The Chromatin Architectural Protein CTCF Is Critical for Cell Survival upon Irradiation-Induced DNA Damage
Abstract
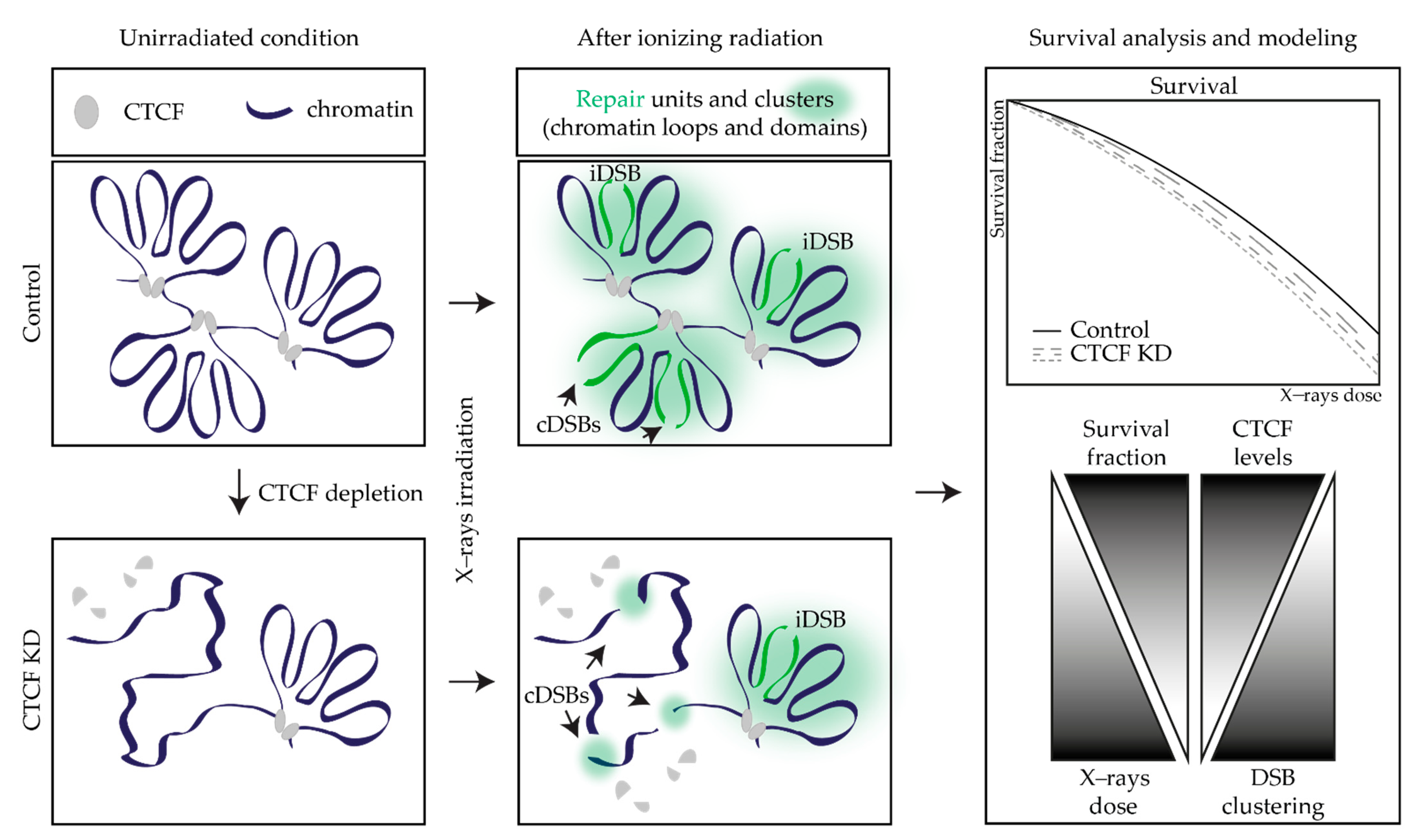
1. Introduction
2. Results
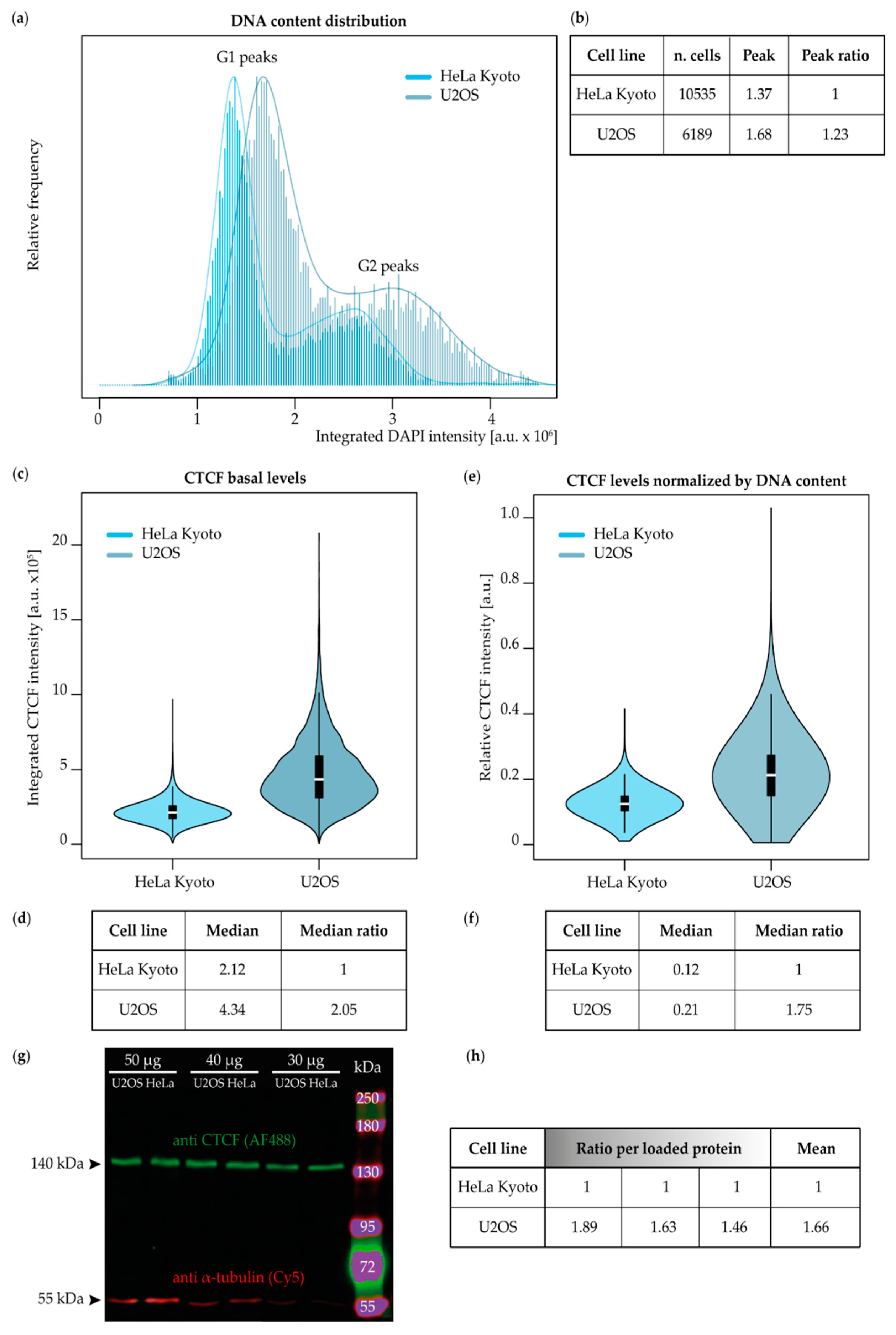
2.1. Validation of the Genome Size, CTCF Basal Levels, and Binding in Different Cancer Cell Lines
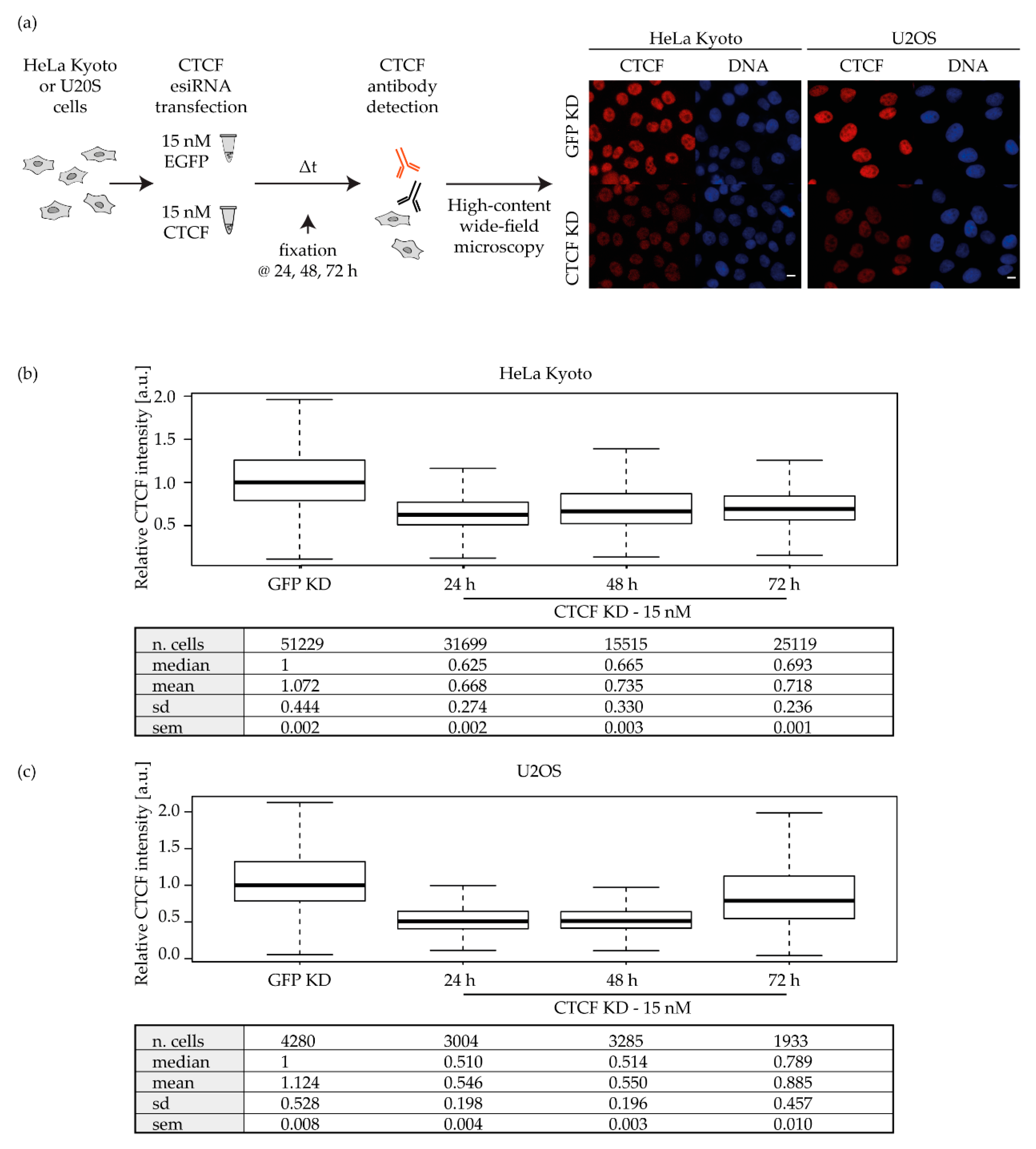
2.2. CTCF Knockdown and Validation in Different Cancer Cell Lines
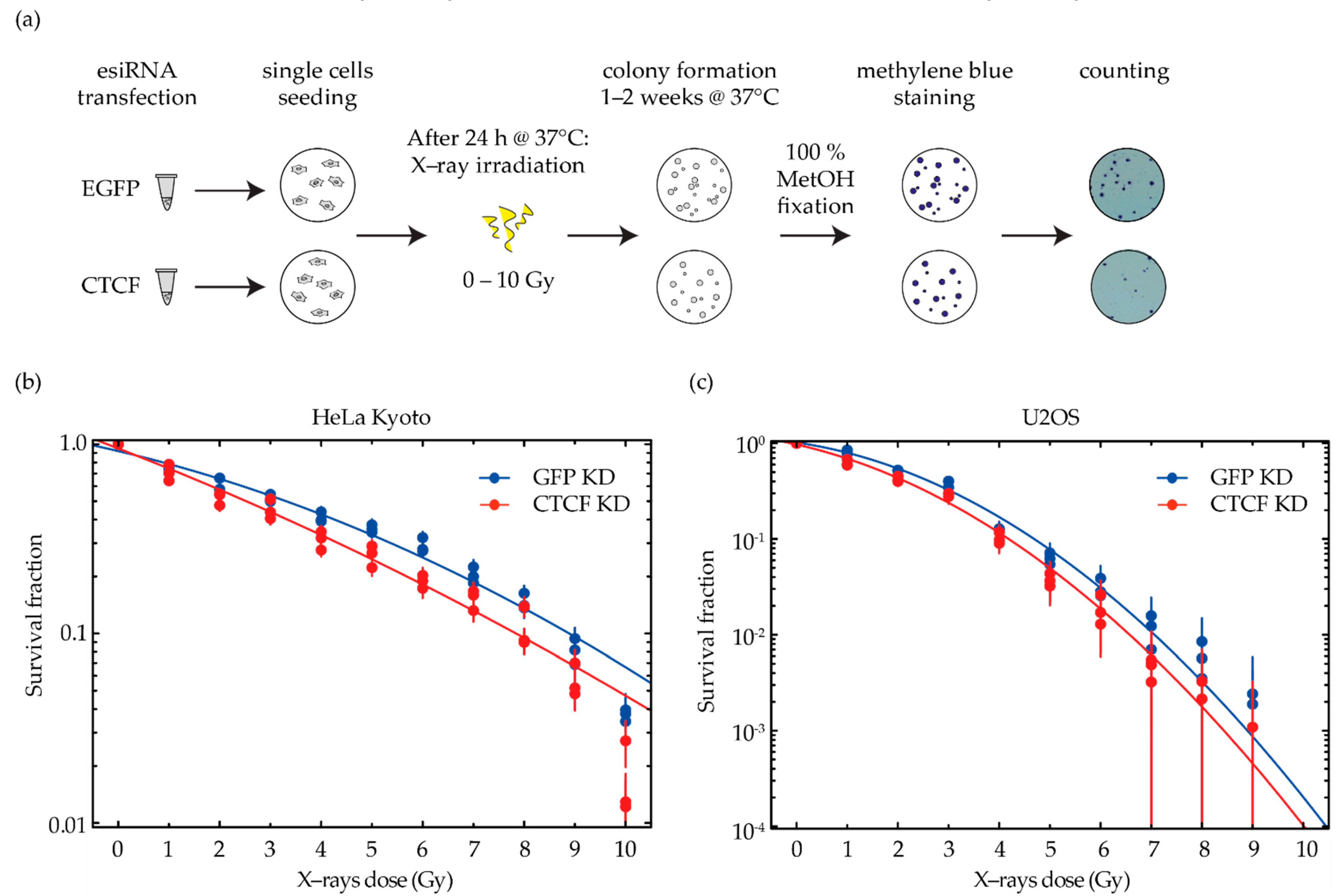
2.3. CTCF Depletion Increases the Radiosensitivity of Cancer Cells in a Cell Line-Dependent Way
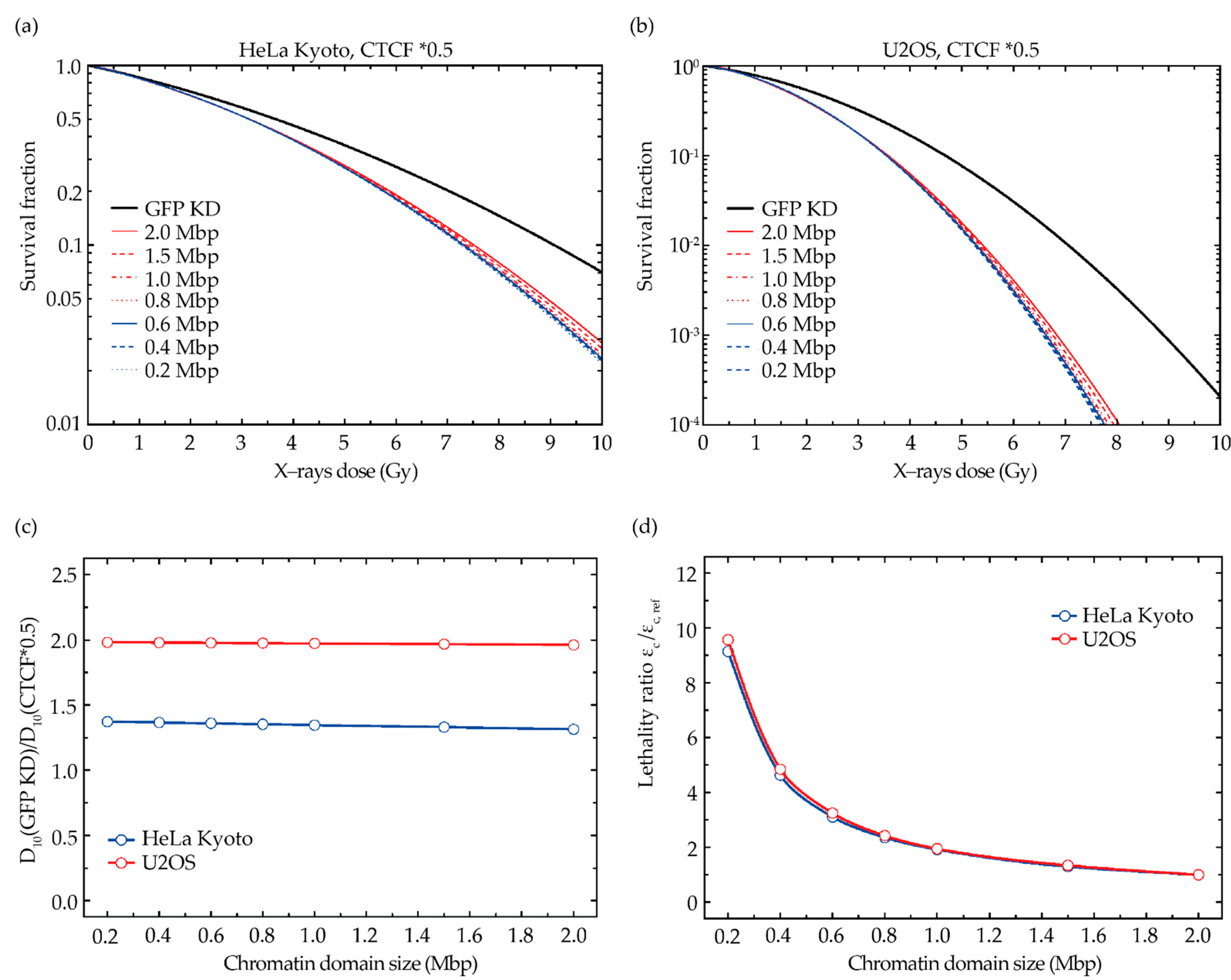
2.4. GLOBLE Model Investigates the Impact of DSB Clustering in Chromatin Domains on Cell Kill
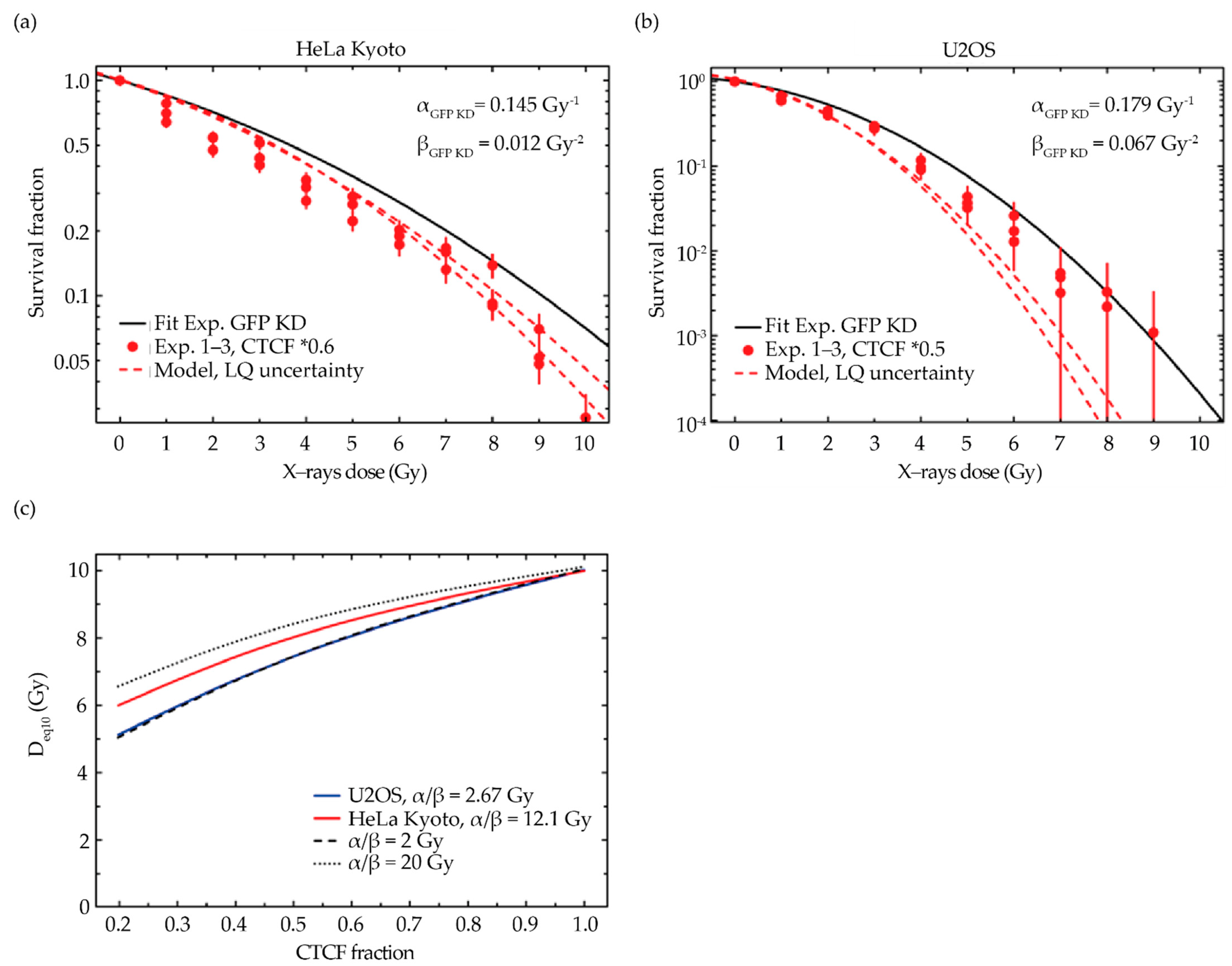
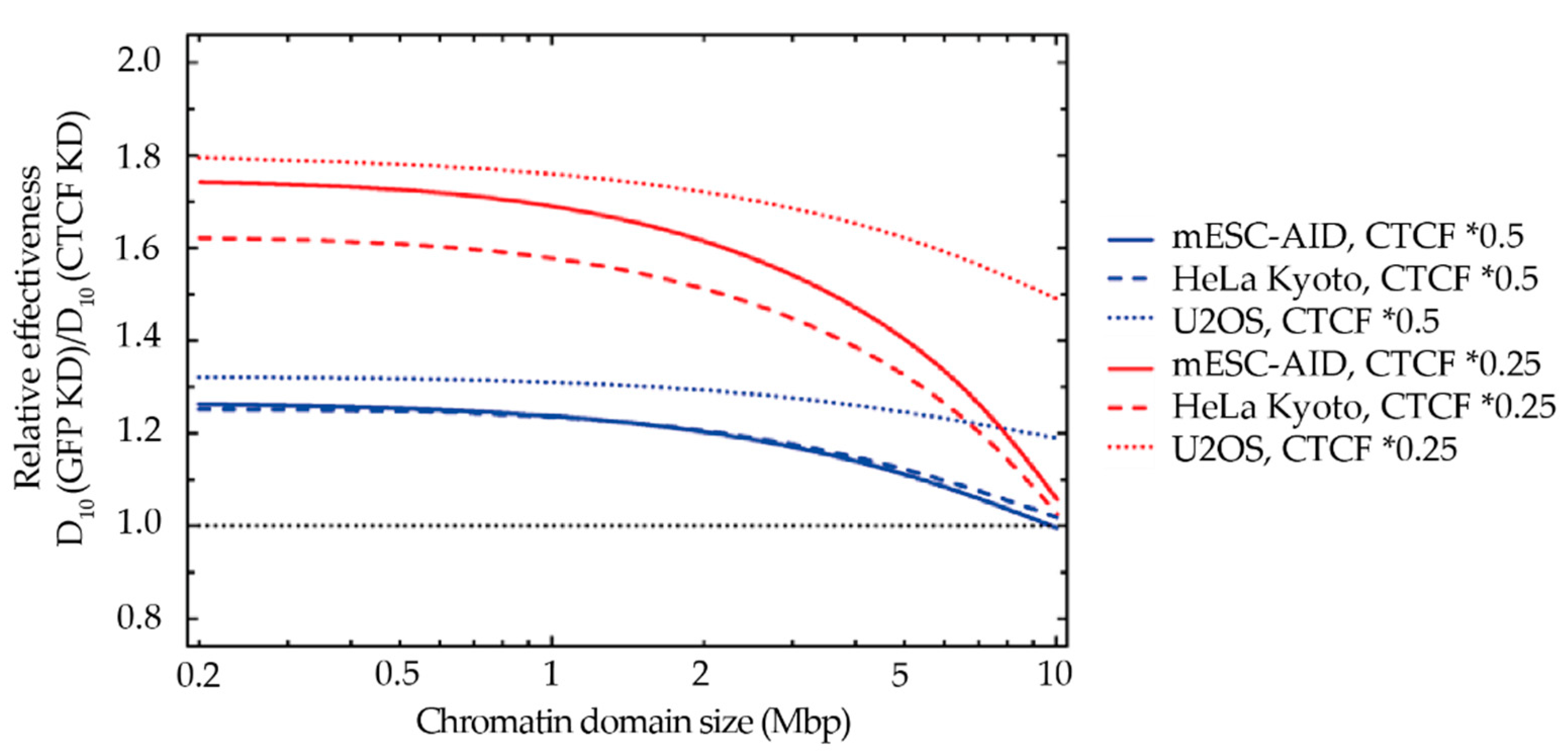
2.5. Comparison of Model Predictions with Experimental Data
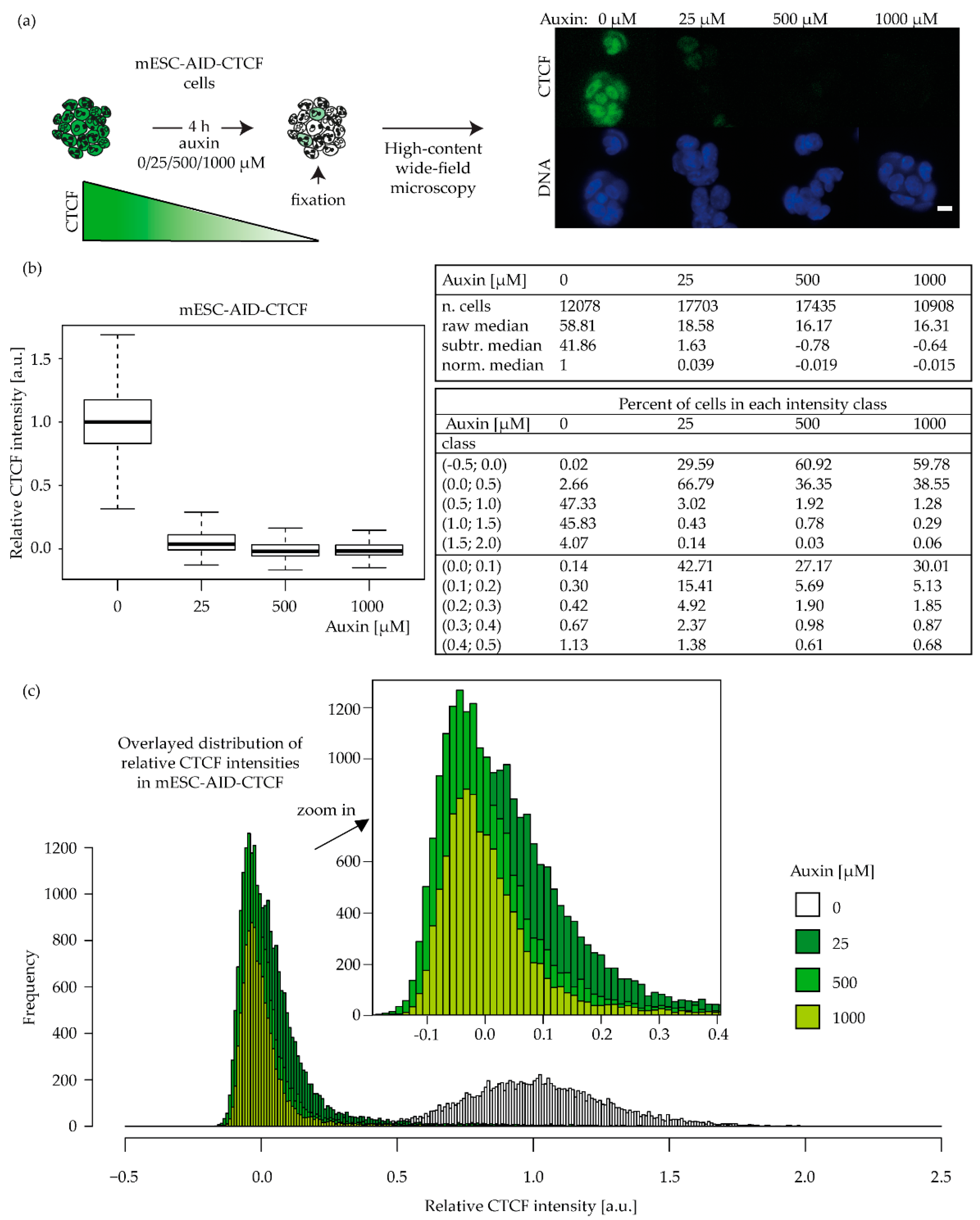
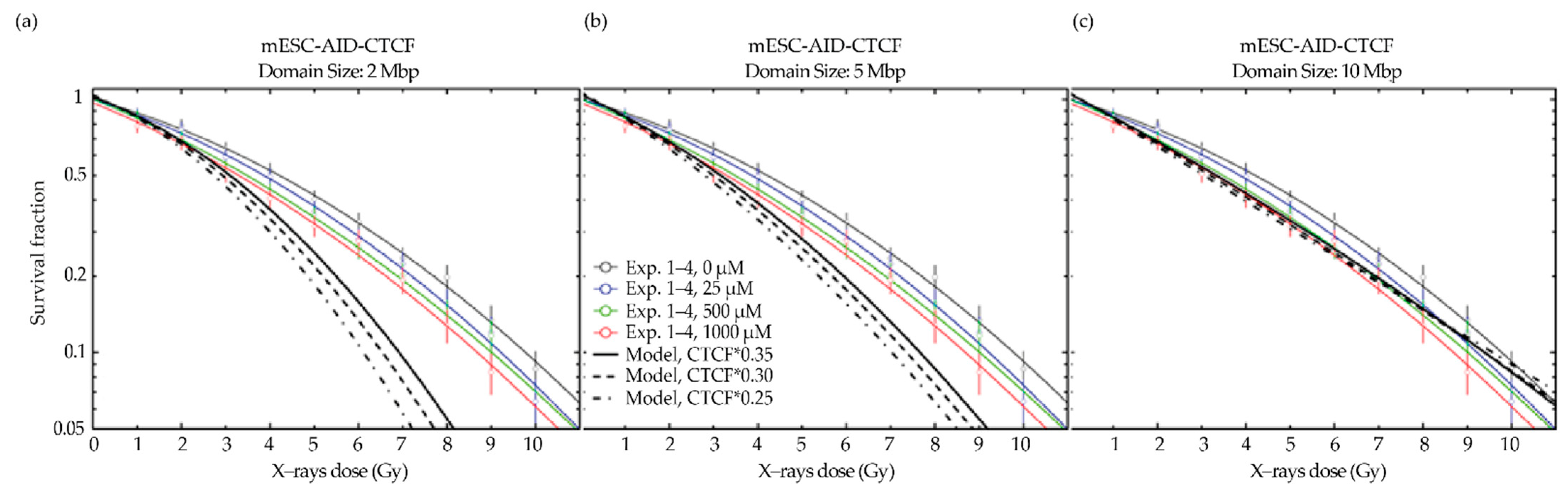
2.6. A CTCF Effect on Radiosensitivity Is also Observed in Mouse ES Cells and Is CTCF Dose Dependent
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. CTCF Knockdown
4.3. CTCF Immunostaining
4.4. CTCF Western Blot
4.5. Microscopy Imaging and Quantification
4.6. Clonogenic Assay
4.7. Modeling of Cell Survival Using the Giant-Loop Binary Lesion (GLOBLE) Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lobanenkov, V.V.; Nicolas, R.H.; Adler, V.V.; Paterson, H.; Klenova, E.M.; Polotskaja, A.V.; Goodwin, G.H. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 1990, 5, 1743–1753. [Google Scholar]
- Klenova, E.M.; Nicolas, R.H.; Paterson, H.F.; Carne, A.F.; Heath, C.M.; Goodwin, G.H.; Neiman, P.E.; Lobanenkov, V.V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 1993, 13, 7612–7624. [Google Scholar] [CrossRef]
- Filippova, G.N.; Fagerlie, S.; Klenova, E.M.; Myers, C.; Dehner, Y.; Goodwin, G.; Neiman, P.E.; Collins, S.J.; Lobanenkov, V.V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996, 16, 2802–2813. [Google Scholar] [CrossRef]
- Phillips, J.E.; Corces, V.G. CTCF: Master weaver of the genome. Cell 2009, 137, 1194–1211. [Google Scholar] [CrossRef] [PubMed]
- Merkenschlager, M.; Nora, E.P. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genom. Hum. Genet. 2016, 17, 17–43. [Google Scholar] [CrossRef] [PubMed]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Barutcu, A.R.; Fritz, A.J.; Zaidi, S.K.; van Wijnen, A.J.; Lian, J.B.; Stein, J.L.; Nickerson, J.A.; Imbalzano, A.N.; Stein, G.S. C-ing the Genome: A Compendium of Chromosome Conformation Capture Methods to Study Higher-Order Chromatin Organization. J. Cell. Physiol. 2016, 231, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- Splinter, E.; Heath, H.; Kooren, J.; Palstra, R.-J.; Klous, P.; Grosveld, F.; Galjart, N.; de Laat, W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006, 20, 2349–2354. [Google Scholar] [CrossRef] [PubMed]
- Heath, H.; Ribeiro de Almeida, C.; Sleutels, F.; Dingjan, G.; van de Nobelen, S.; Jonkers, I.; Ling, K.-W.; Gribnau, J.; Renkawitz, R.; Grosveld, F.; et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008, 27, 2839–2850. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.M.; Rabaia, N.A.; Smith, L.E.; Fagerlie, S.; Gurley, K.; Loukinov, D.; Disteche, C.M.; Collins, S.J.; Kemp, C.J.; Lobanenkov, V.V.; et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS ONE 2012, 7, e34915. [Google Scholar] [CrossRef] [PubMed]
- Zuin, J.; Dixon, J.R.; van der Reijden, M.I.J.A.; Ye, Z.; Kolovos, P.; Brouwer, R.W.W.; van de Corput, M.P.C.; van de Werken, H.J.G.; Knoch, T.A.; van IJcken, W.F.J.; et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA 2014, 111, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, W.; Abdennur, N.; Goloborodko, A.; Pekowska, A.; Fudenberg, G.; Loe-Mie, Y.; Fonseca, N.A.; Huber, W.; Haering, C.H.; Mirny, L.; et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551, 51–56. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huang, S.-C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.-R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin loss eliminates all loop domains. Cell 2017, 171, 305–320. [Google Scholar] [CrossRef]
- Wutz, G.; Várnai, C.; Nagasaka, K.; Cisneros, D.A.; Stocsits, R.R.; Tang, W.; Schoenfelder, S.; Jessberger, G.; Muhar, M.; Hossain, M.J.; et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017, 36, 3573–3599. [Google Scholar] [CrossRef]
- Bintu, B.; Mateo, L.J.; Su, J.-H.; Sinnott-Armstrong, N.A.; Parker, M.; Kinrot, S.; Yamaya, K.; Boettiger, A.N.; Zhuang, X. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 2018, 362. [Google Scholar] [CrossRef]
- Cremer, M.; Brandstetter, K.; Maiser, A.; Rao, S.S.P.; Schmid, V.J.; Guirao-Ortiz, M.; Mitra, N.; Mamberti, S.; Klein, K.N.; Gilbert, D.M.; et al. Cohesin depleted cells rebuild functional nuclear compartments after endomitosis. Nat. Commun. 2020, 11, 6146. [Google Scholar] [CrossRef]
- Nishimura, K.; Fukagawa, T.; Takisawa, H.; Kakimoto, T.; Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 2009, 6, 917–922. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.-L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Nuebler, J.; Fudenberg, G.; Imakaev, M.; Abdennur, N.; Mirny, L.A. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl. Acad. Sci. USA 2018, 115, E6697–E6706. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, H.; Reisser, M.; Wortmann, C.; Gebhardt, J.C.M. Direct Observation of Cell-Cycle-Dependent Interactions between CTCF and Chromatin. Biophys. J. 2017, 112, 2051–2055. [Google Scholar] [CrossRef]
- Hansen, A.S.; Cattoglio, C.; Darzacq, X.; Tjian, R. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus 2018, 9, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Mamberti, S.; Cardoso, M.C. Are the processes of DNA replication and DNA repair reading a common structural chromatin unit? Nucleus 2020, 11, 66–82. [Google Scholar] [CrossRef] [PubMed]
- Borrego-Soto, G.; Ortiz-López, R.; Rojas-Martínez, A. Ionizing radiation-induced DNA injury and damage detection in patients with breast cancer. Genet. Mol. Biol. 2015, 38, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Santivasi, W.L.; Xia, F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid. Redox Signal. 2014, 21, 251–259. [Google Scholar] [CrossRef]
- Huang, R.-X.; Zhou, P.-K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Vignard, J.; Mirey, G.; Salles, B. Ionizing-radiation induced DNA double-strand breaks: A direct and indirect lighting up. Radiother. Oncol. 2013, 108, 362–369. [Google Scholar] [CrossRef]
- Schipler, A.; Iliakis, G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef]
- Natale, F.; Rapp, A.; Yu, W.; Maiser, A.; Harz, H.; Scholl, A.; Grulich, S.; Anton, T.; Hörl, D.; Chen, W.; et al. Identification of the elementary structural units of the DNA damage response. Nat. Commun. 2017, 8, 15760. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999, 146, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Capetillo, O.; Chen, H.-T.; Celeste, A.; Ward, I.; Romanienko, P.J.; Morales, J.C.; Naka, K.; Xia, Z.; Camerini-Otero, R.D.; Motoyama, N.; et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 2002, 4, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Lees-Miller, S.P.; Sakaguchi, K.; Ullrich, S.J.; Appella, E.; Anderson, C.W. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol. Cell. Biol. 1992, 12, 5041–5049. [Google Scholar] [CrossRef]
- Karlsson, K.H.; Stenerlöw, B. Focus formation of DNA repair proteins in normal and repair-deficient cells irradiated with high-LET ions. Radiat. Res. 2004, 161, 517–527. [Google Scholar] [CrossRef]
- Leatherbarrow, E.L.; Harper, J.V.; Cucinotta, F.A.; O’Neill, P. Induction and quantification of gamma-H2AX foci following low and high LET-irradiation. Int. J. Radiat. Biol. 2006, 82, 111–118. [Google Scholar] [CrossRef]
- Bakkenist, C.J.; Kastan, M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003, 421, 499–506. [Google Scholar] [CrossRef]
- Uziel, T.; Lerenthal, Y.; Moyal, L.; Andegeko, Y.; Mittelman, L.; Shiloh, Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003, 22, 5612–5621. [Google Scholar] [CrossRef]
- Lee, J.-H.; Paull, T.T. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 2005, 308, 551–554. [Google Scholar] [CrossRef]
- Singleton, B.K.; Torres-Arzayus, M.I.; Rottinghaus, S.T.; Taccioli, G.E.; Jeggo, P.A. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol. 1999, 19, 3267–3277. [Google Scholar] [CrossRef] [PubMed]
- Sibanda, B.L.; Chirgadze, D.Y.; Ascher, D.B.; Blundell, T.L. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 2017, 355, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef]
- Wang, X.; Ran, T.; Zhang, X.; Xin, J.; Zhang, Z.; Wu, T.; Wang, W.; Cai, G. 3.9 Å structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP. Science 2017, 358, 1206–1209. [Google Scholar] [CrossRef]
- Paull, T.T. Mechanisms of ATM activation. Annu. Rev. Biochem. 2015, 84, 711–738. [Google Scholar] [CrossRef]
- Menolfi, D.; Zha, S. ATM, ATR and DNA-PKcs kinases-the lessons from the mouse models: Inhibition ≠ deletion. Cell Biosci. 2020, 10, 8. [Google Scholar] [CrossRef]
- Paull, T.T.; Rogakou, E.P.; Yamazaki, V.; Kirchgessner, C.U.; Gellert, M.; Bonner, W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000, 10, 886–895. [Google Scholar] [CrossRef]
- Redon, C.; Pilch, D.; Rogakou, E.; Sedelnikova, O.; Newrock, K.; Bonner, W. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 2002, 12, 162–169. [Google Scholar] [CrossRef]
- Bhogal, N.; Jalali, F.; Bristow, R.G. Microscopic imaging of DNA repair foci in irradiated normal tissues. Int. J. Radiat. Biol. 2009, 85, 732–746. [Google Scholar] [CrossRef]
- Schultz, L.B.; Chehab, N.H.; Malikzay, A.; Halazonetis, T.D. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000, 151, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L.; Beishline, K.; Flashner, S.; Azizkhan-Clifford, J. DSB repair pathway choice is regulated by recruitment of 53BP1 through cell cycle-dependent regulation of Sp1. Cell Rep. 2021, 34, 108840. [Google Scholar] [CrossRef] [PubMed]
- Hilmi, K.; Jangal, M.; Marques, M.; Zhao, T.; Saad, A.; Zhang, C.; Luo, V.M.; Syme, A.; Rejon, C.; Yu, Z.; et al. CTCF facilitates DNA double-strand break repair by enhancing homologous recombination repair. Sci. Adv. 2017, 3, e1601898. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, T.; Durante, M.; Scholz, M. Modeling cell survival after photon irradiation based on double-strand break clustering in megabase pair chromatin loops. Radiat. Res. 2012, 178, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, T.; Scholz, U.; Elsässer, T.; Durante, M.; Scholz, M. Systematic analysis of RBE and related quantities using a database of cell survival experiments with ion beam irradiation. J. Radiat. Res. 2013, 54, 494–514. [Google Scholar] [CrossRef]
- Herr, L.; Friedrich, T.; Durante, M.; Scholz, M. A model of photon cell killing based on the spatio-temporal clustering of DNA damage in higher order chromatin structures. PLoS ONE 2014, 9, e83923. [Google Scholar] [CrossRef]
- Erfle, H.; Neumann, B.; Liebel, U.; Rogers, P.; Held, M.; Walter, T.; Ellenberg, J.; Pepperkok, R. Reverse transfection on cell arrays for high content screening microscopy. Nat. Protoc. 2007, 2, 392–399. [Google Scholar] [CrossRef]
- Pontén, J.; Saksela, E. Two established in vitro cell lines from human mesenchymal tumours. Int. J. Cancer 1967, 2, 434–447. [Google Scholar] [CrossRef]
- Chagin, V.O.; Casas-Delucchi, C.S.; Reinhart, M.; Schermelleh, L.; Markaki, Y.; Maiser, A.; Bolius, J.J.; Bensimon, A.; Fillies, M.; Domaing, P.; et al. 4D Visualization of replication foci in mammalian cells corresponding to individual replicons. Nat. Commun. 2016, 7, 11231. [Google Scholar] [CrossRef]
- DepMap Data Explorer. Available online: https://depmap.org/portal/interactive/?yDataset=&yFeature=&x=slice%2Fcopy_number_absolute%2F5524%2Fentity_id (accessed on 18 February 2022).
- Subcellular-CTCF-The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000102974-CTCF/subcellular (accessed on 18 February 2022).
- Guénet, J.L. The mouse genome. Genome Res. 2005, 15, 1729–1740. [Google Scholar] [CrossRef] [PubMed]
- Hufnagl, A.; Herr, L.; Friedrich, T.; Durante, M.; Taucher-Scholz, G.; Scholz, M. The link between cell-cycle dependent radiosensitivity and repair pathways: A model based on the local, sister-chromatid conformation dependent switch between NHEJ and HR. DNA Repair 2015, 27, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Hooper, M.; Hardy, K.; Handyside, A.; Hunter, S.; Monk, M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 1987, 326, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Pękowska, A.; Klaus, B.; Xiang, W.; Severino, J.; Daigle, N.; Klein, F.A.; Oleś, M.; Casellas, R.; Ellenberg, J.; Steinmetz, L.M.; et al. Gain of CTCF-Anchored Chromatin Loops Marks the Exit from Naive Pluripotency. Cell Syst. 2018, 7, 482–495. [Google Scholar] [CrossRef]
- Johnston, P.J.; Olive, P.L.; Bryant, P.E. Higher-order chromatin structure-dependent repair of DNA double-strand breaks: Modeling the elution of DNA from nucleoids. Radiat. Res. 1997, 148, 561–567. [Google Scholar] [CrossRef]
- Tommasino, F.; Friedrich, T.; Scholz, U.; Taucher-Scholz, G.; Durante, M.; Scholz, M. A DNA double-strand break kinetic rejoining model based on the local effect model. Radiat. Res. 2013, 180, 524–538. [Google Scholar] [CrossRef]
- Tommasino, F.; Friedrich, T.; Scholz, U.; Taucher-Scholz, G.; Durante, M.; Scholz, M. Application of the local effect model to predict DNA double-strand break rejoining after photon and high-LET irradiation. Radiat. Prot. Dosim. 2015, 166, 66–70. [Google Scholar] [CrossRef]
- Tommasino, F.; Friedrich, T.; Jakob, B.; Meyer, B.; Durante, M.; Scholz, M. Induction and Processing of the Radiation-Induced Gamma-H2AX Signal and Its Link to the Underlying Pattern of DSB: A Combined Experimental and Modelling Study. PLoS ONE 2015, 10, e0129416. [Google Scholar] [CrossRef][Green Version]
- Ricci, M.A.; Manzo, C.; García-Parajo, M.F.; Lakadamyali, M.; Cosma, M.P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 2015, 160, 1145–1158. [Google Scholar] [CrossRef]
- Efroni, S.; Duttagupta, R.; Cheng, J.; Dehghani, H.; Hoeppner, D.J.; Dash, C.; Bazett-Jones, D.P.; Le Grice, S.; McKay, R.D.G.; Buetow, K.H.; et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2008, 2, 437–447. [Google Scholar] [CrossRef]
- Marks, H.; Kalkan, T.; Menafra, R.; Denissov, S.; Jones, K.; Hofemeister, H.; Nichols, J.; Kranz, A.; Stewart, A.F.; Smith, A.; et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 2012, 149, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Banáth, J.P.; Bañuelos, C.A.; Klokov, D.; MacPhail, S.M.; Lansdorp, P.M.; Olive, P.L. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp. Cell Res. 2009, 315, 1505–1520. [Google Scholar] [CrossRef] [PubMed]
- Saretzki, G.; Armstrong, L.; Leake, A.; Lako, M.; von Zglinicki, T. Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells 2004, 22, 962–971. [Google Scholar] [CrossRef] [PubMed]
- Aladjem, M.I.; Spike, B.T.; Rodewald, L.W.; Hope, T.J.; Klemm, M.; Jaenisch, R.; Wahl, G.M. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 1998, 8, 145–155. [Google Scholar] [CrossRef]
- Cervantes, R.B.; Stringer, J.R.; Shao, C.; Tischfield, J.A.; Stambrook, P.J. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc. Natl. Acad. Sci. USA 2002, 99, 3586–3590. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Chao, C.; Saito, S.; Mazur, S.J.; Murphy, M.E.; Appella, E.; Xu, Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 2005, 7, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Tichy, E.D.; Stambrook, P.J. DNA repair in murine embryonic stem cells and differentiated cells. Exp. Cell Res. 2008, 314, 1929–1936. [Google Scholar] [CrossRef]
- Valerie, K.; Povirk, L.F. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 2003, 22, 5792–5812. [Google Scholar] [CrossRef]
- Sedelnikova, O.A.; Pilch, D.R.; Redon, C.; Bonner, W.M. Histone H2AX in DNA damage and repair. Cancer Biol. Ther. 2003, 2, 233–235. [Google Scholar] [CrossRef]
- Karagiannis, T.C.; El-Osta, A. Double-strand breaks: Signaling pathways and repair mechanisms. Cell. Mol. Life Sci. 2004, 61, 2137–2147. [Google Scholar] [CrossRef]
- Takata, M.; Sasaki, M.S.; Sonoda, E.; Morrison, C.; Hashimoto, M.; Utsumi, H.; Yamaguchi-Iwai, Y.; Shinohara, A.; Takeda, S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998, 17, 5497–5508. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zeng, Z.C.; Bui, T.A.; Sonoda, E.; Takata, M.; Takeda, S.; Iliakis, G. Efficient rejoining of radiation-induced DNA double-strand breaks in vertebrate cells deficient in genes of the RAD52 epistasis group. Oncogene 2001, 20, 2212–2224. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rothkamm, K.; Krüger, I.; Thompson, L.H.; Löbrich, M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003, 23, 5706–5715. [Google Scholar] [CrossRef] [PubMed]
- Savatier, P.; Lapillonne, H.; Jirmanova, L.; Vitelli, L.; Samarut, J. Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol. Biol. 2002, 185, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C.; Meira, L.B. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair 2006, 5, 189–209. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-G.; Cortes, U.; Patnaik, S.; Jasin, M.; Wang, Z.-Q. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 2004, 23, 3872–3882. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.; Richardson, C. Multipotent hematopoietic cells susceptible to alternative double-strand break repair pathways that promote genome rearrangements. Genes Dev. 2007, 21, 1064–1074. [Google Scholar] [CrossRef]
- MacPhail, S.H.; Banáth, J.P.; Yu, T.Y.; Chu, E.H.M.; Lambur, H.; Olive, P.L. Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays. Int. J. Radiat. Biol. 2003, 79, 351–358. [Google Scholar] [CrossRef]
- Banáth, J.P.; Macphail, S.H.; Olive, P.L. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004, 64, 7144–7149. [Google Scholar] [CrossRef]
- Wada, S.; Van Khoa, T.; Kobayashi, Y.; Funayama, T.; Ogihara, K.; Ueno, S.; Ito, N. Prediction of cellular radiosensitivity from DNA damage induced by gamma-rays and carbon ion irradiation in canine tumor cells. J. Vet. Med. Sci. 2005, 67, 1089–1095. [Google Scholar] [CrossRef][Green Version]
- Mirzayans, R.; Severin, D.; Murray, D. Relationship between DNA double-strand break rejoining and cell survival after exposure to ionizing radiation in human fibroblast strains with differing ATM/p53 status: Implications for evaluation of clinical radiosensitivity. Int. J. Radiat. Oncol. Biol. Phys. 2006, 66, 1498–1505. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.F. The complexity of DNA damage: Relevance to biological consequences. Int. J. Radiat. Biol. 1994, 66, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Ottolenghi, A.; Merzagora, M.; Tallone, L.; Durante, M.; Paretzke, H.G.; Wilson, W.E. The quality of DNA double-strand breaks: A Monte Carlo simulation of the end-structure of strand breaks produced by protons and alpha particles. Radiat. Environ. Biophys. 1995, 34, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, B.M.; Bennett, P.V.; Sidorkina, O.; Laval, J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. USA 2000, 97, 103–108. [Google Scholar] [CrossRef]
- Nikjoo, H.; O’Neill, P.; Wilson, W.E.; Goodhead, D.T. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res. 2001, 156, 577–583. [Google Scholar] [CrossRef]
- Johnston, P.J.; MacPhail, S.H.; Banáth, J.P.; Olive, P.L. Higher-order chromatin structure-dependent repair of DNA double-strand breaks: Factors affecting elution of DNA from nucleoids. Radiat. Res. 1998, 149, 533–542. [Google Scholar] [CrossRef]
- Johnston, P.J.; MacPhail, S.H.; Stamato, T.D.; Kirchgessner, C.U.; Olive, P.L. Higher-order chromatin structure-dependent repair of DNA double-strand breaks: Involvement of the V(D)J recombination double-strand break repair pathway. Radiat. Res. 1998, 149, 455–462. [Google Scholar] [CrossRef]
- Friedland, W.; Jacob, P.; Paretzke, H.G.; Ottolenghi, A.; Ballarini, F.; Liotta, M. Simulation of light ion induced DNA damage patterns. Radiat. Prot. Dosim. 2006, 122, 116–120. [Google Scholar] [CrossRef]
- Stenerlöw, B.; Karlsson, K.H.; Cooper, B.; Rydberg, B. Measurement of prompt DNA double-strand breaks in mammalian cells without including heat-labile sites: Results for cells deficient in nonhomologous end joining. Radiat. Res. 2003, 159, 502–510. [Google Scholar] [CrossRef]
- Singh, S.K.; Wang, M.; Staudt, C.; Iliakis, G. Post-irradiation chemical processing of DNA damage generates double-strand breaks in cells already engaged in repair. Nucleic Acids Res. 2011, 39, 8416–8429. [Google Scholar] [CrossRef]
- Singh, S.K.; Bencsik-Theilen, A.; Mladenov, E.; Jakob, B.; Taucher-Scholz, G.; Iliakis, G. Reduced contribution of thermally labile sugar lesions to DNA double strand break formation after exposure to heavy ions. Radiat. Oncol. 2013, 8, 77. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Georgakilas, A.G.; O’Neill, P.; Stewart, R.D. Induction and repair of clustered DNA lesions: What do we know so far? Radiat. Res. 2013, 180, 100–109. [Google Scholar] [CrossRef] [PubMed]
- gd Online Documentation. Available online: http://bio.gsi.de/DOCS/gd.html (accessed on 23 February 2022).


| Cell Line | HeLa Kyoto | U2OS | mESC-AID-CTCF |
|---|---|---|---|
| Genome size [Mbp] | 9.7 × 103 | 11.93 × 103 | 5.2× 103 |
| Domain size [Mbp] | 2 | 2 | 2 |
| α [Gy−1] | 0.145± 0.014 | 0.179 ± 0.034 | 0.111 ± 0.025 |
| β [Gy−2] | 0.012± 0.002 | 0.067 ± 0.0067 | 0.013 ± 0.0029 |
| α/β [Gy] | 12.1 | 2.67 | 8.54 |
| εi | 0.0030 | 0.0030 | 0.0043 |
| εc | 0.0555 | 0.2310 | 0.1085 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamberti, S.; Pabba, M.K.; Rapp, A.; Cardoso, M.C.; Scholz, M. The Chromatin Architectural Protein CTCF Is Critical for Cell Survival upon Irradiation-Induced DNA Damage. Int. J. Mol. Sci. 2022, 23, 3896. https://doi.org/10.3390/ijms23073896
Mamberti S, Pabba MK, Rapp A, Cardoso MC, Scholz M. The Chromatin Architectural Protein CTCF Is Critical for Cell Survival upon Irradiation-Induced DNA Damage. International Journal of Molecular Sciences. 2022; 23(7):3896. https://doi.org/10.3390/ijms23073896
Chicago/Turabian StyleMamberti, Stefania, Maruthi K. Pabba, Alexander Rapp, M. Cristina Cardoso, and Michael Scholz. 2022. "The Chromatin Architectural Protein CTCF Is Critical for Cell Survival upon Irradiation-Induced DNA Damage" International Journal of Molecular Sciences 23, no. 7: 3896. https://doi.org/10.3390/ijms23073896
APA StyleMamberti, S., Pabba, M. K., Rapp, A., Cardoso, M. C., & Scholz, M. (2022). The Chromatin Architectural Protein CTCF Is Critical for Cell Survival upon Irradiation-Induced DNA Damage. International Journal of Molecular Sciences, 23(7), 3896. https://doi.org/10.3390/ijms23073896






