J.R.; Bai, A.; Xavier, R.J.; Korzenik, J.; Gevers, D.; Ting, A.; Robson, S.C.; Moss, A.C. Increased intestinal microbial diversity following fecal microbiota transplant for active Crohn’s disease. Inflamm. Bowel Dis. 2016, 22, 2182–2190. [Google Scholar] [CrossRef]
- Paramsothy, S.; Nielsen, S.; Kamm, M.A.; Deshpande, N.P.; Faith, J.J.; Clemente, J.C.; Paramsothy, R.; Walsh, A.J.; van den Bogaerde, J.; Samuel, D. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology 2019, 156, 1440–1454.e2. [Google Scholar] [CrossRef] [PubMed]
- McNulty, N.P.; Yatsunenko, T.; Hsiao, A.; Faith, J.J.; Muegge, B.D.; Goodman, A.L.; Henrissat, B.; Oozeer, R.; Cools-Portier, S.; Gobert, G.; et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 2011, 3, 106ra106. [Google Scholar] [CrossRef]
- Maurice, C.F.; Haiser, H.J.; Turnbaugh, P.J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2013, 152, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Kushner, S.R. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002, 184, 4658–4665. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deutscher, M.P. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006, 34, 659–666. [Google Scholar] [CrossRef]
- O’Neil, D.; Glowatz, H.; Schlumpberger, M. Ribosomal RNA depletion for efficient use of RNA-seq capacity. Curr. Protoc. Mol. Biol. 2013, 103, 4–19. [Google Scholar] [CrossRef]
- Giannoukos, G.; Ciulla, D.M.; Huang, K.; Haas, B.J.; Izard, J.; Levin, J.Z.; Livny, J.; Earl, A.M.; Gevers, D.; Ward, D.V.; et al. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 2012, 13, r43. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Wurtzel, O.; Singh, K.; Froula, J.L.; Yilmaz, S.; Tringe, S.G.; Wang, Z.; Chen, F.; Lindquist, E.A.; Sorek, R. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat. Methods 2010, 7, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Quince, C.; Faith, J.J.; McHardy, A.C.; Yatsunenko, T.; Niazi, F.; Affourtit, J.; Egholm, M.; Henrissat, B.; Knight, R. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. USA 2010, 107, 7503–7508. [Google Scholar] [CrossRef] [PubMed]
- Blattman, S.B.; Jiang, W.; Oikonomou, P.; Tavazoie, S. Prokaryotic single-cell RNA sequencing by in situ combinatorial indexing. Nat. Microbiol. 2020, 5, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Kuchina, A.; Brettner, L.M.; Paleologu, L.; Roco, C.M.; Rosenberg, A.B.; Carignano, A.; Kibler, R.; Hirano, M.; DePaolo, R.W.; Seelig, G. Microbial single-cell RNA sequencing by split-pool barcoding. Science 2021, 371, 6531. [Google Scholar] [CrossRef]
- Bucar, F.; Wube, A.; Schmid, M. Natural product isolation--how to get from biological material to pure compounds. Nat. Prod. Rep. 2013, 30, 525–545. [Google Scholar] [CrossRef]
- Colosimo, D.A.; Kohn, J.A.; Luo, P.M.; Piscotta, F.J.; Han, S.M.; Pickard, A.J.; Rao, A.; Cross, J.R.; Cohen, L.J.; Brady, S.F. Mapping Interactions of Microbial Metabolites with Human G-Protein-Coupled Receptors. Cell Host Microbe 2019, 26, 273–282.e7. [Google Scholar] [CrossRef] [PubMed]
| Discovery | Company/Institute | Product Name | Mechanism | Product Type | NCT Number | Status |
|---|---|---|---|---|---|---|
| Nucleic acid-based production | Second Genome | SG-2-0776 | Promotes mucosal healing | Protein | Not found | Proceeding to Phase 2 |
| Taxonomy based/probiotic | Seres Therapeutics | Seres-287 | Reduces gut inflammation | LBP | NCT03759041 | Phase 2 active |
| Vedanta | VE202 | Treg induction | LBP | NCT03723746, NCT03931447 | Phase 1 completed | |
| 4D Pharma | Thetanix | Antagonizes NF-κB | LBP | NCT02704728 | Phase 1 completed | |
| Enterome | EB-8018/Sibofimloc | FimH inhibition to block AIEC | Small molecule | NCT03709628, NCT03943446 | Phase 2 recruiting | |
| Intralytix | EcoActive | Depletes AIEC | Bacteriophage | NCT03808103 | Phase 1 recruiting | |
| BiomX | BX003 (BX002) | Targets Klebsiella pneumoniae | Bacteriophage cocktail | NCT04737876 | Phase 1 completed | |
| Nordisk rebalance | Profermin | Reduces dysbiosis | Probiotic strains + fiber | NCT01245465, NCT01193894 | Phase 2 and Phase 3 completed | |
| Metabolite based | Brigham and Women’s Hospital | Butyrate with Hydroxocobalamin | Calprotectin reduction | Short-chain fatty acid | NCT04259060 | Phase 2 |
| University of Padova | Micro-encapsulated sodium Butyrate | Treg and IL10 activation | Short-chain fatty acid | NCT04879914 | Not applicable | |
| Stanford University | Ursodeoxycholic acid | Inflammatory marker reduction | Secondary bile acid | NCT03724175 | Phase 2/3 recruiting | |
| University Medical Centre Groningen | Vitamin B2 | Increase in the amount of F. prausnitzii | Vitamin metabolite | NCT02538354 | Completed | |
| University Medical Centre Groningen | Vitamin B3 | Reduction in inflammation and oxidative stress | Vitamin metabolite | NCT04913467 | Phase 3 |
| Sequencing Method | Strengths | Limitations |
|---|---|---|
| 16S rRNA | Low cost for high throughput | Limited taxonomic resolution |
| Lower complexity of bioinformatics analysis | Amplification bias | |
| Established taxonomic reference databases available | Does not capture gene content | |
| Shotgun Metagenomic Sequencing | Sequences any organism present but only at high read depth | Higher cost |
| Identifies functional potential and gene content | Higher complexity of bioinformatics analysis | |
| Host DNA contamination | ||
| Shotgun Metatranscriptomic Sequencing | Detects and quantifies microbial gene expression | Instability of mRNA |
| Identifies active functional genes and pathways rather than DNA abundance of dead bacteria/silent genes | Host RNA contamination | |
| Identifies the relative importance and gene activity of similarly abundant microbial species | High abundance of ribosomal RNA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bekkers, M.; Stojkovic, B.; Kaiko, G.E. Mining the Microbiome and Microbiota-Derived Molecules in Inflammatory Bowel Disease. Int. J. Mol. Sci. 2021, 22, 11243. https://doi.org/10.3390/ijms222011243
Bekkers M, Stojkovic B, Kaiko GE. Mining the Microbiome and Microbiota-Derived Molecules in Inflammatory Bowel Disease. International Journal of Molecular Sciences. 2021; 22(20):11243. https://doi.org/10.3390/ijms222011243
Chicago/Turabian StyleBekkers, Matthijs, Bojan Stojkovic, and Gerard E. Kaiko. 2021. "Mining the Microbiome and Microbiota-Derived Molecules in Inflammatory Bowel Disease" International Journal of Molecular Sciences 22, no. 20: 11243. https://doi.org/10.3390/ijms222011243
APA StyleBekkers, M., Stojkovic, B., & Kaiko, G. E. (2021). Mining the Microbiome and Microbiota-Derived Molecules in Inflammatory Bowel Disease. International Journal of Molecular Sciences, 22(20), 11243. https://doi.org/10.3390/ijms222011243






