Dual Inhibitors of AChE and BACE-1 for Reducing Aβ in Alzheimer’s Disease: From In Silico to In Vivo
Abstract
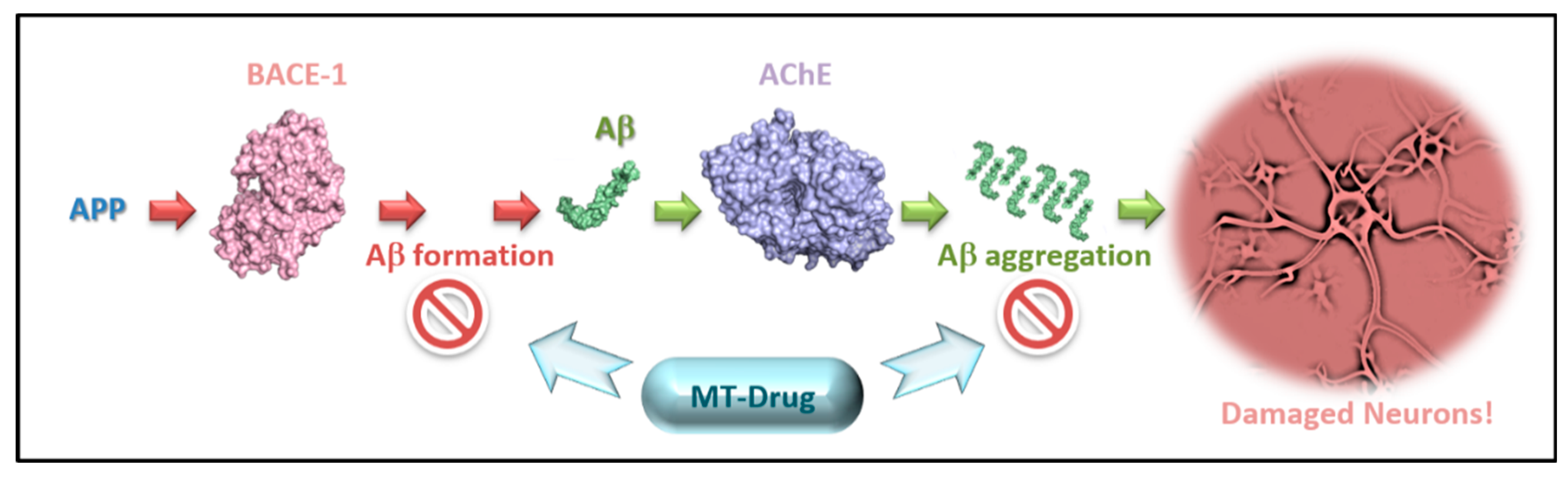
:1. Introduction
2. Methods
2.1. Preparation of the Classification Models and Screening
2.1.1. Initial Preparation and Filtration of Datasets
2.1.2. Preparation of the Training Sets
2.1.3. Model Construction and Evaluation
2.1.4. Screening External Databases with ISE Models
2.2. Docking Preparations and Screening by Docking
2.2.1. General
2.2.2. Selection of a Crystal Structure for Docking to BACE-1
2.2.3. Selection of a Crystal Structure for Docking to AChE
2.2.4. Crystal Structures Preparation for Docking
2.2.5. Docking and Analysis of the Results
2.3. Choice of Molecules by Weighted Scores Based on ISE Indexing and Docking Results
2.4. Biological Examination
2.4.1. BACE-1 In Vitro Single Point Inhibition Measurement
2.4.2. BACE-1 IC50 Determination of Hits
2.4.3. AChE In Vitro Single Point Inhibition Measurement
2.4.4. AChE IC50 Determination of Hits
2.4.5. Animal Experiment
2.4.6. Detection of Amyloid Precursors by ELISA Method
3. Results and Discussion
3.1. Comparison, Evaluation and the Best Physico-Chemical Properties of BACE-1 and AChE Models
3.2. Results from Screening DrugBank
3.3. Selection of Best BACE-1 and AChE Structures for Docking
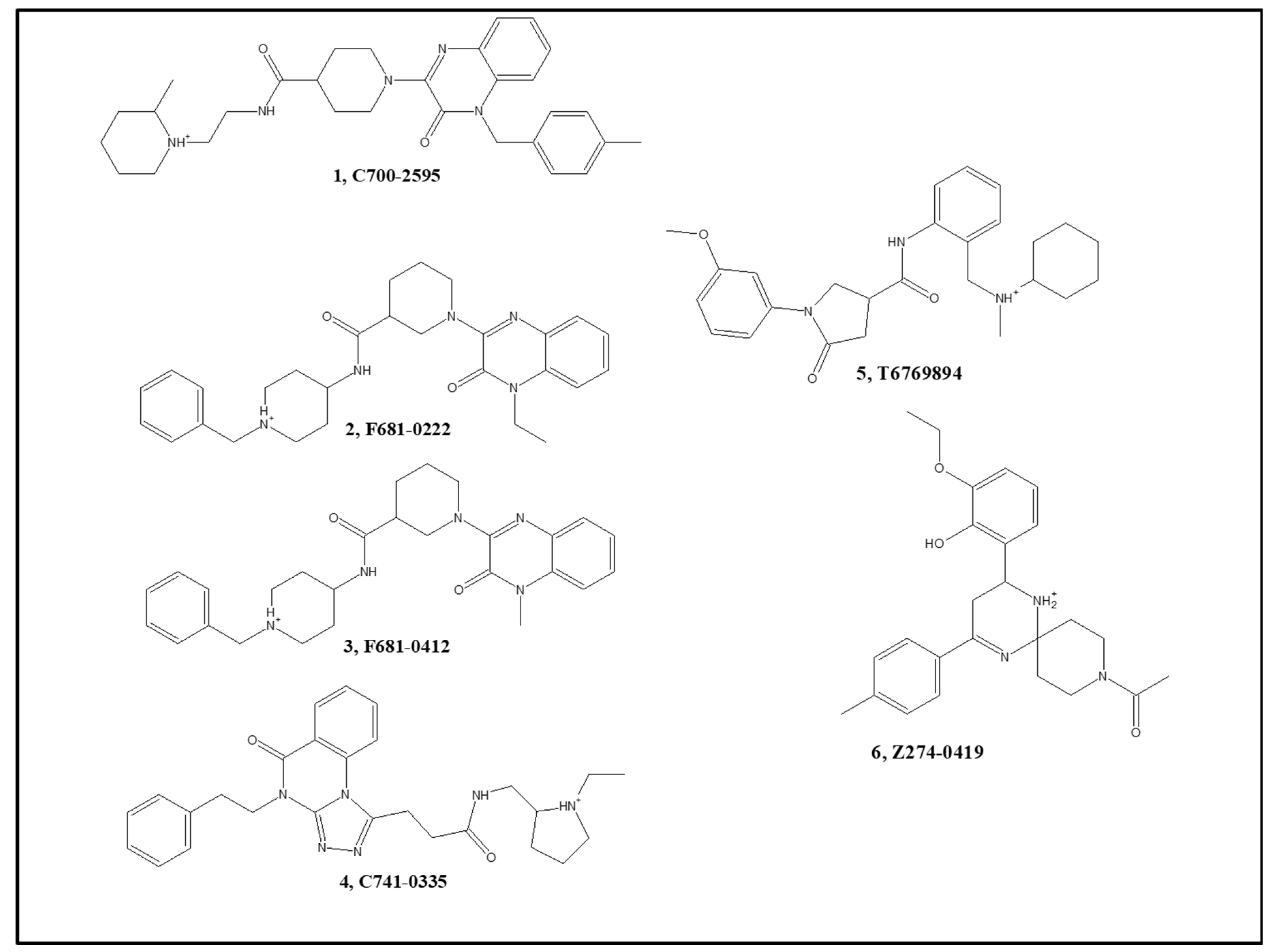
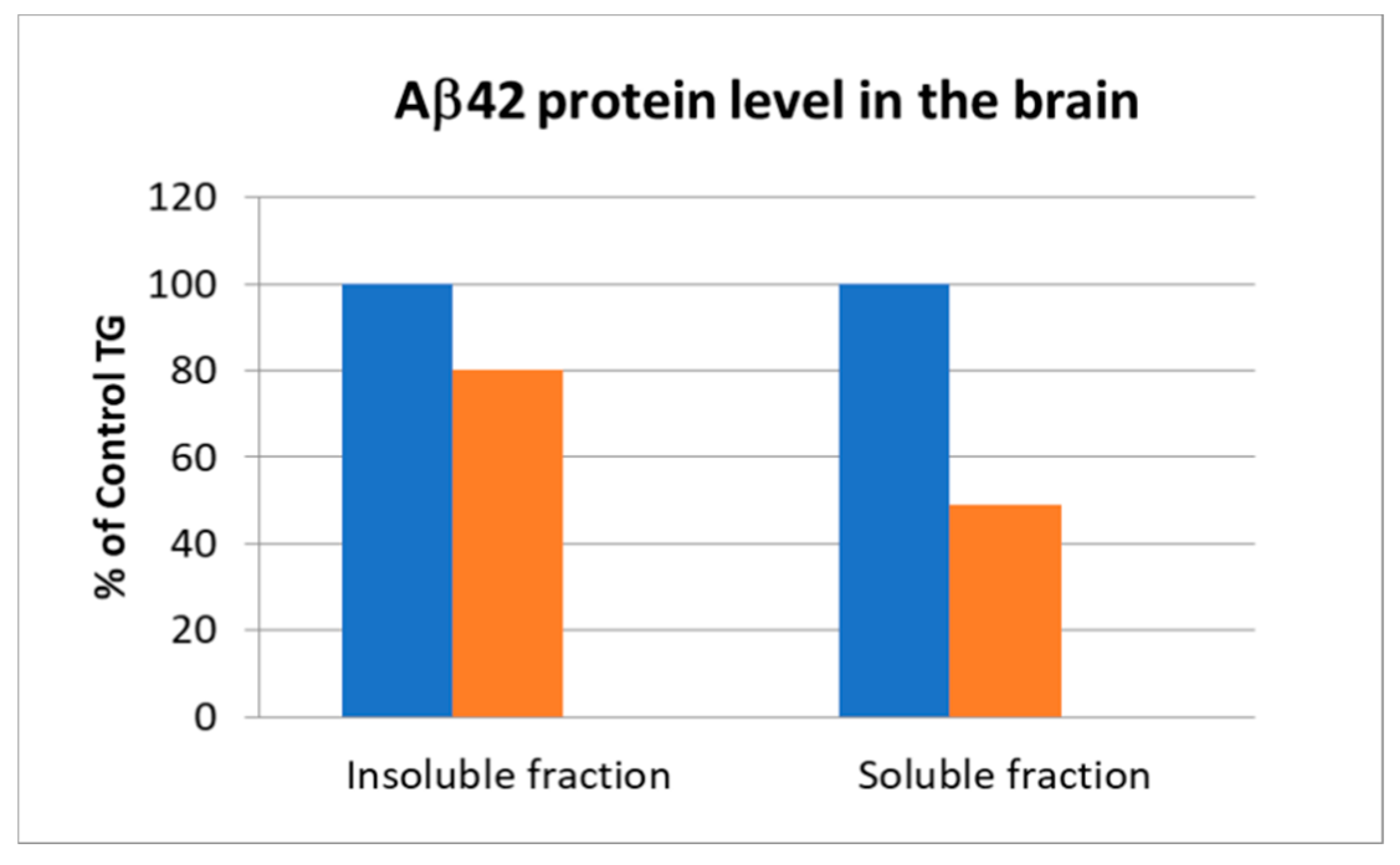
3.4. In Vitro Activity of Hits and In Vivo Results of a Dual Hit
3.5. New Scaffolds for AChE/BACE-1 Inhibitors
3.6. Analysis of Computational vs. Experimental Results
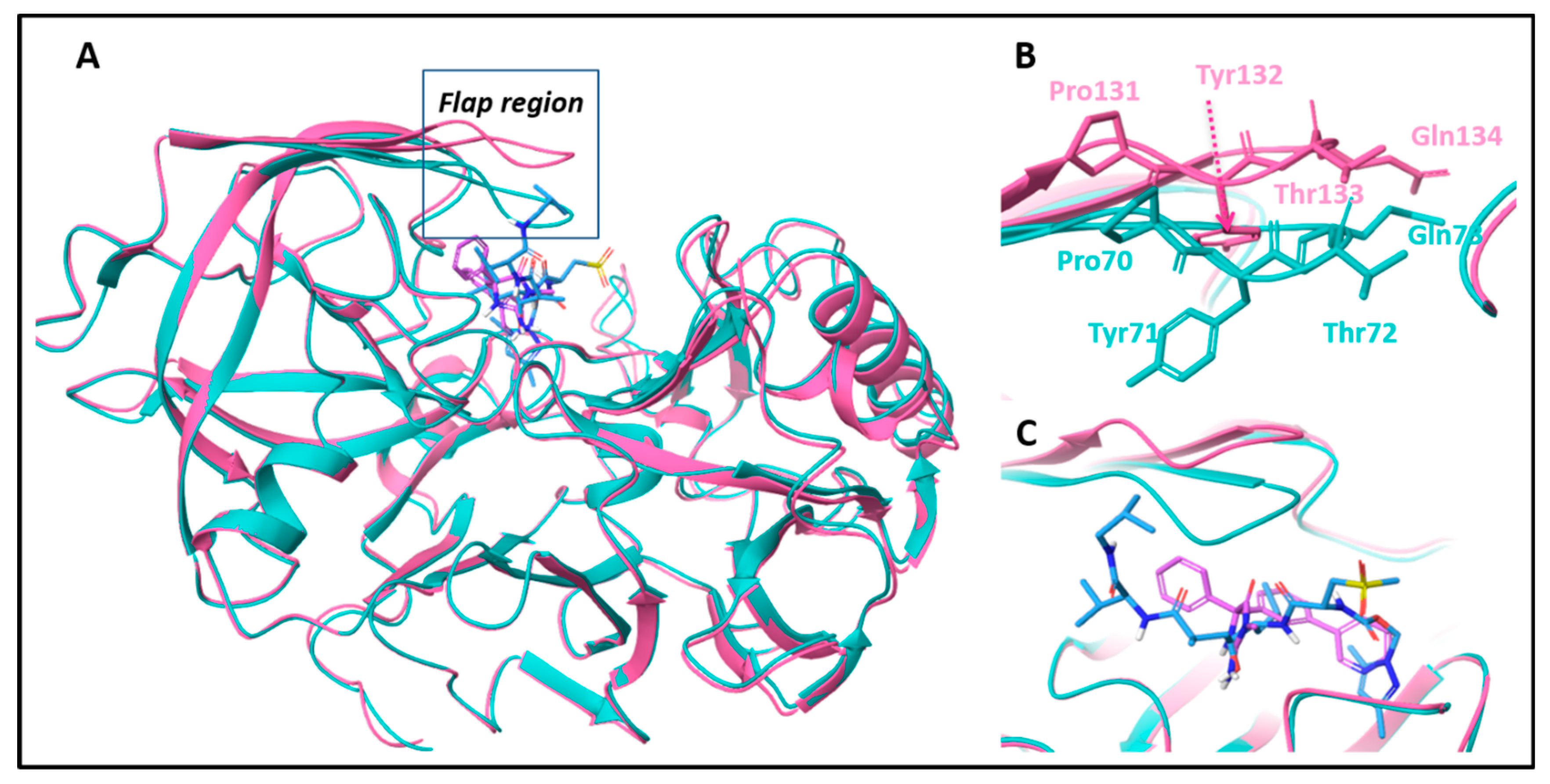
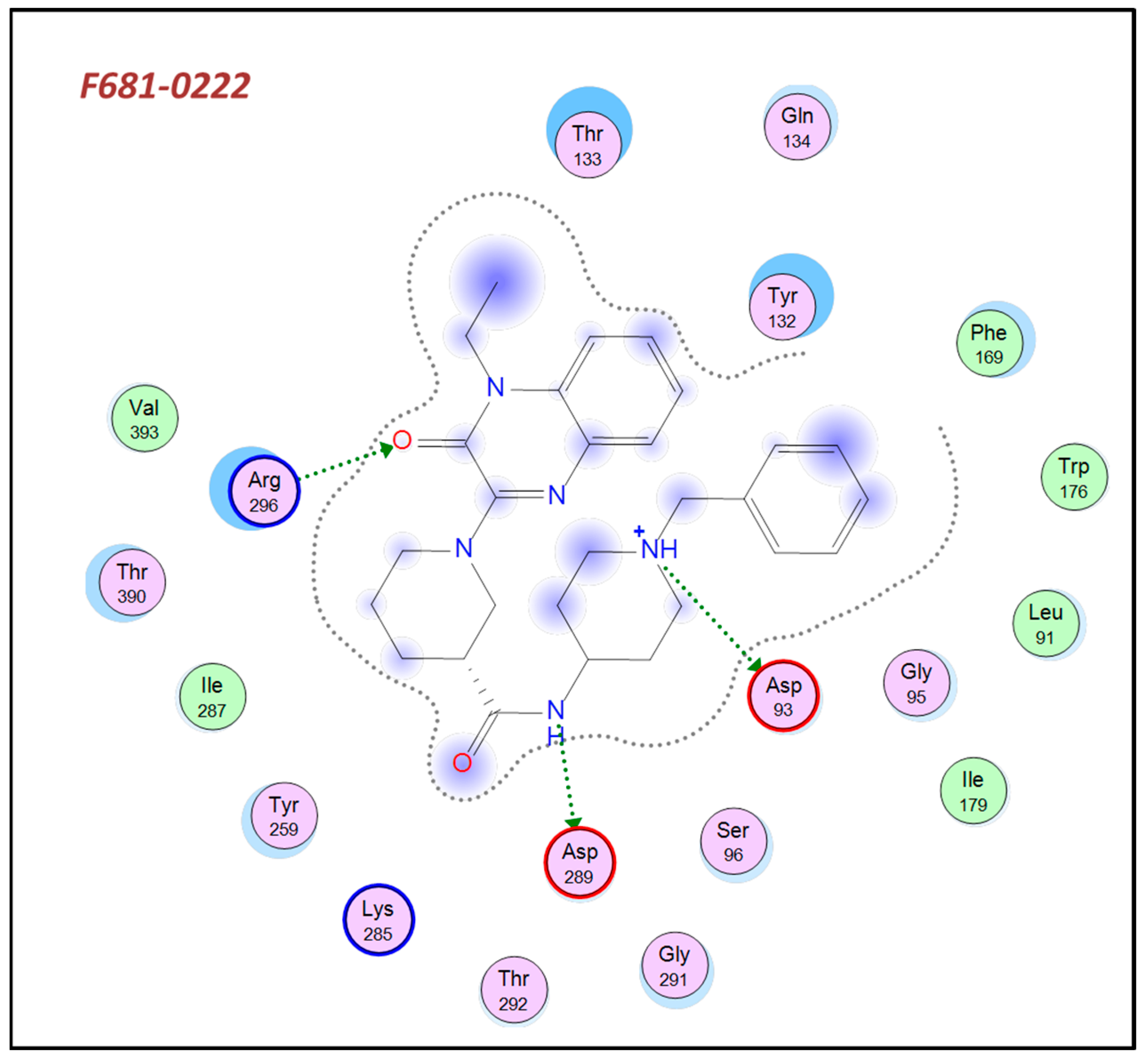
3.7. Examination of Docking Poses in AChE and BACE-1
3.8. Multi-Targeting Finds Different Affinities for Different Targets
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| Aβ | Amyloid-β |
| APP | Amyloid precursor protein |
| AChE | Acetylcholinesterase |
| BACE-1 | Beta-site APP cleaving enzyme 1 (β-secretase) |
| NMDA | N-methyl D-aspartate |
| MTDL | Multi-target directed ligand |
| PAS | Peripheral anionic site |
| ISE | Iterative stochastic elimination |
| MCC | Matthews’s correlation coefficient |
| AUC | Area under curve |
| ROC | Receiver operating characteristic |
| TI | Tanimoto similarity index |
References
- Available online: http://www.nia.nih.gov/alzheimers/topics/symptoms (accessed on 31 January 2022).
- Leon, R.; Garcia, A.G.; Marco-Contelles, J. Recent advances in the multitarget-directed ligands approach for the treatment of Alzheimer’s disease. Med. Res. Res. Rev. 2013, 33, 139–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plascencia-Villa, G.; Perry, G. Status and future directions of clinical trials in Alzheimer’s disease. Int. Rev. Neurobiol. 2020, 154, 3–50. [Google Scholar] [PubMed]
- Savelieff, M.G.; Nam, G.; Kang, J.; Lee, H.J.; Lee, M.; Lim, M.H. Development of Multifunctional Molecules as Potential Therapeutic Candidates for Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis in the Last Decade. Chem. Rev. 2019, 119, 1221–1322. [Google Scholar] [CrossRef]
- Bortolami, M.; Rocco, D.; Messore, A.; Di Santo, R.; Costi, R.; Madia, V.N.; Scipione, L.; Pandolfi, F. Acetylcholinesterase inhibitors for the treatment of Alzheimer’s disease—A patent review (2016–present). Expert Opin. Ther. Pat. 2021, 31, 399–420. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Lee, G.; Ritter, A.; Sabbagh, M.; Zhong, K. Alzheimer’s disease drug development pipeline: 2020. Alzheimers Dement 2020, 6, e12050. [Google Scholar] [CrossRef]
- Devenish, S.R.A. The current landscape in Alzheimer’s disease research and drug discovery. Drug Discov. Today 2020, 25, 943–945. [Google Scholar] [CrossRef]
- Huang, Y.; Mucke, L. Alzheimer mechanisms and therapeutic strategies. Cell 2012, 148, 1204–1222. [Google Scholar] [CrossRef] [Green Version]
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chetelat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer disease. Nat. Rev. Dis. Primers 2021, 7, 33. [Google Scholar] [CrossRef]
- Mikulca, J.A.; Nguyen, V.; Gajdosik, D.A.; Teklu, S.G.; Giunta, E.A.; Lessa, E.A.; Tran, C.H.; Terak, E.C.; Raffa, R.B. Potential novel targets for Alzheimer pharmacotherapy: II. Update on secretase inhibitors and related approaches. J. Clin. Pharm. Ther. 2014, 39, 25–37. [Google Scholar] [CrossRef]
- Alvarez, A.; Opazo, C.; Alarcon, R.; Garrido, J.; Inestrosa, N.C. Acetylcholinesterase promotes the aggregation of amyloid-beta-peptide fragments by forming a complex with the growing fibrils. J. Mol. Biol. 1997, 272, 348–361. [Google Scholar] [CrossRef]
- Inestrosa, N.C.; Alvarez, A.; Perez, C.A.; Moreno, R.D.; Vicente, M.; Linker, C.; Casanueva, O.I.; Soto, C.; Garrido, J. Acetylcholinesterase accelerates assembly of amyloid-beta-peptides into Alzheimer’s fibrils: Possible role of the peripheral site of the enzyme. Neuron 1996, 16, 881–891. [Google Scholar] [CrossRef] [Green Version]
- Proschak, E.; Stark, H.; Merk, D. Polypharmacology by Design: A Medicinal Chemist’s Perspective on Multitargeting Compounds. J. Med. Chem. 2019, 62, 420–444. [Google Scholar] [CrossRef] [PubMed]
- Dormán, G.; Flachner, B.; Hajdú, I.; András, C. Target identification and polypharmacology of nutraceuticals. In Nutraceuticals, 2nd ed.; Ramesh, C., Gupta, R.L., Srivastava, A., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 263–286. [Google Scholar]
- Oldfield, E.; Feng, X. Resistance-resistant antibiotics. Trends Pharmacol. Sci. 2014, 35, 664–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, J.U. Polypharmacology—Foe or friend? J. Med. Chem. 2013, 56, 8955–8971. [Google Scholar] [CrossRef] [PubMed]
- Prati, F.; Uliassi, E.; Bolognesi, M.L. Two diseases, one approach: Multitarget drug discovery in Alzheimer’s and neglected tropical diseases. Medchemcomm 2014, 5, 853–861. [Google Scholar] [CrossRef]
- Benek, O.; Korabecny, J.; Soukup, O. A Perspective on Multi-target Drugs for Alzheimer’s Disease. Trends Pharmacol. Sci. 2020, 41, 434–445. [Google Scholar] [CrossRef]
- Morphy, R.; Kay, C.; Rankovic, Z. From magic bullets to designed multiple ligands. Drug Discov. Today 2004, 9, 641–651. [Google Scholar] [CrossRef]
- Chaudhari, R.; Fong, L.W.; Tan, Z.; Huang, B.; Zhang, S. An up-to-date overview of computational polypharmacology in modern drug discovery. Expert Opin. Drug Discov. 2020, 15, 1025–1044. [Google Scholar] [CrossRef]
- Ma, X.H.; Shi, Z.; Tan, C.; Jiang, Y.; Go, M.L.; Low, B.C.; Chen, Y.Z. In-silico approaches to multi-target drug discovery: Computer aided multi-target drug design, multi-target virtual screening. Pharm. Res. 2010, 27, 739–749. [Google Scholar] [CrossRef]
- Racchi, M.; Mazzucchelli, M.; Porrello, E.; Lanni, C.; Govoni, S. Acetylcholinesterase inhibitors: Novel activities of old molecules. Pharmacol. Res. 2004, 50, 441–451. [Google Scholar] [CrossRef]
- McDade, E.; Voytyuk, I.; Aisen, P.; Bateman, R.J.; Carrillo, M.C.; De Strooper, B.; Haass, C.; Reiman, E.M.; Sperling, R.; Tariot, P.N.; et al. The case for low-level BACE1 inhibition for the prevention of Alzheimer disease. Nat. Rev. Neurol. 2021, 17, 703–714. [Google Scholar] [CrossRef]
- Hemming, M.L.; Elias, J.E.; Gygi, S.P.; Selkoe, D.J. Identification of beta-Secretase (BACE1) Substrates Using Quantitative Proteomics. PLoS ONE 2009, 4, e8477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csermely, P.; Korcsmaros, T.; Kiss, H.J.; London, G.; Nussinov, R. Structure and dynamics of molecular networks: A novel paradigm of drug discovery: A comprehensive review. Pharmacol. Ther. 2013, 138, 333–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Strooper, B. Lessons from a failed gamma-secretase Alzheimer trial. Cell 2014, 159, 721–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, J.P.S.; Albuquerque, H.M.T.; Cardoso, S.M.; Silva, A.M.S.; Silva, V.L.M. Dual-target compounds for Alzheimer’s disease: Natural and synthetic AChE and BACE-1 dual-inhibitors and their structure-activity relationship (SAR). Eur. J. Med. Chem. 2021, 221, 113492. [Google Scholar] [CrossRef]
- Zhang, P.F.; Xu, S.T.; Zhu, Z.Y.; Xu, J.Y. Multi-target design strategies for the improved treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2019, 176, 228–247. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.; Zhao, X.; Wang, J. Design, Synthesize and Bio-Evaluate 1,2-Dihydroisoquinolin-3(4H)-One Derivates as Acetylcholinesterase and β-Secretase Dual Inhibitors in Treatment with Alzheimer’s Disease. J. Biosci. Med. 2016, 4, 112–123. [Google Scholar]
- Camps, P.; Formosa, X.; Galdeano, C.; Munoz-Torrero, D.; Ramirez, L.; Gomez, E.; Isambert, N.; Lavilla, R.; Badia, A.; Clos, M.V.; et al. Pyrano[3,2-c]quinoline-6-Chlorotacrine Hybrids as a Novel Family of Acetylcholinesterase- and beta-Amyloid-Directed Anti-Alzheimer Compounds. J. Med. Chem. 2009, 52, 5365–5379. [Google Scholar] [CrossRef]
- Galdeano, C.; Viayna, E.; Sola, I.; Formosa, X.; Camps, P.; Badia, A.; Clos, M.V.; Relat, J.; Ratia, M.; Bartolini, M.; et al. Huprine-Tacrine Heterodimers as Anti-Amyloidogenic Compounds of Potential Interest against Alzheimer’s and Prion Diseases. J. Med. Chem. 2012, 55, 661–669. [Google Scholar] [CrossRef]
- Sun, Q.; Peng, D.Y.; Yang, S.G.; Zhu, X.L.; Yang, W.C.; Yang, G.F. Syntheses of coumarin-tacrine hybrids as dual-site acetylcholinesterase inhibitors and their activity against butylcholinesterase, Abeta aggregation, and beta-secretase. Bioorganic Med. Chem. 2014, 22, 4784–4791. [Google Scholar] [CrossRef]
- Viayna, E.; Sola, I.; Bartolini, M.; De Simone, A.; Tapia-Rojas, C.; Serrano, F.G.; Sabate, R.; Juarez-Jimenez, J.; Perez, B.; Luque, F.J.; et al. Synthesis and Multitarget Biological Profiling of a Novel Family of Rhein Derivatives as Disease-Modifying Anti-Alzheimer Agents. J. Med. Chem. 2014, 57, 2549–2567. [Google Scholar] [CrossRef] [PubMed]
- Berg, L.; Andersson, C.D.; Artursson, E.; Hornberg, A.; Tunemalm, A.K.; Linusson, A.; Ekstrom, F. Targeting Acetylcholinesterase: Identification of Chemical Leads by High Throughput Screening, Structure Determination and Molecular Modeling. PLoS ONE 2011, 6, e26039. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Fallarero, A.; Jarvinen, P.; Karlsson, D.; Johnson, M.S.; Vuorela, P.M.; Mohan, C.G. Discovery of dual binding site acetylcholinesterase inhibitors identified by pharmacophore modeling and sequential virtual screening techniques. Bioorganic Med. Chem. Lett. 2011, 21, 1105–1112. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.H.; Wu, J.W.; Liu, H.L.; Zhao, J.H.; Liu, K.T.; Chuang, C.K.; Lin, H.Y.; Tsai, W.B.; Ho, Y. The discovery of potential acetylcholinesterase inhibitors: A combination of pharmacophore modeling, virtual screening, and molecular docking studies. J. Biomed. Sci. 2011, 18, 8. [Google Scholar] [CrossRef] [Green Version]
- Mishra, N.; Basu, A. Exploring Different Virtual Screening Strategies for Acetylcholinesterase Inhibitors. Biomed. Res. Int 2013, 2013, 236850. [Google Scholar] [CrossRef] [Green Version]
- Vitorovic-Todorovic, M.D.; Cvijetic, I.N.; Juranic, I.O.; Drakulic, B.J. The 3D-QSAR study of 110 diverse, dual binding, acetylcholinesterase inhibitors based on alignment independent descriptors (GRIND-2). The effects of conformation on predictive power and interpretability of the models. J. Mol. Graph. Model. 2012, 38, 194–210. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, J.L.; Fernandez-Nieto, F.; Castro, M.; Catto, M.; Paleo, M.R.; Porto, S.; Sardina, F.J.; Brea, J.M.; Carotti, A.; Villaverde, M.C.; et al. Computer-aided structure-based design of multitarget leads for Alzheimer’s disease. J. Chem. Inf. Modeling 2015, 55, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.S.; Le, M.T.; Tran, T.D.; Tran, T.H.; Thai, K.M. Design of Curcumin and Flavonoid Derivatives with Acetylcholinesterase and Beta-Secretase Inhibitory Activities Using in Silico Approaches. Molecules 2020, 25, 3644. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [Green Version]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Modeling 2012, 52, 1757–1768. [Google Scholar] [CrossRef]
- Weaver, S.; Gleeson, M.P. The importance of the domain of applicability in QSAR modeling. J. Mol. Graph. Model. 2008, 26, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group Inc. Molecular Operating Environment (MOE); Chemical Computing Group Inc.: Montreal, QC, Canada, 2011. [Google Scholar]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kotter, T.; Meinl, T.; Ohl, P.; Sieb, C.; Thiel, K.; Wiswedel, B. KNIME: The Konstanz Information Miner; KNIME AG Talacker: Zurich, Switzerland, 2009. [Google Scholar]
- Posner, B.A.; Xi, H.; Mills, J.E. Enhanced HTS hit selection via a local hit rate analysis. J. Chem. Inf. Modeling 2009, 49, 2202–2210. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.W. Comparison of the predicted and observed secondary structure of the T4 phage lysozyme. Biochim. Biophys. Acta Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Boughorbel, S.; Jarray, F.; El-Anbari, M. Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PLoS ONE 2017, 12, e0177678. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [Green Version]
- Halimu, C.; Kasem, A.; Newaz, S.H.S. Empirical Comparison of Area under ROC curve (AUC) and Matthew Correlation Coefficient (MCC) for Evaluating Machine Learning Algorithms on Imbalanced Datasets for Binary Classification. In Proceedings of the 3rd International Conference on Machine Learning and Soft Computing (ICMLSC 2019), Da Lat, Vietnam, 25–28 January 2019; pp. 1–6. [Google Scholar]
- Available online: http://www.enamine.net/ (accessed on 4 January 2012).
- Available online: http://www.chemdiv.com/ (accessed on 20 June 2014).
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.F.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: http://www.ibscreen.com/ (accessed on 12 March 2014).
- Available online: http://www.princetonbio.com/ (accessed on 13 March 2014).
- Available online: http://www.ac-discovery.com/ (accessed on 13 March 2014).
- Laskowski, R.A.; Hutchinson, E.G.; Michie, A.D.; Wallace, A.C.; Jones, M.L.; Thornton, J.M. PDBsum: A Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997, 22, 488–490. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- OMEGA 2.5.1; Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Modeling 2010, 50, 572–584. [Google Scholar] [CrossRef]
- OEDOCKING 3.0.1: OpenEye Scientific Software, Inc., Santa Fe, NM, USA. Available online: http://www.eyesopen.com (accessed on 12 August 2022).
- Cheung, J.; Gary, E.N.; Shiomi, K.; Rosenberry, T.L. Structures of Human Acetylcholinesterase Bound to Dihydrotanshinone I and Territrem B Show Peripheral Site Flexibility. ACS Med. Chem. Lett. 2013, 4, 1091–1096. [Google Scholar] [CrossRef] [Green Version]
- Cheung, J.; Rudolph, M.J.; Burshteyn, F.; Cassidy, M.S.; Gary, E.N.; Love, J.; Franklin, M.C.; Height, J.J. Structures of Human Acetylcholinesterase in Complex with Pharmacologically Important Ligands. J. Med. Chem. 2012, 55, 10282–10286. [Google Scholar] [CrossRef] [PubMed]
- Barman, A.; Prabhakar, R. Protonation States of the Catalytic Dyad of beta-Secretase (BACE1) in the Presence of Chemically Diverse Inhibitors: A Molecular Docking Study. J. Chem. Inf. Modeling 2012, 52, 1275–1287. [Google Scholar] [CrossRef] [PubMed]
- Kacker, P.; Masetti, M.; Mangold, M.; Bottegoni, G.; Cavalli, A. Combining Dyad Protonation and Active Site Plasticity in BACE-1 Structure-Based Drug Design. J. Chem. Inf. Modeling 2012, 52, 1079–1085. [Google Scholar] [CrossRef]
- Rajamani, R.; Reynolds, C.H. Modeling the protonation states of the catalytic aspartates in beta-secretase. J. Med. Chem. 2004, 47, 5159–5166. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, J.; Kriz, Z.; Kuca, K.; Jun, D.; Koca, J. Influence of the Acetylcholinesterase Active Site Protonation on Omega Loop and Active Site Dynamics. J. Biomol. Struct. Dyn. 2010, 28, 393–403. [Google Scholar] [CrossRef]
- Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996, 261, 470–489. [Google Scholar] [CrossRef] [Green Version]
- Kramer, B.; Rarey, M.; Lengauer, T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins 1999, 37, 228–241. [Google Scholar] [CrossRef]
- Bolognesi, M.L.; Chiriano, G.; Bartolini, M.; Mancini, F.; Bottegoni, G.; Maestri, V.; Czvitkovich, S.; Windisch, M.; Cavalli, A.; Minarini, A.; et al. Synthesis of Monomeric Derivatives to Probe Memoquin’s Bivalent Interactions. J. Med. Chem. 2011, 54, 8299–8304. [Google Scholar] [CrossRef]
- Fernandez-Bachiller, M.I.; Perez, C.; Monjas, L.; Rademann, J.; Rodriguez-Franco, M.I. New Tacrine-4-Oxo-4H-chromene Hybrids as Multifunctional Agents for the Treatment of Alzheimer’s Disease, with Cholinergic, Antioxidant, and beta-Amyloid-Reducing Properties. J. Med. Chem. 2012, 55, 1303–1317. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, T.; Yeung, J.C.; Vasefi, M.S.; Beazely, M.A.; Rao, P.P. Development and evaluation of multifunctional agents for potential treatment of Alzheimer’s disease: Application to a pyrimidine-2,4-diamine template. Bioorganic Med. Chem. Lett. 2012, 22, 4707–4712. [Google Scholar] [CrossRef]
- Piazzi, L.; Cavalli, A.; Colizzi, F.; Belluti, F.; Bartolini, M.; Mancini, F.; Recanatini, M.; Andrisano, V.; Rampa, A. Multi-target-directed coumarin derivatives: hAChE and BACE1 inhibitors as potential anti-Alzheimer compounds. Bioorganic Med. Chem. Lett. 2008, 18, 423–426. [Google Scholar] [CrossRef]
- Lin, X.; Koelsch, G.; Wu, S.; Downs, D.; Dashti, A.; Tang, J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc. Natl. Acad. Sci. USA 2000, 97, 1456–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stachel, S.J.; Coburn, C.A.; Steele, T.G.; Jones, K.G.; Loutzenhiser, E.F.; Gregro, A.R.; Rajapakse, H.A.; Lai, M.T.; Crouthamel, M.C.; Xu, M.; et al. Structure-based design of potent and selective cell-permeable inhibitors of human beta-secretase (BACE-1). J. Med. Chem. 2004, 47, 6447–6450. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, H.; Yamanishi, Y.; Iimura, Y.; Kawakami, Y. Donepezil hydrochloride (E2020) and other acetylcholinesterase inhibitors. Curr. Med. Chem. 2000, 7, 303–339. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L.; Courtney, K.D.; Andres, V., Jr.; Feather-Stone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharm. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Fang, J.S.; Li, Y.J.; Liu, R.; Pang, X.C.; Li, C.; Yang, R.Y.; He, Y.Y.; Lian, W.W.; Liu, A.L.; Du, G.H. Discovery of Multitarget-Directed Ligands against Alzheimer’s Disease through Systematic Prediction of Chemical Protein Interactions. J. Chem. Inf. Modeling 2015, 55, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Shang, E.; Yuan, Y.; Chen, X.; Liu, Y.; Pei, J.; Lai, L. De novo design of multitarget ligands with an iterative fragment-growing strategy. J. Chem. Inf. Modeling 2014, 54, 1235–1241. [Google Scholar] [CrossRef]
- Basu, A.; Sohn, Y.S.; Alyan, M.; Nechushtai, R.; Domb, A.J.; Goldblum, A. Discovering Novel and Diverse Iron-Chelators in Silico. J. Chem. Inf. Modeling 2016, 56, 2476–2485. [Google Scholar] [CrossRef]
- Hassan, M.; Raza, H.; Abbasi, M.A.; Moustafa, A.A.; Seo, S.Y. The exploration of novel Alzheimer’s therapeutic agents from the pool of FDA approved medicines using drug repositioning, enzyme inhibition and kinetic mechanism approaches. Biomed. Pharm. 2019, 109, 2513–2526. [Google Scholar] [CrossRef]
- Lan, X.Q.; Kiyota, T.; Hanamsagar, R.; Huang, Y.L.; Andrews, S.; Peng, H.; Zheng, J.C.; Swindells, S.; Carlson, G.A.; Ikezu, T. The Effect of HIV Protease Inhibitors on Amyloid-beta Peptide Degradation and Synthesis in Human Cells and Alzheimer’s Disease Animal Model. J. Neuroimmune Pharm. 2012, 7, 412–423. [Google Scholar] [CrossRef] [Green Version]
- Sharma, B.; Singh, N.; Singh, M.; Jaggi, A.S. Exploitation of HIV protease inhibitor Indinavir as a memory restorative agent in experimental dementia. Pharm. Biochem. Behav. 2008, 89, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.D.; Wu, C.L.; Lin, T.K.; Chuang, Y.C.; Yang, D.I. Renin inhibitor aliskiren exerts neuroprotection against amyloid beta-peptide toxicity in rat cortical neurons. Neurochem. Int. 2012, 61, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Dodds, H.M.; Rivory, L.P. The mechanism for the inhibition of acetylcholinesterases by irinotecan (CPT-11). Mol. Pharmacol. 1999, 56, 1346–1353. [Google Scholar] [CrossRef] [PubMed]
- Cosconati, S.; Marinelli, L.; Di Leva, F.S.; La Pietra, V.; De Simone, A.; Mancini, F.; Andrisano, V.; Novellino, E.; Goodsell, D.S.; Olson, A.J. Protein Flexibility in Virtual Screening: The BACE-1 Case Study. J. Chem. Inf. Modeling 2012, 52, 2697–2704. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.X.; Zhang, W.; Li, L.; Salvador, L.A.; Chen, T.T.; Chen, W.Y.; Felsenstein, K.M.; Ladd, T.B.; Price, A.R.; Golde, T.E.; et al. Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases. J. Med. Chem. 2012, 55, 10749–10765. [Google Scholar] [CrossRef] [PubMed]
- Hamada, Y.; Kiso, Y. Advances in the identification of β-secretase inhibitors. Expert Opin. Drug Dis. 2013, 8, 709–731. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Kumaragurubaran, N.; Hong, L.; Lei, H.; Hussain, K.A.; Liu, C.F.; Devasamudram, T.; Weerasena, V.; Turner, R.; Koelsch, G.; et al. Design, synthesis and X-ray structure of protein-ligand complexes: Important insight into selectivity of memapsin 2 (beta-secretase) inhibitors. J. Am. Chem. Soc. 2006, 128, 5310–5311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cumming, J.N.; Smith, E.M.; Wang, L.Y.; Misiaszek, J.; Durkin, J.; Pan, J.P.; Iserloh, U.; Wu, Y.S.; Zhu, Z.N.; Strickland, C.; et al. Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor. Bioorganic Med. Chem. Lett. 2012, 22, 2444–2449. [Google Scholar] [CrossRef]
- Coburn, C.A.; Stachel, S.J.; Li, Y.M.; Rush, D.M.; Steele, T.G.; Chen-Dodson, E.; Holloway, M.K.; Xu, M.; Huang, Q.; Lai, M.T.; et al. Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases. J. Med. Chem. 2004, 47, 6117–6119. [Google Scholar] [CrossRef]
- Shimizu, H.; Tosaki, A.; Kaneko, K.; Hisano, T.; Sakurai, T.; Nukina, N. Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production. Mol. Cell Biol 2008, 28, 3663–3671. [Google Scholar] [CrossRef] [Green Version]
- Bahadur, S.; Saxena, M. Syntheses and Biological-Activities of Some New 4(3h)-Quinazolinones. Arch. Pharm. 1983, 316, 964–968. [Google Scholar] [CrossRef]
- Fuchs, K.; Heine, N.; Eickmeier, C.; Handschuh, S.; Dorner-Ciossek, C.; Hoerer, S. Substituted Amino-Quinazolinones, Medicaments Comprising Said Compound, Their Use and Their Method of Manufacture. U.S. Patent 8,664,388, 4 March 2014. [Google Scholar]
- Jiang, X.Y.; Wu, K.Y.; Bai, R.R.; Zhang, P.F.; Zhang, Y. Functionalized quinoxalinones as privileged structures with broad- ranging pharmacological activities. Eur. J. Med. Chem. 2022, 229, 114085. [Google Scholar] [CrossRef]
- Zeb, A.; Hameed, A.; Khan, L.; Khan, I.; Dalvandi, K.; Choudhary, M.I.; Basha, F.Z. Quinoxaline derivatives: Novel and selective butyrylcholinesterase inhibitors. Med. Chem. 2014, 10, 724–729. [Google Scholar] [CrossRef]
- Harel, M.; Hyatt, J.L.; Brumshtein, B.; Morton, C.L.; Yoon, K.J.; Wadkins, R.M.; Silman, I.; Sussman, J.L.; Potter, P.M. The crystal structure of the complex of the anticancer prodrug 7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothecin (CPT-11) with Torpedo californica acetylcholinesterase provides a molecular explanation for its cholinergic action. Mol. Pharmacol. 2005, 67, 1874–1881. [Google Scholar] [CrossRef] [Green Version]
- Darras, F.H.; Wehle, S.; Huang, G.; Sotriffer, C.A.; Decker, M. Amine substitution of quinazolinones leads to selective nanomolar AChE inhibitors with ‘inverted’ binding mode. Bioorganic Med. Chem. 2014, 22, 4867–4881. [Google Scholar] [CrossRef]
- Anighoro, A.; Bajorath, J.; Rastelli, G. Polypharmacology: Challenges and Opportunities in Drug Discovery. J. Med. Chem. 2014, 57, 7874–7887. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, M.L.; Cavalli, A. Multitarget Drug Discovery and Polypharmacology. ChemMedChem 2016, 11, 1190–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardo, L.J.; Lee, F.Y.; Chen, P.; Norris, D.; Barrish, J.C.; Behnia, K.; Castaneda, S.; Cornelius, L.A.; Das, J.; Doweyko, A.M.; et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 2004, 47, 6658–6661. [Google Scholar] [CrossRef] [PubMed]


| Model Type | Actives vs. Randoms (AvI) | High Actives vs. Low Actives (HvL) | ||||||
|---|---|---|---|---|---|---|---|---|
| Model No. | 1 | 2 a | 3 | 4 | 5 | 6 | 7 | 8 a |
| Tanimoto | 1.0 | 0.9 | 0.8 | 0.7 | 1.0 | 0.9 | 0.8 | 0.7 |
| Actives IC50 (nM) cutoff | <10,000 | <10,000 | <10,000 | <10,000 | <100 | <100 | <100 | <100 |
| no. Actives | 1316 | 617 | 344 | 194 | 438 | 177 | 93 | 51 |
| Inactives IC50 (nM) cutoff | randoms | randoms | randoms | randoms | >1000 | >1000 | >1000 | >1000 |
| no. Inactives | 46,743 | 46,743 | 46,743 | 46,743 | 747 | 463 | 307 | 214 |
| Average b best filter MCC | 0.79 | 0.79 | 0.76 | 0.76 | 0.56 | 0.60 | 0.66 | 0.71 |
| Average b AUC | 0.95 | 0.95 | 0.95 | 0.94 | 0.82 | 0.78 | 0.83 | 0.77 |
| Model Type | Actives vs. Randoms (AvI) | High Actives vs. Low Actives (HvL) | |||
|---|---|---|---|---|---|
| Model No. | 1a | 2 | 3 | 4 a | 5 a |
| Tanimoto | 1.0 | 1.0 | 0.8 | 0.9 | 0.9 |
| Actives IC50 (nM) cutoff | <10,000 | <100 | <100 | <100 | <100 |
| no. Actives | 428 | 211 | 65 | 109 | 109 |
| Inactives IC50 (nM) cutoff | randoms | >1000 | >1000 | >1000 | >3000 |
| no. Inactives | 37,467 | 197 | 65 | 112 | 86 |
| Average b best filter MCC | 0.82 | 0.65 | 0.71 | 0.69 | 0.71 |
| Average b AUC | 0.97 | 0.88 | 0.78 | 0.86 | 0.86 |
| Drug Name (DrugBank 4.0 [53] Name) | Target(s) a | AChE Indexes | BACE-1 Indexes | |||
|---|---|---|---|---|---|---|
| Model 1 | Model 4 | Model 5 | Model 2 | Model 8 | ||
| Indinavir (DB00224) | HIV-1 protease | 0.51 | −0.47 | −0.52 | 0.85 | 0.83 |
| Saquinavir (DB01232) | HIV-1 protease | 0.40 | −0.39 | −0.40 | 0.85 | 0.44 |
| Aliskiren (DB01258) | Renin | 0.73 | 0.19 | 0.20 | 0.83 | 0.81 |
| Nelfinavir (DB00220) | HIV-1 protease | 0.87 | 0.00 | 0.12 | 0.83 | 0.10 |
| Terfenadine (DB00342) | M-ACh-R b | 0.67 | −0.20 | −0.40 | 0.81 | −0.75 |
| Darifenacin (DB00496) | M-ACh-R | 0.88 | −0.47 | −0.68 | 0.58 | −0.31 |
| Tubocurarine (DB01199) | AChE, N-ACh-Rc | 0.54 | −0.51 | −0.41 | 0.58 | −0.68 |
| Metocurine (DB01336) | N-ACh-R, M-ACh-R | 0.54 | −0.38 | −0.44 | 0.27 | −0.76 |
| Irinotecan (DB00762) | DNA- topoisomerase, cholinesterase AChE [84] | 0.33 | 0.26 | 0.14 | 0.18 | 0.65 |
| DrugBank Name | AChE Indexes | BACE-1 Indexes | AChE Poses a | BACE-1 Poses b | Target c | |||
|---|---|---|---|---|---|---|---|---|
| Model 1 | Model 4 | Model 5 | Model 2 | Model 8 | ||||
| DB01721 | 0.33 | −0.46 | −0.66 | 0.85 | 0.75 | 6 | 6 | Gag-Pol polyprotein |
| DB02009 | 0.84 | −0.45 | −0.66 | 0.85 | 0.79 | 4 | 27 | Gag-Pol polyprotein |
| DB04424 | 0.78 | −0.73 | −0.83 | 0.58 | 0.66 | 4 | 16 | Trypsin-1 |
| DB04614 | 0.88 | 0.46 | 0.39 | 0.85 | 0.09 | 9 | 2 | AChE |
| DB04615 | 0.88 | 0.46 | 0.39 | 0.85 | 0.09 | 8 | 7 | AChE |
| DB08749 | 0.85 | 0.53 | 0.34 | 0.75 | −0.15 | 3 | 30 | BACE-1 |
| Cmp. No. | Cmp. Name | AChE % Inhibition at 10 µM | AChE IC50 (µM) | BACE-1 % Inhibition at 100 µM | BACE-1 IC50 (µM) |
|---|---|---|---|---|---|
| 1 | C700-2595 | 65 | 8.2 | 19 | n.m. a |
| 2 | F681-0222 | 64 | 4.4 | 69 | 63 |
| 3 | F681-0412 | 36 | 6.7 | 66 | 55 |
| 4 | C741-0335 | 76 | 9.7 | 6 | n.m. |
| 5 | T6769894 b | 19 | n.m. | 82 | 60 |
| 6 | Z274-0419 | −12 | n.m. | 76 | 150 |
| Cmp. No. | AChE Indexes | BACE-1 Indexes | AChE Poses a | BACE-1 Poses b | |||
|---|---|---|---|---|---|---|---|
| Model 5 | Model 4 | Model 1 | Model 8 | Model 2 | |||
| 1 | 0.63 | 0.62 | 0.78 | 0.58 | 0.18 | 8 | 1 (1) |
| 2 | 0.6 | 0.62 | 0.78 | 0.56 | 0.18 | 9 | 2 (1) |
| 3 | 0.61 | 0.63 | 0.78 | 0.54 | 0.18 | 2 | 4 (2) |
| 4 | 0.37 | 0.44 | 0.78 | 0.64 | 0.18 | 4 | 2 (1) |
| 5 | 0.6 | 0.77 | 0.8 | 0.57 | 0.16 | 8 | 5 (2) |
| 6 | 0.09 | 0.13 | 0.49 | 0.59 | 0.6 | 2 | 8 (3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stern, N.; Gacs, A.; Tátrai, E.; Flachner, B.; Hajdú, I.; Dobi, K.; Bágyi, I.; Dormán, G.; Lőrincz, Z.; Cseh, S.; et al. Dual Inhibitors of AChE and BACE-1 for Reducing Aβ in Alzheimer’s Disease: From In Silico to In Vivo. Int. J. Mol. Sci. 2022, 23, 13098. https://doi.org/10.3390/ijms232113098
Stern N, Gacs A, Tátrai E, Flachner B, Hajdú I, Dobi K, Bágyi I, Dormán G, Lőrincz Z, Cseh S, et al. Dual Inhibitors of AChE and BACE-1 for Reducing Aβ in Alzheimer’s Disease: From In Silico to In Vivo. International Journal of Molecular Sciences. 2022; 23(21):13098. https://doi.org/10.3390/ijms232113098
Chicago/Turabian StyleStern, Noa, Alexandra Gacs, Enikő Tátrai, Beáta Flachner, István Hajdú, Krisztina Dobi, István Bágyi, György Dormán, Zsolt Lőrincz, Sándor Cseh, and et al. 2022. "Dual Inhibitors of AChE and BACE-1 for Reducing Aβ in Alzheimer’s Disease: From In Silico to In Vivo" International Journal of Molecular Sciences 23, no. 21: 13098. https://doi.org/10.3390/ijms232113098






