Divergences of the RLR Gene Families across Lophotrochozoans: Domain Grafting, Exon–Intron Structure, Expression, and Positive Selection
Abstract
1. Introduction
2. Results
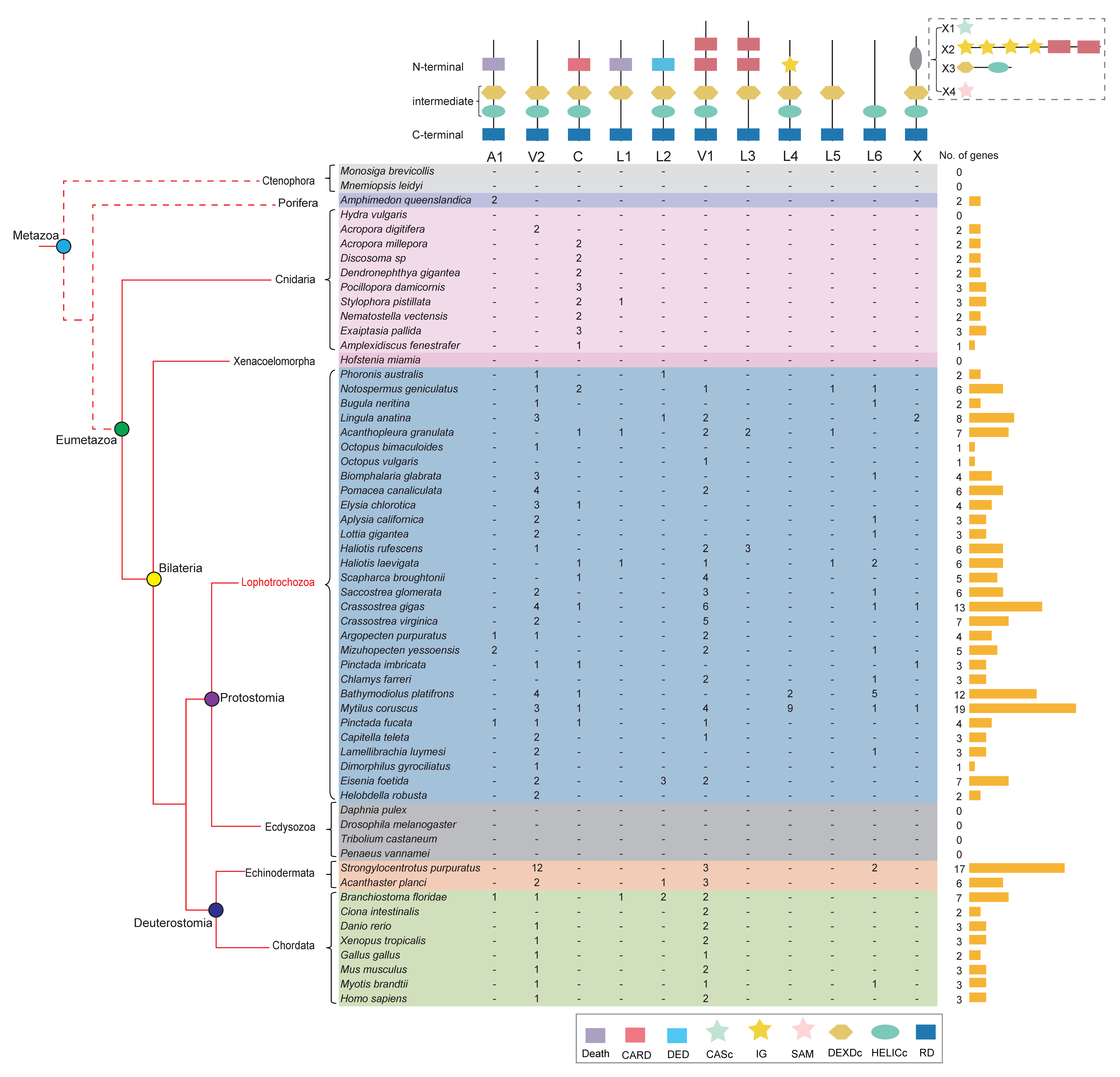
2.1. Identification of the RIG-I-like Receptor Repertoires
2.2. RLR Domain Annotation
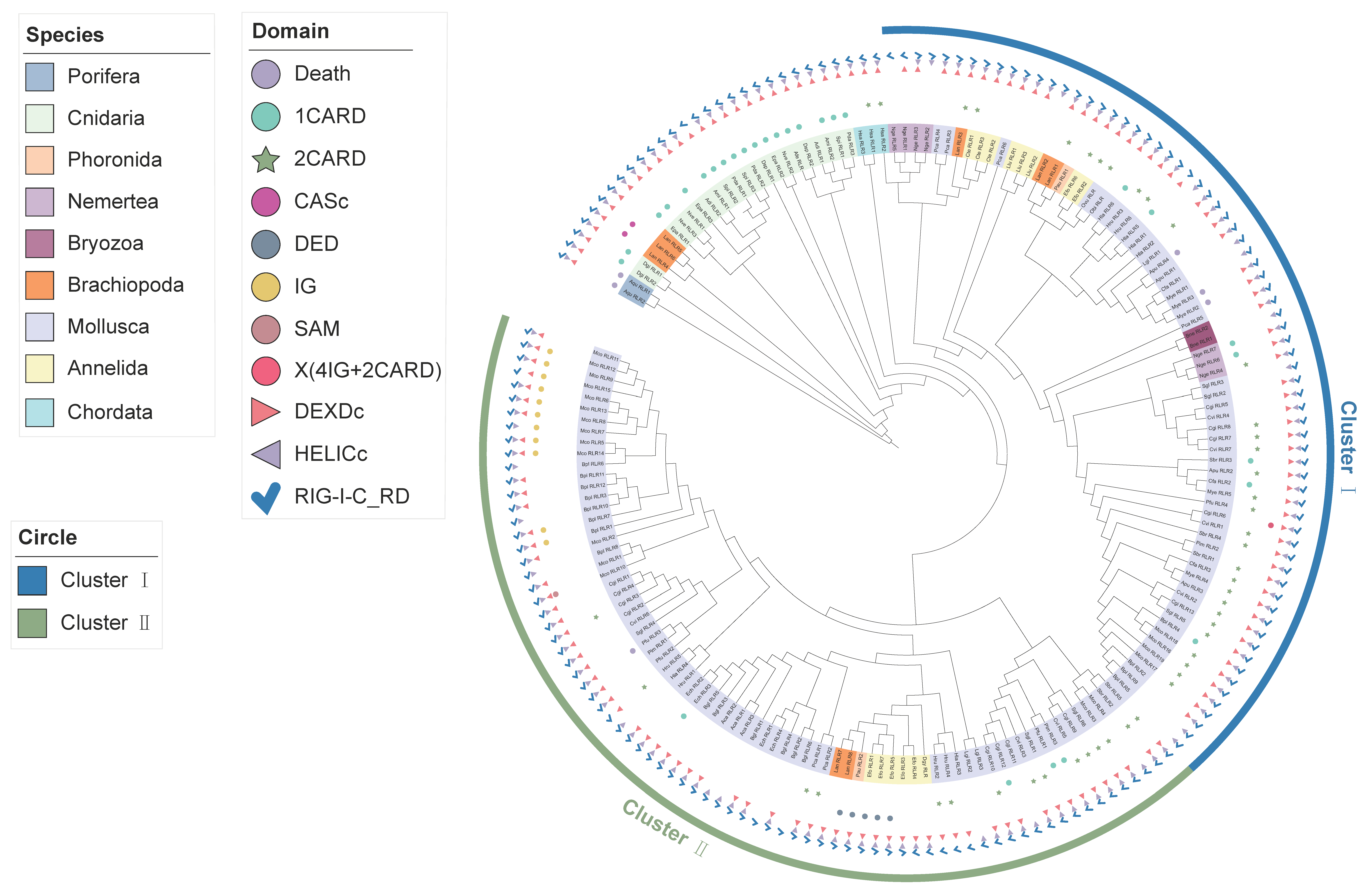
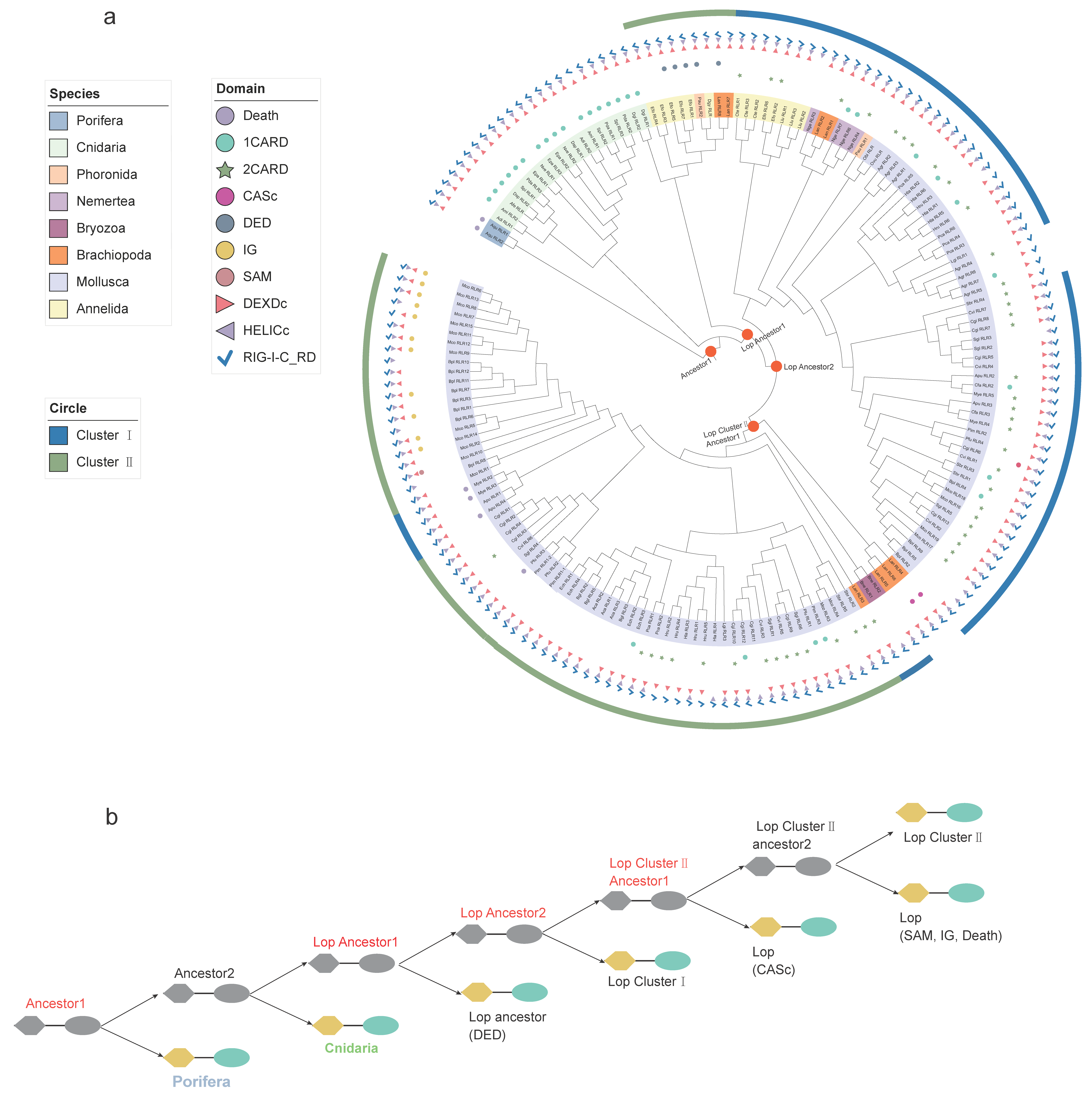
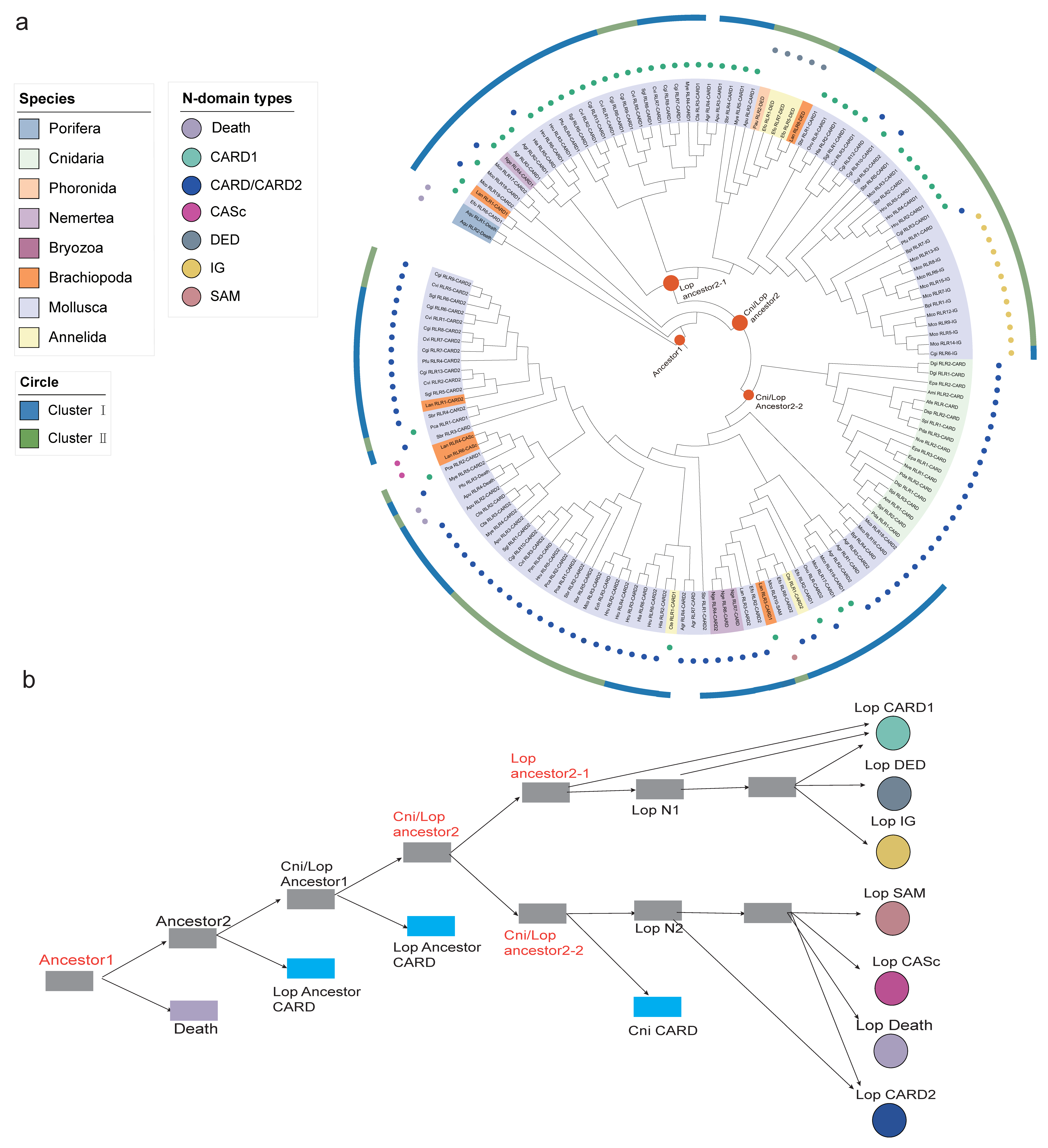
2.3. Phylogenetic Distribution of Three Discrete Domains
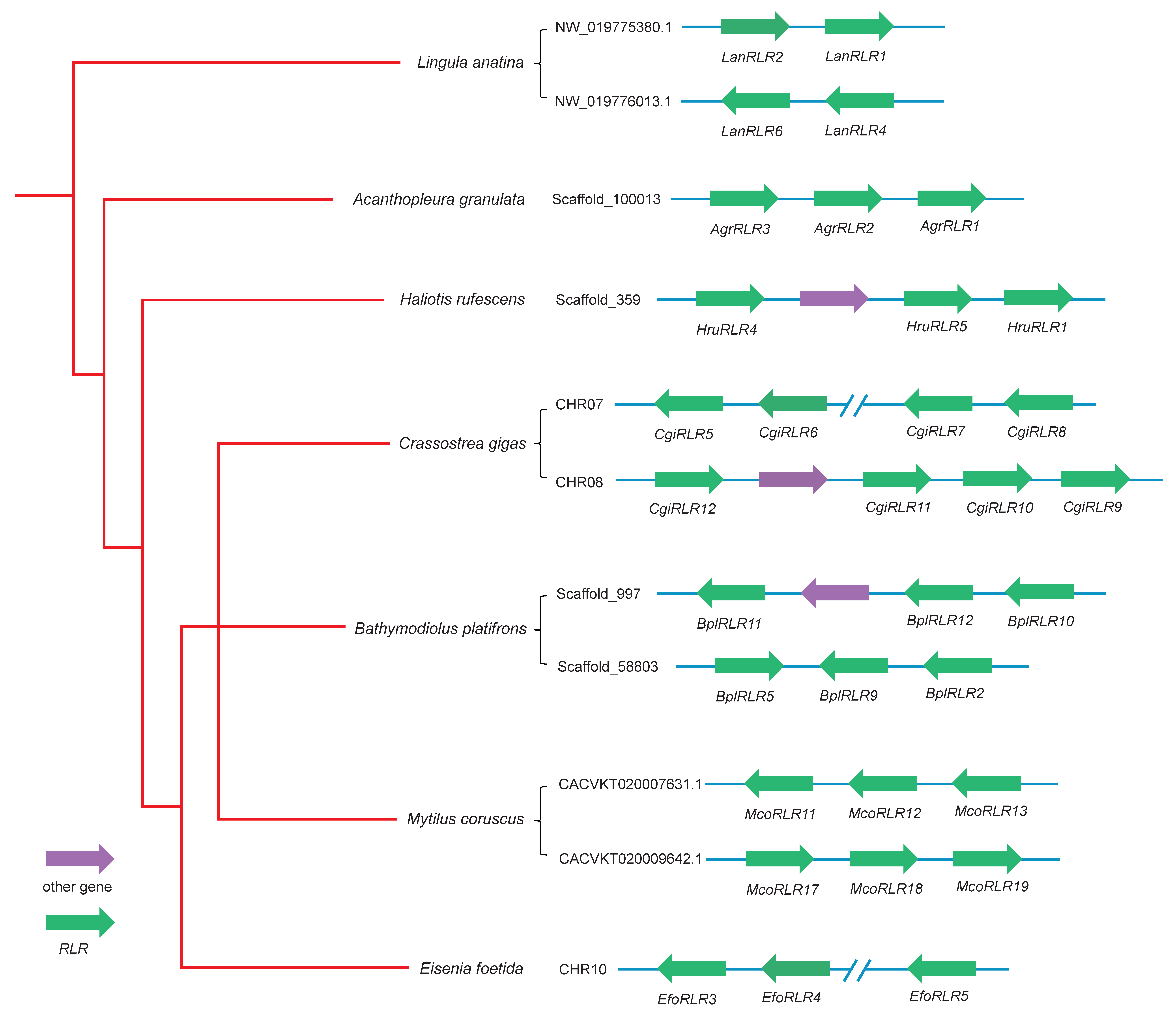
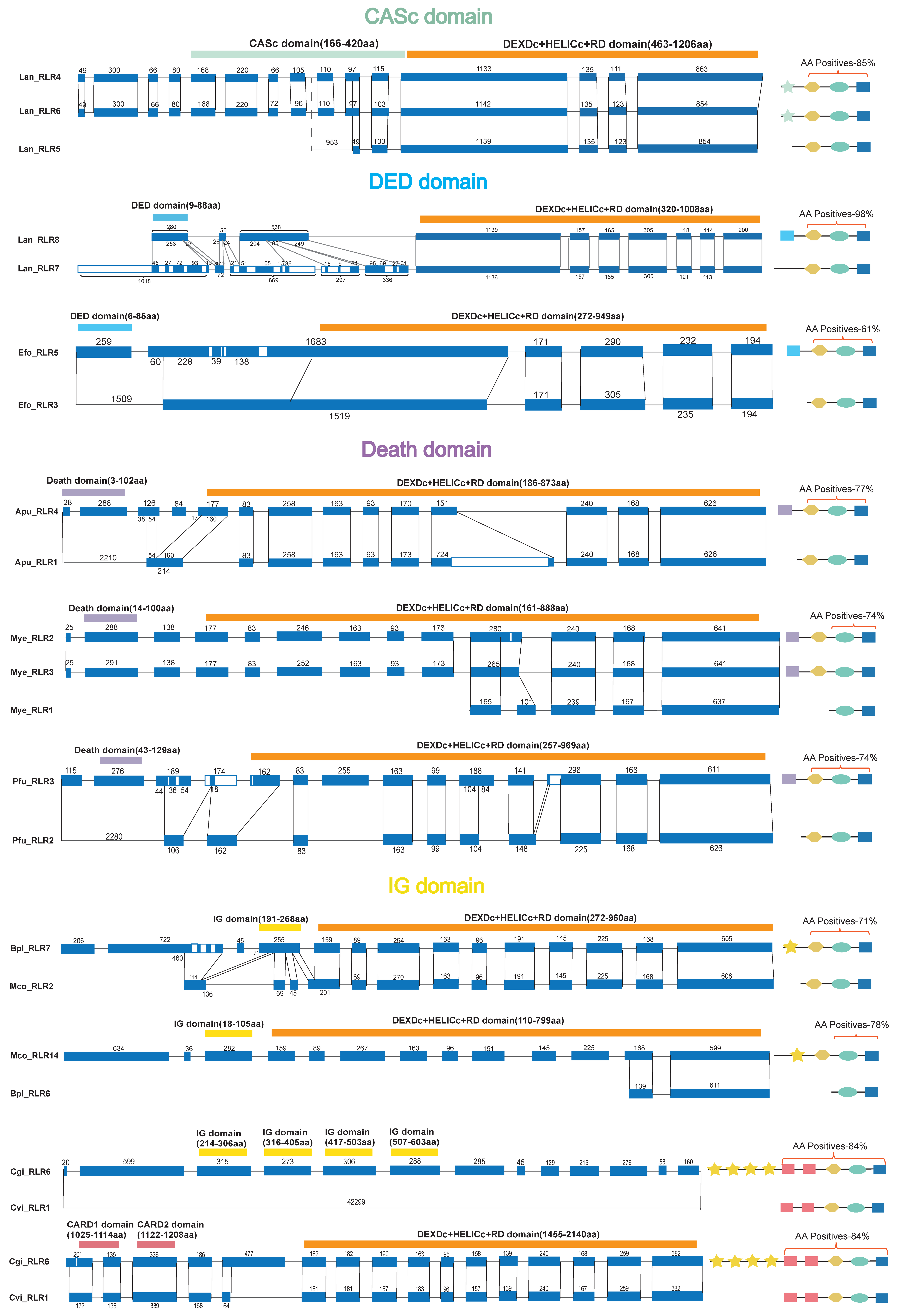
2.4. Intron–Exon Structure Analysis of Lophotrochozoan RLRs with Diverse N-Terminal Domains
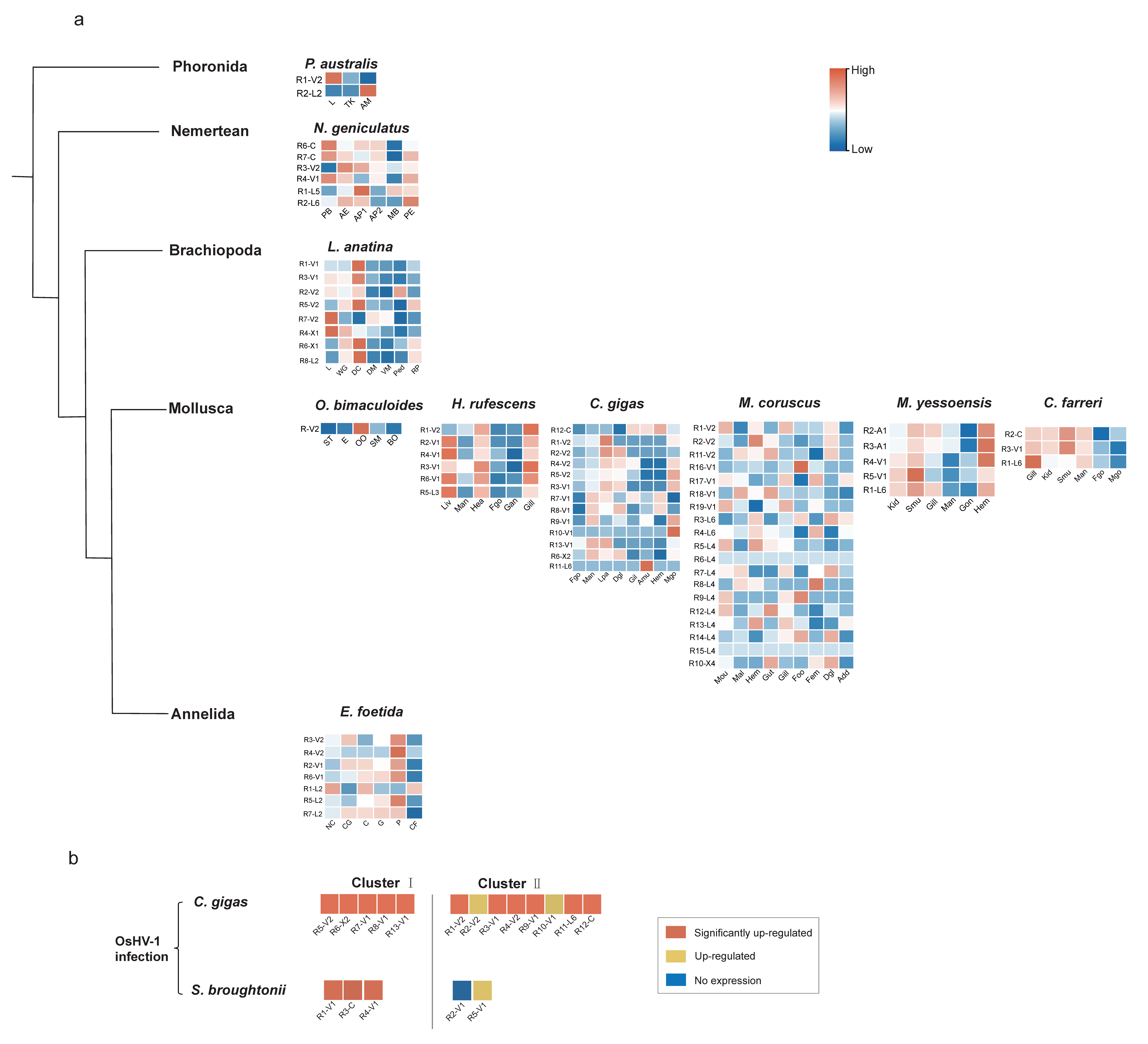
2.5. Expression Profiles of the Lophotrochozoan RLRs
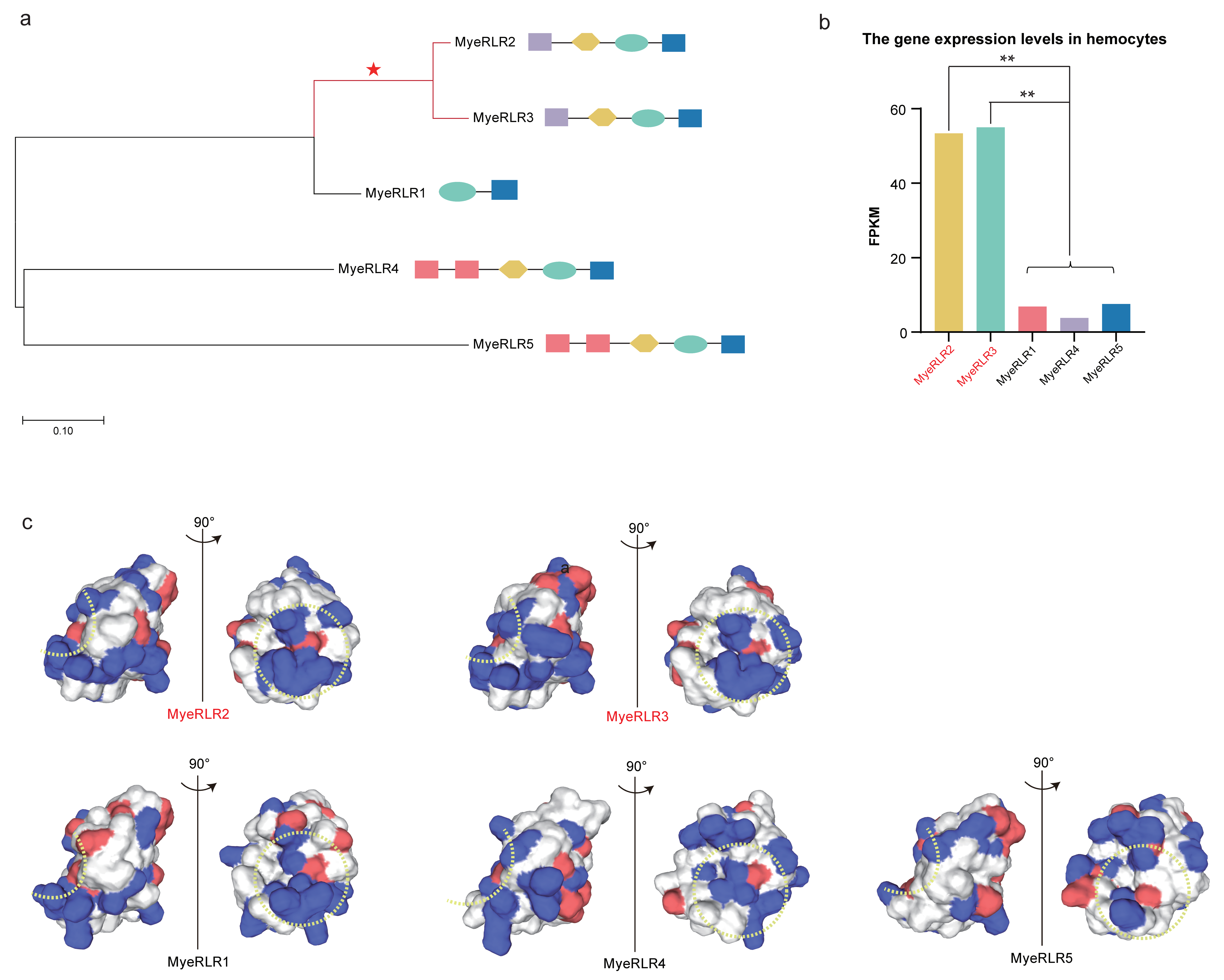
2.6. Evidence of Positive Selection in Lophotrochozoan RLRs with Diverse N-Terminal Domains
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. RLR Gene Identification and Phylogenetic Analysis
4.3. Identification of RLR Homologs and Analysis of Gene Structures
4.4. Transcriptomic Analysis of Gene Expression
4.5. Positive Selection Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koonin, E.V.; Dolja, V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013, 3, 546–557. [Google Scholar] [CrossRef] [PubMed]
- TenOever, B.R. The Evolution of Antiviral Defense Systems. Cell Host Microbe 2016, 19, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Halanych, K.M. Lophotrochozoa, Diversification of. In Encyclopedia of Evolutionary Biology; Kliman, R.M., Ed.; Academic Press: London, UK, 2016; pp. 405–408. [Google Scholar]
- Salzet, M.; Tasiemski, A.; Cooper, E. Innate immunity in lophotrochozoans: The annelids. Curr. Pharm. Des. 2006, 12, 3043–3050. [Google Scholar] [CrossRef]
- Zhang, L.; Li, L.; Zhu, Y.; Zhang, G.; Guo, X. Transcriptome analysis reveals a rich gene set related to innate immunity in the Eastern oyster (Crassostrea virginica). Mar. Biotechnol. 2014, 16, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; He, Y.; Zhang, L.; Lelong, C.; Jouaux, A. Immune and stress responses in oysters with insights on adaptation. Fish Shellfish Immunol. 2015, 46, 107–119. [Google Scholar] [CrossRef]
- Wang, L.; Song, X.; Song, L. The oyster immunity. Dev. Comp. Immunol. 2018, 80, 99–118. [Google Scholar] [CrossRef]
- Rast, J.P.; Smith, L.C.; Loza-Coll, M.; Hibino, T.; Litman, G.W. Genomic insights into the immune system of the sea urchin. Science 2006, 314, 952–956. [Google Scholar] [CrossRef]
- Huang, S.; Yuan, S.; Guo, L.; Yu, Y.; Li, J.; Wu, T.; Liu, T.; Yang, M.; Wu, K.; Liu, H.; et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008, 18, 1112–1126. [Google Scholar] [CrossRef]
- Zhang, L.; Li, L.; Guo, X.; Litman, G.W.; Dishaw, L.J.; Zhang, G. Massive expansion and functional divergence of innate immune genes in a protostome. Sci. Rep. 2015, 5, 8693. [Google Scholar] [CrossRef]
- Hoffmann, J.A., Jr.; Kafatos, F.C.; Janeway, C.A.; Ezekowitz, R.A. Phylogenetic perspectives in innate immunity. Science 1999, 284, 1313–1318. [Google Scholar] [CrossRef]
- Rosenstiel, P.; Philipp, E.E.; Schreiber, S.; Bosch, T.C. Evolution and function of innate immune receptors--nsights from marine invertebrates. J. Innate Immun. 2009, 1, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef]
- Chan, J.; Wang, L.; Li, L.; Mu, K.; Bushek, D.; Xu, Y.; Guo, X.; Zhang, G.; Zhang, L. Transcriptomic Response to Perkinsus marinus in Two Crassostrea Oysters Reveals Evolutionary Dynamics of Host-Parasite Interactions. Front. Genet. 2021, 12, 795706. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Gale, M., Jr. Immune signaling by RIG-I-like receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Fujita, T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009, 227, 54–65. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef]
- Sarkar, D.; Desalle, R.; Fisher, P.B. Evolution of MDA-5/RIG-I-dependent innate immunity: Independent evolution by domain grafting. Proc. Natl. Acad. Sci. USA 2008, 105, 17040–17045. [Google Scholar] [CrossRef]
- Gerdol, M.; Luo, Y.J.; Satoh, N.; Pallavicini, A. Genetic and molecular basis of the immune system in the brachiopod Lingula anatina. Dev. Comp. Immunol. 2018, 82, 7–30. [Google Scholar] [CrossRef]
- Lewandowska, M.; Sharoni, T.; Admoni, Y.; Aharoni, R.; Moran, Y. Functional Characterization of the Cnidarian Antiviral Immune Response Reveals Ancestral Complexity. Mol. Biol. Evol. 2021, 38, 4546–4561. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M., Jr.; Akira, S.; et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Uematsu, S.; Matsui, K.; Tsujimura, T.; Takeda, K.; Fujita, T.; Takeuchi, O.; et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 2005, 23, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Gitlin, L.; Barchet, W.; Gilfillan, S.; Cella, M.; Beutler, B.; Flavell, R.A.; Diamond, M.S.; Colonna, M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 2006, 103, 8459–8464. [Google Scholar] [CrossRef]
- Kang, D.C.; Gopalkrishnan, R.V.; Wu, Q.; Jankowsky, E.; Pyle, A.M.; Fisher, P.B. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. USA 2002, 99, 637–642. [Google Scholar] [CrossRef]
- Su, Z.Z.; Sarkar, D.; Emdad, L.; Barral, P.M.; Fisher, P.B. Central role of interferon regulatory factor-1 (IRF-1) in controlling retinoic acid inducible gene-I (RIG-I) expression. J. Cell. Physiol. 2007, 213, 502–510. [Google Scholar] [CrossRef]
- Saito, T.; Hirai, R.; Loo, Y.M.; Owen, D.; Johnson, C.L.; Sinha, S.C.; Akira, S.; Fujita, T.; Gale, M., Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 2007, 104, 582–587. [Google Scholar] [CrossRef]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef]
- Meylan, E.; Curran, J.; Hofmann, K.; Moradpour, D.; Binder, M.; Bartenschlager, R.; Tschopp, J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 2005, 437, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.B.; Sun, L.; Ea, C.K.; Chen, Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 2005, 122, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Yanai, H.; Negishi, H.; Asagiri, M.; Sato, M.; Mizutani, T.; Shimada, N.; Ohba, Y.; Takaoka, A.; Yoshida, N.; et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 2005, 434, 772–777. [Google Scholar] [CrossRef]
- Huang, B.; Zhang, L.; Du, Y.; Xu, F.; Li, L.; Zhang, G. Characterization of the Mollusc RIG-I/MAVS Pathway Reveals an Archaic Antiviral Signalling Framework in Invertebrates. Sci. Rep. 2017, 7, 8217. [Google Scholar] [CrossRef] [PubMed]
- Bamming, D.; Horvath, C.M. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 2009, 284, 9700–9712. [Google Scholar] [CrossRef]
- Satoh, T.; Kato, H.; Kumagai, Y.; Yoneyama, M.; Sato, S.; Matsushita, K.; Tsujimura, T.; Fujita, T.; Akira, S.; Takeuchi, O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. USA 2010, 107, 1512–1517. [Google Scholar] [CrossRef]
- Venkataraman, T.; Valdes, M.; Elsby, R.; Kakuta, S.; Caceres, G.; Saijo, S.; Iwakura, Y.; Barber, G.N. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 2007, 178, 6444–6455. [Google Scholar] [CrossRef]
- Li, X.; Ranjith-Kumar, C.T.; Brooks, M.T.; Dharmaiah, S.; Herr, A.B.; Kao, C.; Li, P. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 2009, 284, 13881–13891. [Google Scholar] [CrossRef]
- Zou, J.; Chang, M.; Nie, P.; Secombes, C.J. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 2009, 9, 85. [Google Scholar] [CrossRef]
- Mukherjee, K.; Korithoski, B.; Kolaczkowski, B. Ancient origins of vertebrate-specific innate antiviral immunity. Mol. Biol. Evol. 2014, 31, 140–153. [Google Scholar] [CrossRef]
- Majzoub, K.; Wrensch, F.; Baumert, T.F. The Innate Antiviral Response in Animals: An Evolutionary Perspective from Flagellates to Humans. Viruses 2019, 11, 758. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Huang, Z.Y.; Cao, C.C.; Chen, C.S.; Chen, Y.X.; Fan, D.D.; He, J.; Hou, H.L.; Hu, L.; Hu, X.T.; et al. Genome of the Chinese tree shrew. Nat. Commun. 2013, 4, 1426. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yu, D.; Fan, Y.; Peng, L.; Wu, Y.; Yao, Y.G. Loss of RIG-I leads to a functional replacement with MDA5 in the Chinese tree shrew. Proc. Natl. Acad. Sci. USA 2016, 113, 10950–10955. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Satta, Y. Functional Evolution of Avian RIG-I-Like Receptors. Genes 2018, 9, 456. [Google Scholar] [CrossRef]
- Chen, J.; Shang, S.; Wu, X.; Zhong, H.; Zhao, C.; Wei, Q.; Zhang, H.; Xia, T.; Chen, Y.; Zhang, H.; et al. Genomic analysis and adaptive evolution of the RIG-I-like and NOD-like receptors in reptiles. Int. J. Biol. Macromol. 2019, 134, 1045–1051. [Google Scholar] [CrossRef]
- Liu, M.; Grigoriev, A. Protein domains correlate strongly with exons in multiple eukaryotic genomes--vidence of exon shuffling? Trends Genet. TIG 2004, 20, 399–403. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Lu, C.; Ranjith-Kumar, C.T.; Hao, L.; Kao, C.C.; Li, P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2010, 39, 1565–1575. [Google Scholar] [CrossRef]
- Xu, L.G.; Wang, Y.Y.; Han, K.J.; Li, L.Y.; Zhai, Z.; Shu, H.B. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 2005, 19, 727–740. [Google Scholar] [CrossRef]
- Wajant, H. Death receptors. Essays Biochem. 2003, 39, 53–71. [Google Scholar]
- Bhardwaj, A.; Aggarwal, B.B. Receptor-mediated choreography of life and death. J. Clin. Immunol. 2003, 23, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Widmann, C. Antiapoptotic signaling generated by caspase-induced cleavage of RasGAP. Mol. Cell. Biol. 2001, 21, 5346–5358. [Google Scholar] [CrossRef] [PubMed]
- Salvesen, G.S.; Abrams, J.M. Caspase activation—stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene 2004, 23, 2774–2784. [Google Scholar] [CrossRef] [PubMed]
- Jiménez Fernández, D.; Lamkanfi, M. Inflammatory caspases: Key regulators of inflammation and cell death. Biol. Chem. 2015, 396, 193–203. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef]
- Kofoed, E.M.; Vance, R.E. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 2011, 477, 592–595. [Google Scholar] [CrossRef]
- Van Lookeren Campagne, M.; Verschoor, A. Pathogen clearance and immune adherence “revisited”: Immuno-regulatory roles for CRIg. Semin. Immunol. 2018, 37, 4–11. [Google Scholar] [CrossRef]
- Barclay, A.N. Membrane proteins with immunoglobulin-like domains--a master superfamily of interaction molecules. Semin. Immunol. 2003, 15, 215–223. [Google Scholar] [CrossRef]
- Zhang, S.M.; Adema, C.M.; Kepler, T.B.; Loker, E.S. Diversification of Ig superfamily genes in an invertebrate. Science 2004, 305, 251–254. [Google Scholar] [CrossRef]
- Zhang, Q.; Zmasek, C.M.; Godzik, A. Domain architecture evolution of pattern-recognition receptors. Immunogenetics 2010, 62, 263–272. [Google Scholar] [CrossRef]
- Wu, B.; Huan, T.; Gong, J.; Zhou, P.; Bai, Z. Domain combination of the vertebrate-like TLR gene family: Implications for their origin and evolution. J. Genet. 2011, 90, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Rajalingam, R.; Parham, P.; Abi-Rached, L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J. Immunol. 2004, 172, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Freitas, E.M.; Witt, C.S.; Christiansen, F.T. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics 2000, 51, 268–280. [Google Scholar] [CrossRef]
- Hamada, M.; Shoguchi, E.; Shinzato, C.; Kawashima, T.; Miller, D.J.; Satoh, N. The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Mol. Biol. Evol. 2013, 30, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Gao, B.; Zhu, S. Characterization of a chimeric antimicrobial peptide uncovers evolutionary significance of exon-shuffling. Biochem. Biophys. Res. Commun. 2012, 428, 360–364. [Google Scholar] [CrossRef]
- Ranji, A.; Boris-Lawrie, K. RNA helicases: Emerging roles in viral replication and the host innate response. RNA Biol. 2010, 7, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Zambounis, A.; Elias, M.; Sterck, L.; Maumus, F.; Gachon, C.M. Highly dynamic exon shuffling in candidate pathogen receptors… what if brown algae were capable of adaptive immunity? Mol. Biol. Evol. 2012, 29, 1263–1276. [Google Scholar] [CrossRef]
- Kambris, Z.; Hoffmann, J.A.; Imler, J.L.; Capovilla, M. Tissue and stage-specific expression of the Tolls in Drosophila embryos. Gene Expr. Patterns GEP 2002, 2, 311–317. [Google Scholar] [CrossRef]
- Leulier, F.; Lemaitre, B. Toll-like receptors--taking an evolutionary approach. Nat. Rev. Genet. 2008, 9, 165–178. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Langmead BJGb: Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, S.; Chan, J.; Xu, Y.; Wu, S.; Zhang, L. Divergences of the RLR Gene Families across Lophotrochozoans: Domain Grafting, Exon–Intron Structure, Expression, and Positive Selection. Int. J. Mol. Sci. 2022, 23, 3415. https://doi.org/10.3390/ijms23073415
Yao S, Chan J, Xu Y, Wu S, Zhang L. Divergences of the RLR Gene Families across Lophotrochozoans: Domain Grafting, Exon–Intron Structure, Expression, and Positive Selection. International Journal of Molecular Sciences. 2022; 23(7):3415. https://doi.org/10.3390/ijms23073415
Chicago/Turabian StyleYao, Shanshan, Jiulin Chan, Yue Xu, Shimei Wu, and Linlin Zhang. 2022. "Divergences of the RLR Gene Families across Lophotrochozoans: Domain Grafting, Exon–Intron Structure, Expression, and Positive Selection" International Journal of Molecular Sciences 23, no. 7: 3415. https://doi.org/10.3390/ijms23073415
APA StyleYao, S., Chan, J., Xu, Y., Wu, S., & Zhang, L. (2022). Divergences of the RLR Gene Families across Lophotrochozoans: Domain Grafting, Exon–Intron Structure, Expression, and Positive Selection. International Journal of Molecular Sciences, 23(7), 3415. https://doi.org/10.3390/ijms23073415






