Abstract
Acute myocardial infarction (AMI) is a pandemic in which conventional risk factors are inadequate to detect who is at risk early in the asymptomatic stage. Although gene variants in genes related to cholesterol, which may increase the risk of AMI, have been identified, no studies have systematically screened the genes involved in this pathway. In this study, we included 105 patients diagnosed with AMI with an elevation of the ST segment (STEMI) and treated with primary percutaneous coronary intervention (PPCI). Using next-generation sequencing, we examined the presence of rare variants in 40 genes proposed to be involved in lipid metabolism and we found that 60% of AMI patients had a rare variant in the genes involved in the cholesterol pathway. Our data show the importance of considering the wide scope of the cholesterol pathway in order to assess the genetic risk related to AMI.
1. Introduction
Acute myocardial infarction (AMI) is defined as myocardial cell death due to prolonged ischemia []. It is the most severe type of coronary artery disease (CAD) and one of the main causes of death in developed countries [].
Epidemiological studies have identified the following three major categories of risk factors for AMI: unchangeable factors (age, gender, and family history); variable factors (smoking, alcohol intake, lack of exercise, poor diet, high blood pressure, diabetes, dyslipidemia, and metabolism syndrome); and emerging factors (abnormal levels of C-reactive protein (CRP), fibrinogen, coronary artery calcification (CAC), homocysteine, and lipoprotein(a)) []. The environmental and lifestyle components of this triad of AMI have been well-documented since the 1960s, starting with the Framingham study []. However, it Is important to note that genetic predisposition has been stated to account for 40–50% of the variability in the development of CAD [,].
However, to date, the range of genes underlying the heritable component of AMI is not fully known. According to Musunuru et al. (2016) [], 25% of AMI loci are lipid-related, including the following: LDLR; PCSK9; APOB; SORT1; ABCG5/G8; LPA; ABO; TCF21; SH2B3; APOE; APOA1/A5; LPL; TRIB1; and LIPA. These data reinforce the role of cholesterol homeostasis in the genetic landscape of AMI. According to the Kyoto Encyclopedia of Genes and Genomes (KEGG), the main cholesterol pathway includes more than 40 genes that operate on many planes, including cholesterol uptake, efflux, transport, storage, utilization, and/or excretion [].
Most studies [,,,,,,,,] have focused on the analysis of a limited number of genes in the cholesterol pathway, converging on the secretion of the HDL and LDL molecules, without considering the complexity of the pathway as a whole. Most of these studies have focused on single nucleotide polymorphisms (SNPs) [,,,,] without taking into account the study of rare variants in the whole pathway. An ambitious strategy of studying the rare variants in the whole pathway is important as a rare variant can be considered, not the cause of the disease, but an additional risk factor [,,].
Thus, in this paper we aimed to study potential rare variants in the cholesterol pathway genes in patients who presented AMI in order to obtain a broader spectrum of variants that may be involved.
2. Results
2.1. Characteristics of the Study Participants
The principal characteristics of the 105 patients included in the study are summarized in Table 1. The data are also shown to be disaggregated between patients with mutation and without mutation. There was not a significant difference between the groups in any of these variables.

Table 1.
Clinical data and demographic characteristics of the studied population.
2.2. Classification of Variants Identified in the Cholesterol Pathway
The 40 genes of the cholesterol metabolism pathway (Figure 1) presented a mean coverage of 117.06 ± 22.14-fold (Figure S1).
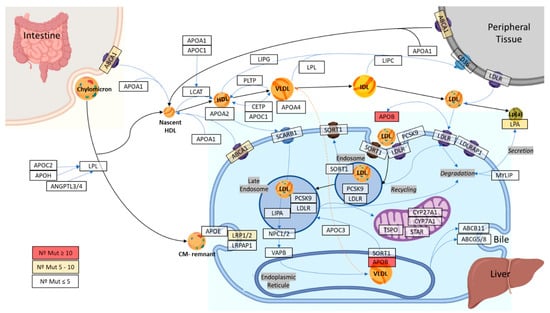
Figure 1.
Schematic view of the 40 selected genes in the cholesterol metabolism pathway.
After the analysis of the sequencing data, 474 unique genetic variants in the codifying region, and ±10 intronic bases of the 40 genes related to cholesterol metabolism, were identified. Considering the variants in the 105 patients, the total number of variants was 8805. On average, each patient carried around 83 variants in the analyzed region.
Next, the filtering strategy described in the methods section was applied (Figure 2) in order to discriminate between the relevance of each variant. First, 259 common variants, with a frequency of higher than 0.01 in the population, were filtered out, leaving 215 rare variants. Second, the synonymous and intronic variants in more than ±5 bases on the splicing sites were excluded, reducing the variants to 129 (86 filtered out). Those 129 variants were classified using Hass et al.’s classification [], with slight modifications which are presented in the methods section.
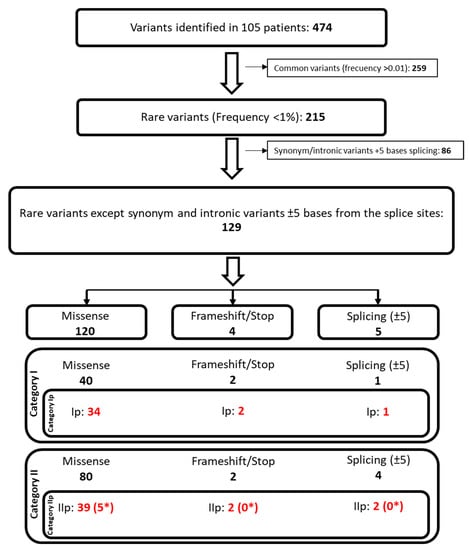
Figure 2.
Schematic view of the followed steps for the filtering and classification of the variants identified in the patients of study. * Variants non-previously described. Red: category Ip (known mutation) and IIp (potential mutation) variants.
From the 129 selected variants, 43 had already been described in the Human Gene Mutation Database (HGMD) [], classified following our strategy as category I (Table 2, [,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,]). A total of 37 of the 43 variants were related to a lipid metabolism disorder or to cardiopathy. Consequently, they were classified as category Ip (known mutation). The mutations categorized as Ip were present in 18 genes, including the top three genes with the most mutations in our patients, namely APOB (n = 11), ABCA1 (n = 4), and LDLR (n = 3) (Table 2).

Table 2.
Category Ip (known mutation).
The remaining 86 variants were classified as category II. After the assessment of their potential impact on the protein by in silico prediction tools, 46 variants were classified as potentially damaging and, consequently, were classified as category IIp (potential mutation) (Table 3). In this group, the genes APOB (n = 7), LRP1 (n = 6), and LRP2 (n = 5) were the most represented.

Table 3.
Category IIp (potential mutation).
Five new missense variants (IIp) were described, namely ABCA1-p.A2058G, ABCG5-p.P431A, APOB-p.D2193Y, APOB-p.A4119D, and PCSK9-p.A300V (Table 3). As for all IIp variants, at least three out of the five in silico tools predicted an impact on protein function (Mutation taster, SNAP2, SIFT, PP2, PhD-SNP).
2.3. Distribution of the Variants of Interest in the Genes
A total of 80 mutations and potential mutations (category Ip and IIp) were identified in the studied population. These variants were present in 72% of the genes studied (29 out of 40) (Figure 3). The distribution of variants in the genes was uneven. APOB was the gene in which more of these variants were identified (n = 18). Then, genes LRP2, ABCA1, LPA, and LRP1 had between six to eight variants. Finally, the 24 remaining genes had between one to three variants of category Ip (known mutation) and IIp (potential mutation) mutations (Figure 3). These group had 44% of the variants identified in our population.
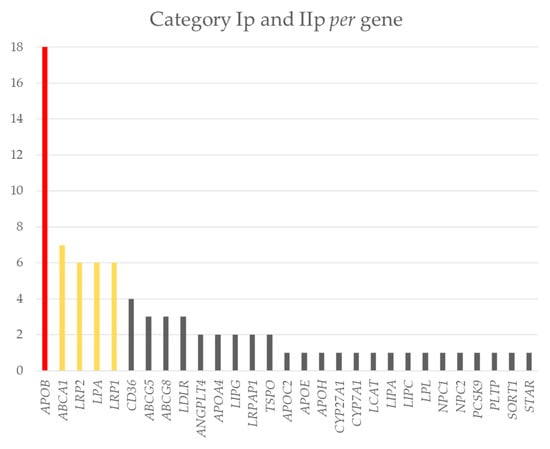
Figure 3.
Bar graph displaying the number of mutations and potential mutations (Y axis) per gene (X axis). Red: more than 8 variants; yellow: between 8–6 variants; grey: less than 6 variants.
2.4. Distribution of the Variants of Interest in the Patients
The distribution of the 80 variants included in category Ip (known mutation) and IIp (potential mutation) showed that approximately 60% of the patients (n = 63) had a mutation or potential mutation in one of the genes analyzed (Figure 4). The 37 variants of category Ip (known mutation) appeared 50 times in 43 patients, and 7 of these patients had more than one variant. The 43 variants of category IIp (potential mutation) appeared 44 times in 37 patients of which 7 had at least two variants.
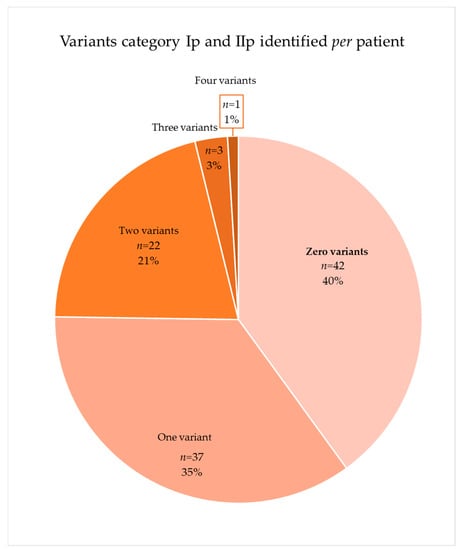
Figure 4.
Distribution of patients with category Ip (known mutation) and IIp (potential mutation).
3. Discussion
In the present study, we examined the presence of rare variants in 40 genes proposed to be involved in lipid metabolism in AMI patients, and we identified mutations in the genes of this pathway in 60% of patients. Therefore, our data highlight the importance of analyzing rare variants in patients with AMI in the cholesterol pathway.
The importance of the cholesterol pathway has been stressed in GWAS studies that show the association between loci in lipid-related genes and susceptibility to AMI [,,,,,,,]. The association between SNPs in genes, such as APOB [], LDLR [,,], PCSCK9 [], and AMI has been described, while other studies have reported a weak association between variants in ABCA1 and both the incidence of AMI and the risk of symptomatic CAD [].
Although common sequence variants have been extensively studied in large genome-wide association studies, it is also important to understand the contribution of rare variants to the susceptibility of AMI [,]. For this purpose, the selection of the genes implicated in the cholesterol pathway and the strategy used to identify rare variants associated with AMI were two key points of this study.
One of the genes studied the most in order to identify low-frequency lipid-associated variants with AMI is LDLR, which codifies the low-density lipoprotein receptor. Approximately 5% of patients with CAD and AMI under the age of 60 years carry heterozygous LDLR mutations [,,]. These mutations are present and equally distributed throughout the gene in the form of exonic substitutions, small exonic rearrangements, large rearrangements, promoter variants, intronic variants, variants in the 3′ untranslated sequence, point mutations, splice site mutations, and large deletions []. In our study, two missense variants, p.G269D and p.G592E, were identified in one patient each while a third variant, p.T726I, was identified in two patients. These data imply a frequency of variants in the LDLR gene of 0.038, similar to those described in the literature. These three variants were previously described in patients with AMI []. However, the impact on the protein is not fully understood because in one study, the authors were not able to observe a disruptive effect on LDL uptake []. Meanwhile, in other research [], it was demonstrated that patients who carried the mutation presented levels of 50% for LDLR expression, LDL-LDLR binding, and LDLR uptake.
However, in our study, the major gene in which variants were found was APOB, which codified the primary apolipoprotein, including 100 chylomicrons, as well as VLDL, Lp(a), IDL, and LDL particles []. In fact, 18 variants were identified in the APOB gene in 19 patients, which implies a frequency of mutations in this gene in our cohort of 0.181. It is important to note that one patient carried three variants of the APOB gene (p.P145S, p.T741N, p.L1212M) and that the variants p.T3826M and p.R1128H were present in two and three patients, respectively. Of the 18 variants found in the APOB gene, 11 were previously described as being mainly associated with hypercholesterolemia and hypertriglyceridemia (category Ip (known mutation)), while 7 were only listed in databases, such as Gnomad, with a frequency of less than 0.01 (Category IIp (potential mutation)). Surprisingly, 9 out of 11 of the variants in category Ip (known mutation) were not previously associated with AMI [,,,,,,,]. However, two of them, namely p.P2821L and p.S2429T, have been described in patients with cardiovascular artery diseases [,,]. Moreover, our study is in concordance with previous studies that suggest that rare deleterious mutations in APOB, such as R3500Q/W, confer a higher risk of ischemic cardiovascular disease in mutation carriers []. In fact, the importance of APOB levels has been stressed because the Copenhagen City Heart Study showed that apolipoprotein B levels are associated with ischemic diseases [].
The most important finding of this study was that 60% of AMI patients had a rare variant in genes involved in the cholesterol pathway. This fact confirms the importance of not focusing only on the gene with a higher presence of rare variants, such as LDLR in the literature or APOB in our case, but rather on the 40 selected genes of the cholesterol pathway. In our study, we have found more than three mutations in LRP2, ABCA1 [], LPA, LRP1, CD36 genes. Additionally, we found between one and three rare variants in genes such as LDLR and PCSK9 which, in other studies, had a high impact on their association with AMI [,,,]. In fact, the low frequency of variants in the LDLR and PCSK9 genes in the group is surprising and supports the idea of the importance of analyzing rare variants in the entire pathway of cholesterol genes.
It is important to highlight that 11 genes of the 40 included, in which we did not identify rare variants, have been described in the literature to have an association with AMI [,,,,,,,,,,]. Therefore, these genes should be further analyzed in following studies in order to increase the 60% of patients with rare variants detected in this study.
The main limitation of this study is that it relies on the fact that it was a descriptive study, not an association study. The sample size is small for an association study of rare variants that present a frequency lower than 0.01. We have not been able to associate the variants with cholesterol levels because in many cases there was no previous data on cholesterol levels prior to infarction due to AMI being the first event. Moreover, because a functional validation of each variant was not performed, this method may have led to a misclassification in some cases.
AMI is a pandemic in which the risk of patients can be assessed; however, conventional risk factors are inadequate to detect who is at risk early in the asymptomatic stage []. Genetic risks, which can be determined at birth, could help to identify patients at higher risk of presenting AMI []. Our data show that rare variants in genes related to cholesterol are present in AMI patients. In future clinical settings where genomic sequencing might be available for all patients, the evaluation of genetic risks would be improved by incorporating variants of the genes of the cholesterol pathway.
4. Materials and Methods
4.1. Study Population
In this study, 105 patients diagnosed with AMI with an elevation of the ST segment (STEMI) and treated with primary percutaneous coronary intervention (PPCI) in A Coruña University Hospital (Spain) between July 2017 to January 2021, were included.
Written informed consent was obtained from every patient included in the study. The protocol of this study was in accordance with the principles of the Declaration of Helsinki, and the “Comité de Ética de la Investigación de Galicia” (ref: 2016/299) also approved it. Blood samples were collected at the time of the hemodynamic procedure and stored at −80 °C until analysis. All samples were included in the biobank of the National Biobank Network of “Instituto de Salud Carlos III” (C.0002483, 2013/109).
A chi-squared test was performed for the following variables: sex; dyslipidemia; hypertension; diabetes; and tobacco, as these variables were considered dichotomous. For age, an ANOVA test was performed.
4.2. Selection of Genes Related to Cholesterol Metabolism
To establish the main genes involved in the cholesterol metabolism pathway, the KEGG map04979 and WikiPathways WP4522 databases were analyzed. Therefore, 40 genes related to the cholesterol metabolism pathway were selected. All genes selected are involved in cholesterol uptake, efflux, transport, storage, utilization, and/or excretion.
4.3. Next-Generation Sequencing
Targeted resequencing was performed using two different kits: the TruSight One sequencing kit of Illumina (San Diego, California, USA) (n = 22) and the Exome Research Panel v.2 of IDT (Leuven, Belgium) (n = 83). The sequencing reactions were conducted on a NextSeq500 platform of Illumina (San Diego, California, USA). The TruSight One kit yielded the sequencing of approximately 5000 genes and the Exome Research Panel v.2 yielded the sequencing of around 20,000 genes. Both strategies included the selected 40 genes related to cholesterol metabolism (Table S1).
The strategy for exome sequence data analysis included several computational tools. The raw data files in the binary base call (BCL) format, generated by the NextSeq Sequencing System, were demultiplexed and converted to a standard FASTQ file by bcl2fastq Conversion Software v2.17. The coverage (Figure S1) and Q-score were analyzed in order to establish the quality of the sequence. Based on the guidelines of the American College of Medical Genetics and Genomics, if the region analyzed presented a sequencing mean depth <30, the region was considered unsuitable for analysis. Furthermore, the threshold in the Q-score established was 30 (base call accuracy of 99.9%).
Computational biology sequence alignment to the human genome version GRCh37/hg19 was performed with Burrows–Wheeler Aligner software 7.17, and the BAM file output was obtained. Variants were detected using the Genome Analysis Toolkit (GATK), and the output files were VCF files. All exonic regions and ±10 intronic regions of the 40 genes related to cholesterol metabolism were visualized via the Integrative Genome Viewer (IGV).
4.4. Variant Annotation and Classification
All variants identified in this study were searched in the Genome Aggregation Database (gnomAD) [] and the Single Nucleotide Polymorphism Database (dbSNP) [] in order to establish frequency in the general population. Moreover, to identify previous genotype–phenotype associations, all variants were checked in HGMD [].
After the annotation of all variants, variant filtering was performed using an adapted variant filtering and prioritization strategy described by Akinrinade et al. [] (Figure 2). In detail, the first filter used was the frequency of the variants was described in the Gnomad and/or dbSNP to be higher than 0.01. This frequency was selected following the classical classification of common variants. The second filtering excluded the synonymous and intronic variants located farther than five bases from the acceptor/donor splice sites.
The remaining variants were classified into well-defined categories described by Hass et al. [] with slight modifications. We defined two categories with a subcategory each for the determination of the likelihood of being disease-relevant mutations (Figure 2): (a) Category I consisted of variants previously described in the literature (HGMD) to be associated with pathologies. Variants of this group associated with lipid metabolism disorders or cardiopathies were classified as Ip (known mutation). (b) Category II included all variants that were not previously pathology-associated in the literature. The subcategory IIp (potential mutation) included variants with more than three out of five in silico predictions tools to be pathogenic and all the stop-gained variants.
The in silico prediction tools used differed between the types of variants. The potential effect of the missense variants was predicted using five different tools: Mutation Taster (http://www.mutationtaster.org (accessed on 28 October 2022)) []; SNAP2 (https://www.rostlab.org/services/snap/ (accessed on 28 October 2022)) [], SIFT (http://sift.jcvi.org/www/SIFT_seq_submit2.html (accessed on 28 October 2022)) []; Polyphen2 (http://genetics.bwh.harvard.edu/%20pph2/ (accessed on 28 October 2022)) []; and PhD-SNP (http://snps.uib.es/phd-snp/phdsnp.html (accessed on 28 October 2022)) []. For the synonym and intronic variants located ± 5 bases from the acceptor/donor splice sites, the potential effect was assessed with five different tools: Mutation Taster (http://www.mutationtaster.org (accessed on 28 October 2022)) []; Splice Site Prediction by Neural Network of Berkeley Drosophila Genome Project (https://www.fruitfly.org/seq_tools/splice.html (accessed on 28 October 2022)) []; NetGene2–2.42 (https://services.healthtech.dtu.dk/service.php?NetGene2-2.42 (accessed on 28 October 2022)) []; Alternative Splice Site Predictor (http://wangcomputing.com/assp/index.html (accessed on 28 October 2022)) []; and CADD-Splice (https://cadd.gs.washington.edu/snv (accessed on 28 October 2022)) []. As suggested by Houdayer et al. [], it is considered a potential pathogenic variant if the variation of the score given by the prediction tool, between the wildtype and the variant, is at 20% or is more significant.
Thus, category Ip (known mutation) mutations were considered known mutations and category IIp (potential mutation) mutations were considered potential mutations.
5. Conclusions
A high prevalence of rare variants in the genes of the cholesterol pathway can be found in AMI patients. Our data show the need to consider not only one gene or a small set of genes, but the wide scope of cholesterol pathway genes in order to make a more realistic assessment of the risks related to AMI.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232416127/s1.
Author Contributions
Conceptualization, L.M.-P., L.N., J.M.V.-R. and M.H.-P.; methodology, R.P.-L., F.R.-L., D.L.-V., A.P. and A.M.-N.; software, R.P.-L., L.M.-P. and L.N.; investigation, R.P.-L., L.M.-P., L.N., F.R.-L., D.L.-V., A.P., A.M.-N. and R.C.; data curation, F.R.-L., D.L.-V., R.C. and J.M.V.-R.; writing, R.P.-L., L.M.-P., L.N., J.M.V.-R. and M.H.-P.; funding acquisition, J.M.V.-R. and M.H.-P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from “Instituto de Salud Carlos III” (PI18/01737) and a non-conditional grant from Abott Vascular.
Institutional Review Board Statement
The protocol of this study was in accordance with the principles of the Declaration of Helsinki, and the “Comité de Ética de la Investigación de Galicia” (ref: 2016/299) had approved it. All the samples were included in the biobank of the National Biobank Network of “Instituto de Salud Carlos III” (C.0002483, 2013/109).
Informed Consent Statement
Written informed consent was obtained from every patient included in the study.
Conflicts of Interest
Authors declare no conflict of interest.
References
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D.; Executive Group on behalf of the Joint European Society of Cardiology (ESC)/American College of Cardiology (ACC)/American Heart Association (AHA)/World Heart Federation (WHF) Task Force for the Universal Definition of Myocardial Infarction. Fourth Universal Definition of Myocardial Infarction (2018). J. Am. Coll. Cardiol. 2018, 72, 2231–2264. [Google Scholar] [CrossRef] [PubMed]
- Writing Group Members; Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; Das, S.R.; de Ferranti, S.; Després, J.P.; et al. Heart Disease and Stroke Statistics-2016 Update: A Report from the American Heart Association. Circulation 2016, 133, e38–e360, Erratum in Circulation 2016, 133, e599. [Google Scholar] [CrossRef]
- Feng, B.; Li, H. Genetic Polymorphism of Matrix Metalloproteinase-9 and Susceptibility to Myocardial Infarction: A Meta-Analysis. Dis. Markers 2022, 2022, 5507153. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.; Chang, C.C.; Hadley, T. Genetic Risk Stratification: A Paradigm Shift in Prevention of Coronary Artery Disease. JACC Basic Transl. Sci. 2021, 6, 287–304. [Google Scholar] [CrossRef] [PubMed]
- McPherson, R.; Tybjaerg-Hansen, A. Genetics of Coronary Artery Disease. Circ Res. 2016, 118, 564–578. [Google Scholar] [CrossRef]
- Musunuru, K.; Kathiresan, S. Surprises From Genetic Analyses of Lipid Risk Factors for Atherosclerosis. Circ Res. 2016, 118, 579–585. [Google Scholar] [CrossRef]
- Duan, Y.; Gong, K.; Xu, S.; Zhang, F.; Meng, X.; Han, J. Regulation of cholesterol homeostasis in health and diseases: From mechanisms to targeted therapeutics. Signal Transduct. Target. Ther. 2022, 7, 265. [Google Scholar] [CrossRef]
- Ference, B.A.; Kastelein, J.J.P.; Ray, K.K.; Ginsberg, H.N.; Chapman, M.J.; Packard, C.J.; Laufs, U.; Oliver-Williams, C.; Wood, A.M.; Butterworth, A.S.; et al. Association of Triglyceride-Lowering LPL Variants and LDL-C-Lowering LDLR Variants with Risk of Coronary Heart Disease. JAMA 2019, 321, 364–373. [Google Scholar] [CrossRef]
- Rać, M.E.; Suchy, J.; Kurzawski, G.; Kurlapska, A.; Safranow, K.; Rać, M.; Sagasz-Tysiewicz, D.; Krzystolik, A.; Poncyljusz, W.; Jakubowska, K.; et al. Polymorphism of the CD36 Gene and Cardiovascular Risk Factors in Patients with Coronary Artery Disease Manifested at a Young Age. Biochem. Genet. 2012, 50, 103–111. [Google Scholar] [CrossRef]
- Qi, L.P.; Chen, L.F.; Dang, A.M.; Li, L.Y.; Fang, Q.; Yan, X.W. Association between the ABCA1-565C/T gene promoter polymorphism and coronary heart disease severity and cholesterol efflux in the Chinese Han population. Genet. Test. Mol. Biomark. 2015, 19, 347–352. [Google Scholar] [CrossRef]
- Dalan, A.B.; Toptaş, B.; Buğra, Z.; Polat, N.; Yılmaz-Aydoğan, H.; Çimen, A.; Isbir, T. The effects of endothelial lipase gene (LIPG) variants on inflammation marker levels and atherosclerosis development. Mol. Biol. Rep. 2013, 40, 5143–5149. [Google Scholar] [CrossRef] [PubMed]
- Ranjith, N.; Pegoraro, R.J.; Rom, L. Lipid profiles and associated gene polymorphisms in young Asian Indian patients with acute myocardial infarction and the metabolic syndrome. Metab. Syndr. Relat. Disord. 2009, 7, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Cyrus, C.; Vatte, C.; Al-Nafie, A.; Chathoth, S.; Al-Ali, R.; Al-Shehri, A.; Akhtar, M.S.; Almansori, M.; Al-Muhanna, F.; Keating, B.; et al. The impact of common polymorphisms in CETP and ABCA1 genes with the risk of coronary artery disease in Saudi Arabians. Hum. Genom. 2016, 10, 8. [Google Scholar] [CrossRef]
- Kotaska, K.; Kolarova, J.; Kotrcova, K.; Cepova, J.; Prusa, R. Correlation between common genetic variants and risk factors associated with prediction of cardiovascular diseases in dyslipidemic patients. Genet. Test. Mol. Biomark. 2012, 16, 210–214. [Google Scholar] [CrossRef]
- Kotlęga, D.; Gołąb-Janowska, M.; Masztalewicz, M.; Ciećwież, S.; Nowacki, P. Association between selected gene polymorphisms and statin metabolism, risk of ischemic stroke and cardiovascular disorders. Postepy Hig. Med. Dosw. (Online) 2016, 70, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Trinder, M.; Paquette, M.; Cermakova, L.; Ban, M.R.; Hegele, R.A.; Baass, A.; Brunham, L.R. Polygenic Contribution to Low-Density Lipoprotein Cholesterol Levels and Cardiovascular Risk in Monogenic Familial Hypercholesterolemia. Circ. Genom. Precis. Med. 2020, 13, 515–523. [Google Scholar] [CrossRef]
- Karjalainen, J.P.; Mononen, N.; Hutri-Kähönen, N.; Lehtimäki, M.; Hilvo, M.; Kauhanen, D.; Juonala, M.; Viikari, J.; Kähönen, M.; Raitakari, O.; et al. New evidence from plasma ceramides links apoE polymorphism to greater risk of coronary artery disease in Finnish adults. J. Lipid Res. 2019, 60, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Tajima, T.; Morita, H.; Ito, K.; Yamazaki, T.; Kubo, M.; Komuro, I.; Momozawa, Y. Blood lipid-related low-frequency variants in LDLR and PCSK9 are associated with onset age and risk of myocardial infarction in Japanese. Sci. Rep. 2018, 8, 8107. [Google Scholar] [CrossRef]
- Do, R.; Stitziel, N.O.; Won, H.H.; Jørgensen, A.B.; Duga, S.; Angelica Merlini, P.; Kiezun, A.; Farrall, M.; Goel, A.; Zuk, O.; et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 2015, 518, 102–106. [Google Scholar] [CrossRef]
- Awan, Z.; Choi, H.Y.; Stitziel, N.; Ruel, I.; Bamimore, M.A.; Husa, R.; Gagnon, M.H.; Wang, R.H.; Peloso, G.M.; Hegele, R.A.; et al. APOE p.Leu167del mutation in familial hypercholesterolemia. Atherosclerosis 2013, 231, 218–222. [Google Scholar] [CrossRef]
- Haas, J.; Frese, K.S.; Peil, B.; Kloos, W.; Keller, A.; Nietsch, R.; Feng, Z.; Müller, S.; Kayvanpour, E.; Vogel, B.; et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur. Heart J. 2015, 36, 1123–1135a. [Google Scholar] [CrossRef] [PubMed]
- The Human Gene Mutation Database. Available online: https://www.hgmd.cf.ac.uk/ac/index.php (accessed on 21 July 2022).
- Service, S.K.; Teslovich, T.M.; Fuchsberger, C.; Ramensky, V.; Yajnik, P.; Koboldt, D.C.; Larson, D.E.; Zhang, Q.; Lin, L.; Welch, R.; et al. Re-sequencing expands our understanding of the phenotypic impact of variants at GWAS loci. PLoS Genet. 2014, 10, e1004147. [Google Scholar] [CrossRef]
- Frikke-Schmidt, R.; Nordestgaard, B.G.; Schnohr, P.; Steffensen, R.; Tybjaerg-Hansen, A. Mutation in ABCA1 predicted risk of ischemic heart disease in the Copenhagen City Heart Study Population. J. Am. Coll. Cardiol. 2005, 46, 1516–1520. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dron, J.S.; Wang, J.; Low-Kam, C.; Khetarpal, S.A.; Robinson, J.F.; McIntyre, A.D.; Ban, M.R.; Cao, H.; Rhainds, D.; Dubé, M.P.; et al. Polygenic determinants in extremes of high-density lipoprotein cholesterol. J. Lipid Res. 2017, 58, 2162–2170. [Google Scholar] [CrossRef] [PubMed]
- Geller, A.S.; Polisecki, E.Y.; Diffenderfer, M.R.; Asztalos, B.F.; Karathanasis, S.K.; Hegele, R.A.; Schaefer, E.J. Genetic and secondary causes of severe HDL deficiency and cardiovascular disease. J. Lipid Res. 2018, 59, 2421–2435. [Google Scholar] [CrossRef]
- Bodzioch, M.; Orsó, E.; Klucken, J.; Langmann, T.; Böttcher, A.; Diederich, W.; Drobnik, W.; Barlage, S.; Büchler, C.; Porsch-Ozcürümez, M.; et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 1999, 22, 347–351. [Google Scholar] [CrossRef]
- Sadananda, S.N.; Foo, J.N.; Toh, M.T.; Cermakova, L.; Trigueros-Motos, L.; Chan, T.; Liany, H.; Collins, J.A.; Gerami, S.; Singaraja, R.R.; et al. Targeted next-generation sequencing to diagnose disorders of HDL cholesterol. J. Lipid Res. 2015, 56, 1993–2001. [Google Scholar] [CrossRef]
- Kars, M.E.; Başak, A.N.; Onat, O.E.; Bilguvar, K.; Choi, J.; Itan, Y.; Çağlar, C.; Palvadeau, R.; Casanova, J.L.; Cooper, D.N.; et al. The genetic structure of the Turkish population reveals high levels of variation and admixture. Proc. Natl. Acad. Sci. USA 2021, 118, e2026076118, Erratum in Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Hong, S.H.; Rhyne, J.; Zeller, K.; Miller, M. ABCA1(Alabama): A novel variant associated with HDL deficiency and premature coronary artery disease. Atherosclerosis 2002, 164, 245–250. [Google Scholar] [CrossRef]
- Backman, J.D.; Li, A.H.; Marcketta, A.; Sun, D.; Mbatchou, J.; Kessler, M.D.; Benner, C.; Liu, D.; Locke, A.E.; Balasubramanian, S.; et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature 2021, 599, 628–634. [Google Scholar] [CrossRef]
- Johansen, C.T.; Dubé, J.B.; Loyzer, M.N.; MacDonald, A.; Carter, D.E.; McIntyre, A.D.; Cao, H.; Wang, J.; Robinson, J.F.; Hegele, R.A. LipidSeq: A next-generation clinical resequencing panel for monogenic dyslipidemias. J. Lipid Res. 2014, 55, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Downes, K.; Megy, K.; Duarte, D.; Vries, M.; Gebhart, J.; Hofer, S.; Shamardina, O.; Deevi, S.V.V.; Stephens, J.; Mapeta, R.; et al. Diagnostic high-throughput sequencing of 2396 patients with bleeding, thrombotic, and platelet disorders. Blood 2019, 134, 2082–2091. [Google Scholar] [CrossRef]
- Hansel, B.; Carrié, A.; Brun-Druc, N.; Leclert, G.; Chantepie, S.; Coiffard, A.S.; Kahn, J.F.; Chapman, M.J.; Bruckert, E. Premature atherosclerosis is not systematic in phytosterolemic patients: Severe hypercholesterolemia as a confounding factor in five subjects. Atherosclerosis 2014, 234, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Dron, J.S.; Wang, J.; McIntyre, A.D.; Iacocca, M.A.; Robinson, J.F.; Ban, M.R.; Cao, H.; Hegele, R.A. Six years’ experience with LipidSeq: Clinical and research learnings from a hybrid, targeted sequencing panel for dyslipidemias. BMC Med. Genom. 2020, 13, 23. [Google Scholar] [CrossRef]
- Romeo, S.; Yin, W.; Kozlitina, J.; Pennacchio, L.A.; Boerwinkle, E.; Hobbs, H.H.; Cohen, J.C. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J. Clin. Investig. 2009, 119, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Marmontel, O.; Rollat-Farnier, P.A.; Wozny, A.S.; Charrière, S.; Vanhoye, X.; Simonet, T.; Chatron, N.; Collin-Chavagnac, D.; Nony, S.; Dumont, S.; et al. Development of a new expanded next-generation sequencing panel for genetic diseases involved in dyslipidemia. Clin. Genet. 2020, 98, 589–594. [Google Scholar] [CrossRef]
- Deeb, S.S.; Nevin, D.N.; Iwasaki, L.; Brunzell, J.D. Two novel apolipoprotein A-IV variants in individuals with familial combined hyperlipidemia and diminished levels of lipoprotein lipase activity. Hum. Mutat. 1996, 8, 319–325. [Google Scholar] [CrossRef]
- Radovica-Spalvina, I.; Latkovskis, G.; Silamikelis, I.; Fridmanis, D.; Elbere, I.; Ventins, K.; Ozola, G.; Erglis, A.; Klovins, J. Next-generation-sequencing-based identification of familial hypercholesterolemia-related mutations in subjects with increased LDL-C levels in a latvian population. BMC Med. Genet. 2015, 16, 86. [Google Scholar] [CrossRef][Green Version]
- Alves, A.C.; Benito-Vicente, A.; Medeiros, A.M.; Reeves, K.; Martin, C.; Bourbon, M. Further evidence of novel APOB mutations as a cause of familial hypercholesterolaemia. Atherosclerosis 2018, 277, 448–456. [Google Scholar] [CrossRef]
- Grzymski, J.J.; Elhanan, G.; Morales Rosado, J.A.; Smith, E.; Schlauch, K.A.; Read, R.; Rowan, C.; Slotnick, N.; Dabe, S.; Metcalf, W.J.; et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat. Med. 2020, 26, 1235–1239. [Google Scholar] [CrossRef]
- Hou, Y.C.; Yu, H.C.; Martin, R.; Cirulli, E.T.; Schenker-Ahmed, N.M.; Hicks, M.; Cohen, I.V.; Jönsson, T.J.; Heister, R.; Napier, L.; et al. Precision medicine integrating whole-genome sequencing, comprehensive metabolomics, and advanced imaging. Proc. Natl. Acad. Sci. USA 2020, 117, 3053–3062. [Google Scholar] [CrossRef] [PubMed]
- Rimbert, A.; Daggag, H.; Lansberg, P.; Buckley, A.; Viel, M.; Kanninga, R.; Johansson, L.; Dullaart, R.P.F.; Sinke, R.; Al Tikriti, A.; et al. Low Detection Rates of Genetic FH in Cohort of Patients with Severe Hypercholesterolemia in the United Arabic Emirates. Front. Genet. 2022, 12, 809256. [Google Scholar] [CrossRef] [PubMed]
- Leren, T.P.; Berge, K.E. Identification of mutations in the apolipoprotein B-100 gene and in the PCSK9 gene as the cause of hypocholesterolemia. Clin. Chim. Acta 2008, 397, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.X.; Wu, N.Q.; Sun, D.; Liu, H.H.; Jin, J.L.; Li, S.; Guo, Y.L.; Zhu, C.G.; Gao, Y.; Dong, Q.T.; et al. Application of expanded genetic analysis in the diagnosis of familial hypercholesterolemia in patients with very early-onset coronary artery disease. J. Transl. Med. 2018, 16, 345. [Google Scholar] [CrossRef]
- Johansen, C.T.; Wang, J.; Lanktree, M.B.; Cao, H.; McIntyre, A.D.; Ban, M.R.; Martins, R.A.; Kennedy, B.A.; Hassell, R.G.; Visser, M.E.; et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 2010, 42, 684–687. [Google Scholar] [CrossRef]
- Roa Garrido, J.; Carrasco Salas, P.; Toscano Pérez, C.; Arrobas Velilla, T.; Vázquez Rico, I.; Díaz Fernández, J.F. Genetics and biochemistry of familial hypercholesterolemia in Southwest of the Iberian Peninsula. Particularidades genéticas y bioquímicas de la hipercolesterolemia familiar en el suroeste de la Península Ibérica. Clin. Investig. Arterioscler. 2021, 33, 62–69. [Google Scholar] [CrossRef]
- Noto, D.; Spina, R.; Giammanco, A.; Barbagallo, C.M.; Ganci, A.; Scrimali, C.; Brucato, F.; Misiano, G.; Ciaccio, M.; Caldarella, R.; et al. Diagnosis of familial hypercholesterolemia in a large cohort of Italian genotyped hypercholesterolemic patients. Atherosclerosis 2022, 347, 63–67. [Google Scholar] [CrossRef]
- Batais, M.A.; Almigbal, T.H.; Shaik, N.A.; Alharbi, F.K.; Alharbi, K.K.; Ali Khan, I. Screening of common genetic variants in the APOB gene related to familial hypercholesterolemia in a Saudi population: A case-control study. Medicine 2019, 98, e14247. [Google Scholar] [CrossRef]
- Lancellotti, S.; Di Leo, E.; Penacchioni, J.Y.; Balli, F.; Viola, L.; Bertolini, S.; Calandra, S.; Tarugi, P. Hypobetalipoproteinemia with an apparently recessive inheritance due to a “de novo” mutation of apolipoprotein B. Biochim. Biophys. Acta 2004, 1688, 61–67. [Google Scholar] [CrossRef]
- Khlebus, E.; Kutsenko, V.; Meshkov, A.; Ershova, A.; Kiseleva, A.; Shevtsov, A.; Shcherbakova, N.; Zharikova, A.; Lankin, V.; Tikhaze, A.; et al. Multiple rare and common variants in APOB gene locus associated with oxidatively modified low-density lipoprotein levels. PLoS ONE 2019, 14, e0217620. [Google Scholar] [CrossRef]
- Zeissig, S.; Dougan, S.K.; Barral, D.C.; Junker, Y.; Chen, Z.; Kaser, A.; Ho, M.; Mandel, H.; McIntyre, A.; Kennedy, S.M.; et al. Primary deficiency of microsomal triglyceride transfer protein in human abetalipoproteinemia is associated with loss of CD1 function. J. Clin. Investig. 2010, 120, 2889–2899. [Google Scholar] [CrossRef] [PubMed]
- Hegele, R.A.; Connelly, P.W.; Maguire, G.F.; Huff, M.W.; Leiter, L.; Wolfe, B.M.; Evans, A.J.; Little, J.A. An apolipoprotein CII mutation, CIILys19—Thr’ identified in patients with hyperlipidemia. Dis. Markers 1991, 9, 73–80. [Google Scholar] [PubMed]
- Sanghera, D.K.; Wagenknecht, D.R.; McIntyre, J.A.; Kamboh, M.I. Identification of structural mutations in the fifth domain of apolipoprotein H (beta 2-glycoprotein I) which affect phospholipid binding. Hum. Mol. Genet. 1997, 6, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Eicher, J.D.; Chami, N.; Kacprowski, T.; Nomura, A.; Chen, M.H.; Yanek, L.R.; Tajuddin, S.M.; Schick, U.M.; Slater, A.J.; Pankratz, N.; et al. Platelet-Related Variants Identified by Exomechip Meta-analysis in 157,293 Individuals. Am. J. Hum. Genet. 2016, 99, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Iotchkova, V.; Huang, J.; Morris, J.A.; Jain, D.; Barbieri, C.; Walter, K.; Min, J.L.; Chen, L.; Astle, W.; Cocca, M.; et al. Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat. Genet. 2016, 48, 1303–1312, Erratum in Nat. Genet. 2018, 50, 1752. [Google Scholar] [CrossRef]
- Flesch, B.K.; Scherer, V.; Opitz, A.; Ochmann, O.; Janson, A.; Steitz, M.; Zeiler, T. Platelet CD36 deficiency is present in 2.6% of Arabian individuals and can cause NAIT and platelet refractoriness. Transfusion 2021, 61, 1932–1942. [Google Scholar] [CrossRef]
- Pilo, B.; de Blas, G.; Sobrido, M.J.; Navarro, C.; Grandas, F.; Barrero, F.J.; Moya, M.A.; Jimenez-Escrig, A. Neurophysiological study in cerebrotendinous xanthomatosis. Muscle Nerve 2011, 43, 531–536. [Google Scholar] [CrossRef]
- Mozas, P.; Castillo, S.; Tejedor, D.; Reyes, G.; Alonso, R.; Franco, M.; Saenz, P.; Fuentes, F.; Almagro, F.; Mata, P.; et al. Molecular characterization of familial hypercholesterolemia in Spain: Identification of 39 novel and 77 recurrent mutations in LDLR. Hum. Mutat. 2004, 24, 187. [Google Scholar] [CrossRef]
- Leren, T.P.; Bogsrud, M.P. Molecular genetic testing for autosomal dominant hypercholesterolemia in 29,449 Norwegian index patients and 14,230 relatives during the years 1993–2020. Atherosclerosis 2021, 322, 61–66. [Google Scholar] [CrossRef]
- Hobbs, H.H.; Brown, M.S.; Goldstein, J.L. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum. Mutat. 1992, 1, 445–466. [Google Scholar] [CrossRef]
- Van der Schoot, V.; Haer-Wigman, L.; Feenstra, I.; Tammer, F.; Oerlemans, A.J.M.; van Koolwijk, M.P.A.; van Agt, F.; Arens, Y.H.J.M.; Brunner, H.G.; Vissers, L.E.L.M.; et al. Lessons learned from unsolicited findings in clinical exome sequencing of 16,482 individuals. Eur. J. Hum. Genet. 2022, 30, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Loux, N.; Saint-Jore, B.; Collod, G.; Dairou, F.; Benlian, P.; Truffert, J.; Dastugue, B.; Douste-Blazy, P.; de Gennes, J.L.; Junien, C. Screening for new mutations in the LDL receptor gene in seven French familial hypercholesterolemia families by the single strand conformation polymorphism method. Hum. Mutat. 1992, 1, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Chora, J.R.; Medeiros, A.M.; Alves, A.C.; Bourbon, M. Analysis of publicly available LDLR, APOB, and PCSK9 variants associated with familial hypercholesterolemia: Application of ACMG guidelines and implications for familial hypercholesterolemia diagnosis. Genet. Med. 2018, 20, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Martín-Campos, J.M.; Plana, N.; Figueras, R.; Ibarretxe, D.; Caixàs, A.; Esteve, E.; Pérez, A.; Bueno, M.; Mauri, M.; Roig, R.; et al. Autosomal dominant hypercholesterolemia in Catalonia: Correspondence between clinical-biochemical and genetic diagnostics in 967 patients studied in a multicenter clinical setting. J. Clin. Lipidol. 2018, 12, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Vinje, T.; Wierød, L.; Leren, T.P.; Strøm, T.B. Prevalence of cholesteryl ester storage disease among hypercholesterolemic subjects and functional characterization of mutations in the lysosomal acid lipase gene. Mol. Genet. Metab. 2018, 123, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Singaraja, R.R.; Sivapalaratnam, S.; Hovingh, K.; Dubé, M.P.; Castro-Perez, J.; Collins, H.L.; Adelman, S.J.; Riwanto, M.; Manz, J.; Hubbard, B.; et al. The impact of partial and complete loss-of-function mutations in endothelial lipase on high-density lipoprotein levels and functionality in humans. Circ. Cardiovasc. Genet. 2013, 6, 54–62. [Google Scholar] [CrossRef]
- Motazacker, M.M.; Peter, J.; Treskes, M.; Shoulders, C.C.; Kuivenhoven, J.A.; Hovingh, G.K. Evidence of a polygenic origin of extreme high-density lipoprotein cholesterol levels. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1521–1528, Erratum in Arterioscler. Thromb. Vasc. Biol. 2013, 33, e128. [Google Scholar] [CrossRef]
- Edmondson, A.C.; Brown, R.J.; Kathiresan, S.; Cupples, L.A.; Demissie, S.; Manning, A.K.; Jensen, M.K.; Rimm, E.B.; Wang, J.; Rodrigues, A.; et al. Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J. Clin. Investig. 2009, 119, 1042–1050. [Google Scholar] [CrossRef]
- Cole, J.; Blackhurst, D.M.; Solomon, G.A.E.; Ratanjee, B.D.; Benjamin, R.; Marais, A.D. Atherosclerotic cardiovascular disease in hyperalphalipoproteinemia due to LIPG variants. J. Clin. Lipidol. 2021, 15, 142–150.e2. [Google Scholar] [CrossRef]
- Morgan, B.M.; Brown, A.N.; Deo, N.; Harrop, T.W.R.; Taiaroa, G.; Mace, P.D.; Wilbanks, S.M.; Merriman, T.R.; Williams, M.J.A.; McCormick, S.P.A. Nonsynonymous SNPs in LPA homologous to plasminogen deficiency mutants represent novel null apo(a) alleles. J. Lipid Res. 2020, 61, 432–444. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Natarajan, P.; Klarin, D.; Won, H.H.; Peloso, G.M.; Stitziel, N.O.; Nomura, A.; Zekavat, S.M.; Bick, A.G.; et al. Phenotypic Characterization of Genetically Lowered Human Lipoprotein(a) Levels. J Am Coll Cardiol. 2016, 68, 2761–2772. [Google Scholar] [CrossRef] [PubMed]
- Theis, J.L.; Vogler, G.; Missinato, M.A.; Li, X.; Nielsen, T.; Zeng, X.I.; Martinez-Fernandez, A.; Walls, S.M.; Kervadec, A.; Kezos, J.N.; et al. Patient-specific genomics and cross-species functional analysis implicate LRP2 in hypoplastic left heart syndrome. Elife 2020, 9, e59554. [Google Scholar] [CrossRef] [PubMed]
- Park, W.D.; O’Brien, J.F.; Lundquist, P.A.; Kraft, D.L.; Vockley, C.W.; Karnes, P.S.; Patterson, M.C.; Snow, K. Identification of 58 novel mutations in Niemann-Pick disease type C: Correlation with biochemical phenotype and importance of PTC1-like domains in NPC1. Hum. Mutat. 2003, 22, 313–325. [Google Scholar] [CrossRef]
- Sriretnakumar, V.; Harripaul, R.; Vincent, J.B.; Kennedy, J.L.; So, J. Enrichment of pathogenic variants in genes associated with inborn errors of metabolism in psychiatric populations. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2019, 180, 46–54. [Google Scholar] [CrossRef]
- Picillo, M.; Amboni, M.; Bruni, A.; Maletta, R.; Barone, P. Prevalence of heterozygous mutations in Niemann-Pick type C genes in a cohort of progressive supranuclear palsy. Park. Relat. Disord. 2020, 79, 9–10. [Google Scholar] [CrossRef]
- Abouelhoda, M.; Faquih, T.; El-Kalioby, M.; Alkuraya, F.S. Revisiting the morbid genome of Mendelian disorders. Genome Biol. 2016, 17, 235. [Google Scholar] [CrossRef] [PubMed]
- Samani, N.J.; Hengstenberg, C.; Mangino, M.; Mayer, B.; Dixon, R.J.; Meitinger, T.; Braund, P.; Wichmann, H.E.; Barrett, J.H.; König, I.R.; et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007, 357, 443–453. [Google Scholar] [CrossRef]
- Kathiresan, S.; Voight, B.F.; Purcell, S.; Musunuru, K.; Ardissino, D.; Mannucci, P.M.; Anand, S.; Engert, J.C.; Samani, N.J.; Schunkert, H.; et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009, 41, 334–341. [Google Scholar] [CrossRef]
- Schunkert, H.; König, I.R.; Kathiresan, S.; Reilly, M.P.; Assimes, T.L.; Holm, H.; Preuss, M.; Stewart, A.F.; Barbalic, M.; Gieger, C.; et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011, 43, 333–338. [Google Scholar] [CrossRef]
- Lu, X.; Wang, L.; Chen, S.; He, L.; Yang, X.; Shi, Y.; Cheng, J.; Zhang, L.; Gu, C.C.; Huang, J.; et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat. Genet. 2012, 44, 890–894. [Google Scholar] [CrossRef]
- Deloukas, P.; Kanoni, S.; Willenborg, C.; Farrall, M.; Assimes, T.L.; Thompson, J.R.; Ingelsson, E.; Saleheen, D.; Erdmann, J.; Goldstein, B.A.; et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013, 45, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.P.; Peloso, G.M.; Pirruccello, J.P.; Johansen, C.T.; Dubé, J.B.; Larach, D.B.; Ban, M.R.; Dallinge-Thie, G.M.; Gupta, N.; Boehnke, M.; et al. Targeted exonic sequencing of GWAS loci in the high extremes of the plasma lipids distribution. Atherosclerosis 2016, 250, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Van der Harst, P.; Verweij, N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res. 2018, 122, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.S.; Xie, C.; Paré, G.; Montpetit, A.; Rangarajan, S.; McQueen, M.J.; Cordell, H.J.; Keavney, B.; Yusuf, S.; Hudson, T.J.; et al. Genetic variants associated with myocardial infarction risk factors in over 8000 individuals from five ethnic groups: The Interheart Genetics Study. Circ. Cardiovasc. Genet. 2009, 2, 16–25. [Google Scholar] [CrossRef]
- Brænne, I.; Kleinecke, M.; Reiz, B.; Graf, E.; Strom, T.; Wieland, T.; Fischer, M.; Kessler, T.; Hengstenberg, C.; Meitinger, T.; et al. Systematic analysis of variants related to familial hypercholesterolemia in families with premature myocardial infarction. Eur. J. Hum. Genet. 2016, 24, 191–197. [Google Scholar] [CrossRef]
- Dai, X.; Wiernek, S.; Evans, J.P.; Runge, M.S. Genetics of coronary artery disease and myocardial infarction. World J. Cardiol. 2016, 8, 1–23. [Google Scholar] [CrossRef]
- Cohen, J.C.; Boerwinkle, E.; Mosley, T.H., Jr.; Hobbs, H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006, 354, 1264–1272. [Google Scholar] [CrossRef]
- Song, C.; Pedersen, N.L.; Reynolds, C.A.; Sabater-Lleal, M.; Kanoni, S.; Willenborg, C.; CARDIoGRAMplusC4D Consortium; Syvänen, A.C.; Watkins, H.; Hamsten, A.; et al. Genetic variants from lipid-related pathways and risk for incident myocardial infarction. PLoS ONE 2013, 8, e60454. [Google Scholar] [CrossRef]
- Helgadottir, A.; Gretarsdottir, S.; Thorleifsson, G.; Hjartarson, E.; Sigurdsson, A.; Magnusdottir, A.; Jonasdottir, A.; Kristjansson, H.; Sulem, P.; Oddsson, A.; et al. Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. Nat. Genet. 2016, 48, 634–639. [Google Scholar] [CrossRef]
- Thormaehlen, A.S.; Schuberth, C.; Won, H.H.; Blattmann, P.; Joggerst-Thomalla, B.; Theiss, S.; Asselta, R.; Duga, S.; Merlini, P.A.; Ardissino, D.; et al. Systematic cell-based phenotyping of missense alleles empowers rare variant association studies: A case for LDLR and myocardial infarction. PLoS Genet. 2015, 11, e1004855. [Google Scholar] [CrossRef]
- Alves, A.C.; Azevedo, S.; Benito-Vicente, A.; Graça, R.; Galicia-Garcia, U.; Barros, P.; Jordan, P.; Martin, C.; Bourbon, M. LDLR variants functional characterization: Contribution to variant classification. Atherosclerosis 2021, 329, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Behbodikhah, J.; Ahmed, S.; Elyasi, A.; Kasselman, L.J.; De Leon, J.; Glass, A.D.; Reiss, A.B. Apolipoprotein B and Cardiovascular Disease: Biomarker and Potential Therapeutic Target. Metabolites 2021, 11, 690. [Google Scholar] [CrossRef] [PubMed]
- Dagli-Hernandez, C.; Borges, J.B.; Marçal, E.D.S.R.; de Freitas, R.C.C.; Mori, A.A.; Gonçalves, R.M.; Faludi, A.A.; de Oliveira, V.F.; Ferreira, G.M.; Bastos, G.M.; et al. Genetic Variant ABCC1 rs45511401 Is Associated with Increased Response to Statins in Patients with Familial Hypercholesterolemia. Pharmaceutics 2022, 14, 944. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, A.M.; Alves, A.C.; Bourbon, M. Mutational analysis of a cohort with clinical diagnosis of familial hypercholesterolemia: Considerations for genetic diagnosis improvement. Genet. Med. 2016, 18, 316–324. [Google Scholar] [CrossRef]
- Dewey, F.E.; Murray, M.F.; Overton, J.D.; Habegger, L.; Leader, J.B.; Fetterolf, S.N.; O’Dushlaine, C.; Van Hout, C.V.; Staples, J.; Gonzaga-Jauregui, C.; et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 2016, 354, aaf6814. [Google Scholar] [CrossRef]
- Alves, A.C.; Etxebarria, A.; Soutar, A.K.; Martin, C.; Bourbon, M. Novel functional APOB mutations outside LDL-binding region causing familial hypercholesterolaemia. Hum. Mol. Genet. 2014, 23, 1817–1828. [Google Scholar] [CrossRef]
- Hayat, M.; Kerr, R.; Bentley, A.R.; Rotimi, C.N.; Raal, F.J.; Ramsay, M. Genetic associations between serum low LDL-cholesterol levels and variants in LDLR, APOB, PCSK9 and LDLRAP1 in African populations. PLoS ONE 2020, 15, e0229098, Erratum in PLoS ONE 2021, 16, e0249478. [Google Scholar] [CrossRef]
- Benn, M. Apolipoprotein B levels, APOB alleles, and risk of ischemic cardiovascular disease in the general population, a review. Atherosclerosis 2009, 206, 17–30. [Google Scholar] [CrossRef]
- Cohen, J.; Pertsemlidis, A.; Kotowski, I.K.; Graham, R.; Garcia, C.K.; Hobbs, H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005, 37, 161–165, Erratum in Nat. Genet. 2005, 37, 328. [Google Scholar] [CrossRef]
- Henkel, A.S.; Kavesh, M.H.; Kriss, M.S.; Dewey, A.M.; Rinella, M.E.; Green, R.M. Hepatic overexpression of abcb11 promotes hypercholesterolemia and obesity in mice. Gastroenterology 2011, 141, 1404–1411.e2. [Google Scholar] [CrossRef]
- Su, X.; Peng, D.Q. New insights into ANGPLT3 in controlling lipoprotein metabolism and risk of cardiovascular diseases. Lipids Health Dis. 2018, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.D.; Chen, X.C.; Yang, Y.N.; Gao, X.M.; Ma, X.; Huang, Y.; Li, X.M.; Gai, M.T.; Liu, F.; Pan, S.; et al. Apolipoprotein A1 is associated with SYNTAX score in patients with a non-ST segment elevation myocardial infarction. Lipids Health Dis. 2019, 18, 159. [Google Scholar] [CrossRef] [PubMed]
- Birjmohun, R.S.; Dallinga-Thie, G.M.; Kuivenhoven, J.A.; Stroes, E.S.; Otvos, J.D.; Wareham, N.J.; Luben, R.; Kastelein, J.J.; Khaw, K.T.; Boekholdt, S.M. Apolipoprotein A-II is inversely associated with risk of future coronary artery disease. Circulation 2007, 116, 2029–2035. [Google Scholar] [CrossRef]
- Gautier, T.; Deckert, V.; Aires, V.; Le Guern, N.; Proukhnitzky, L.; Patoli, D.; Lemaire, S.; Maquart, G.; Bataille, A.; Xolin, M.; et al. Human apolipoprotein C1 transgenesis reduces atherogenesis in hypercholesterolemic rabbits. Atherosclerosis 2021, 320, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Aziz, T.A.; Mohamed, R.H.; Hashem, R.M. Association of lipoprotein lipase and apolipoprotein C-III genes polymorphism with acute myocardial infarction in diabetic patients. Mol. Cell. Biochem. 2011, 354, 141–150. [Google Scholar] [CrossRef]
- Meiner, V.; Friedlander, Y.; Milo, H.; Sharon, N.; Ben-Avi, L.; Shpitzen, S.; Leitersdorf, E.; Siscovick, D.S.; Schwartz, S.M. Cholesteryl ester transfer protein (CETP) genetic variation and early onset of non-fatal myocardial infarction. Ann. Hum. Genet. 2008, 72 Pt 6, 732–741. [Google Scholar] [CrossRef]
- Lee, C.; Cui, Y.; Song, J.; Li, S.; Zhang, F.; Wu, M.; Li, L.; Hu, D.; Chen, H. Effects of familial hypercholesterolemia-associated genes on the phenotype of premature myocardial infarction. Lipids Health Dis. 2019, 18, 95. [Google Scholar] [CrossRef]
- Weissglas-Volkov, D.; Calkin, A.C.; Tusie-Luna, T.; Sinsheimer, J.S.; Zelcer, N.; Riba, L.; Tino, A.M.; Ordoñez-Sánchez, M.L.; Cruz-Bautista, I.; Aguilar-Salinas, C.A.; et al. The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL receptor degradation in humans. J. Clin. Investig. 2011, 121, 3062–3071. [Google Scholar] [CrossRef]
- Stanislovaitiene, D.; Lesauskaite, V.; Zaliuniene, D.; Smalinskiene, A.; Gustiene, O.; Zaliaduonyte-Peksiene, D.; Tamosiunas, A.; Luksiene, D.; Petkeviciene, J.; Zaliunas, R. SCARB1 single nucleotide polymorphism (rs5888) is associated with serum lipid profile and myocardial infarction in an age- and gender-dependent manner. Lipids Health Dis. 2013, 12, 24. [Google Scholar] [CrossRef]
- Silbernagel, N.; Walecki, M.; Schäfer, M.K.; Kessler, M.; Zobeiri, M.; Rinné, S.; Kiper, A.K.; Komadowski, M.A.; Vowinkel, K.S.; Wemhöner, K.; et al. The VAMP-associated protein VAPB is required for cardiac and neuronal pacemaker channel function. FASEB J. 2018, 32, 6159–6173. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443, Erratum in Nature 2021, 597, E3–E4. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Akinrinade, O.; Ollila, L.; Vattulainen, S.; Tallila, J.; Gentile, M.; Salmenperä, P.; Koillinen, H.; Kaartinen, M.; Nieminen, M.S.; Myllykangas, S.; et al. Genetics and genotype-phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur. Heart J. 2015, 36, 2327–2337. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, Y.; Rost, B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007, 35, 3823–3835. [Google Scholar] [CrossRef]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P. PhD-SNPg: A webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Res. 2017, 45, W247–W252. [Google Scholar] [CrossRef]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef]
- Hebsgaard, S.M.; Korning, P.G.; Tolstrup, N.; Engelbrecht, J.; Rouzé, P.; Brunak, S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996, 24, 3439–3452. [Google Scholar] [CrossRef]
- Wang, M.; Marín, A. Characterization and prediction of alternative splice sites. Gene 2006, 366, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, P.; Schubach, M.; Shendure, J.; Kircher, M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Houdayer, C.; Dehainault, C.; Mattler, C.; Michaux, D.; Caux-Moncoutier, V.; Pagès-Berhouet, S.; d’Enghien, C.D.; Laugé, A.; Castera, L.; Gauthier-Villars, M.; et al. Evaluation of in silico splice tools for decision-making in molecular diagnosis. Hum. Mutat. 2008, 29, 975–982. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).