Dissection of the Genetic Basis of Yield Traits in Line per se and Testcross Populations and Identification of Candidate Genes for Hybrid Performance in Maize
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Data of the Three Populations
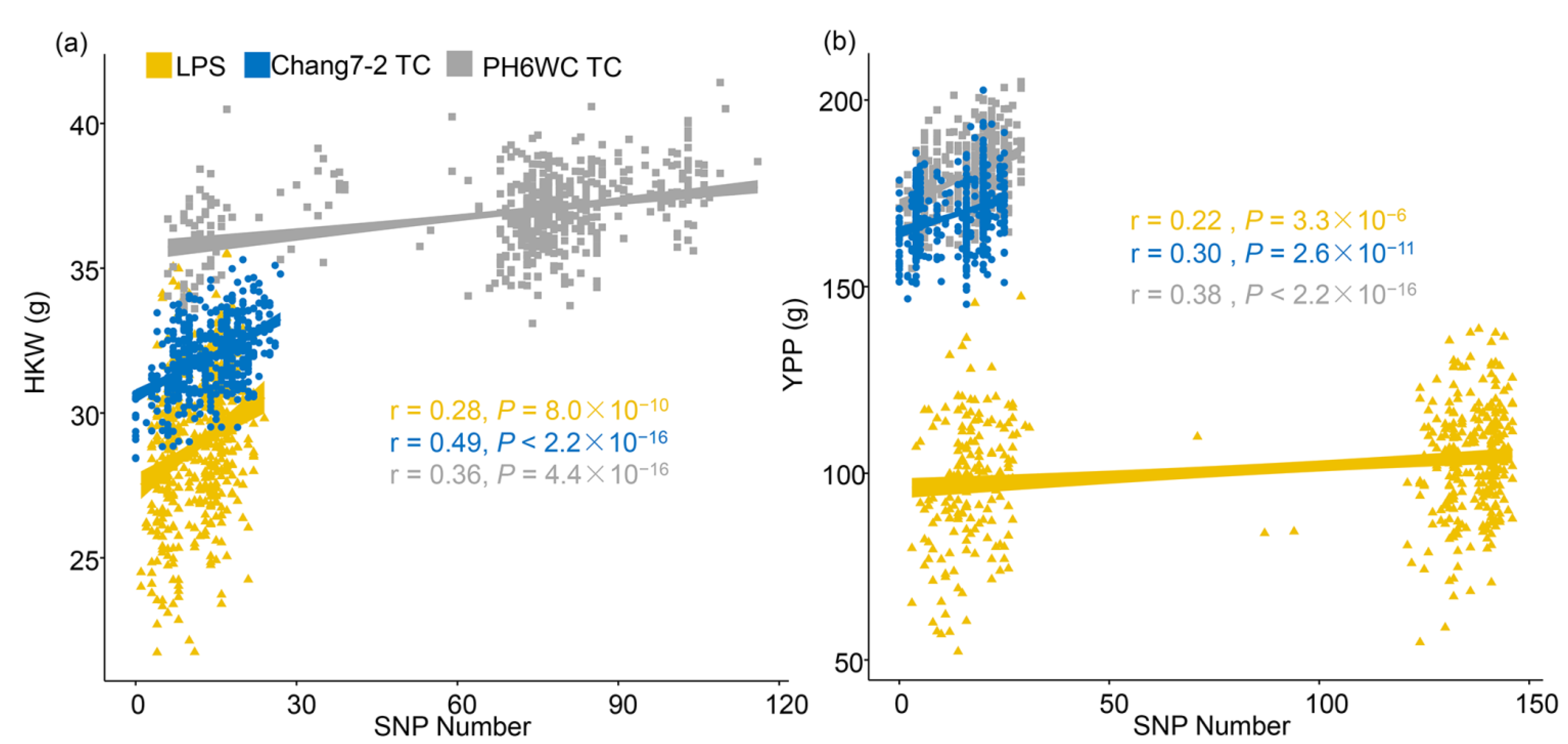
2.2. Genotypic Data Analysis and Genetic Dissection of Yield Traits of the Three Populations
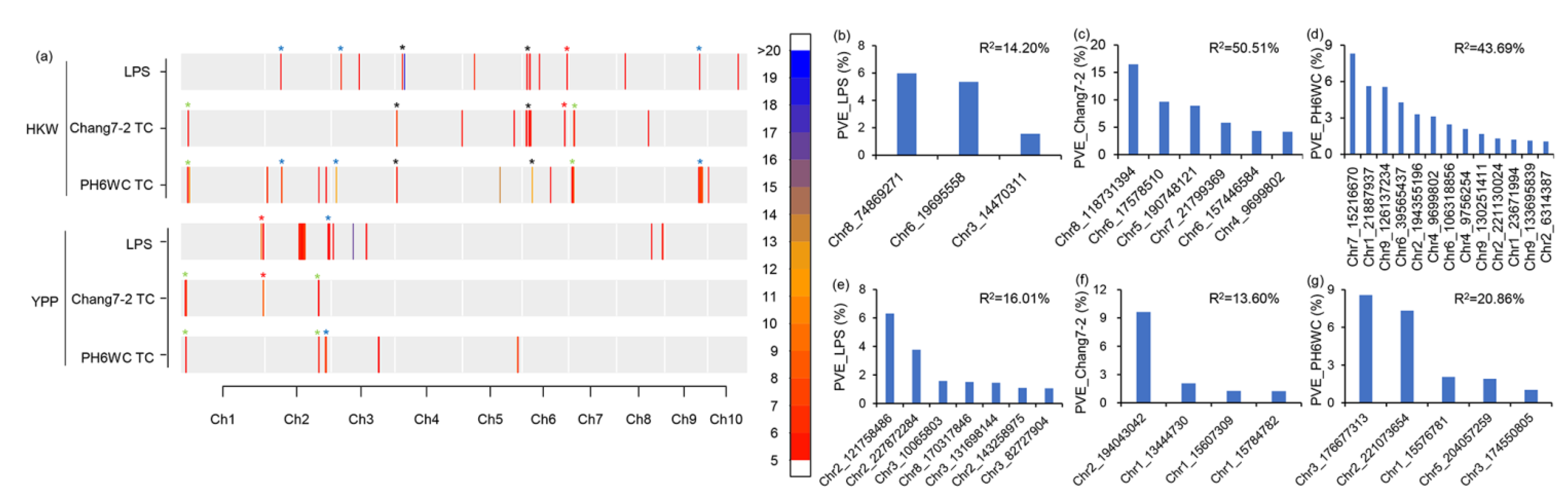
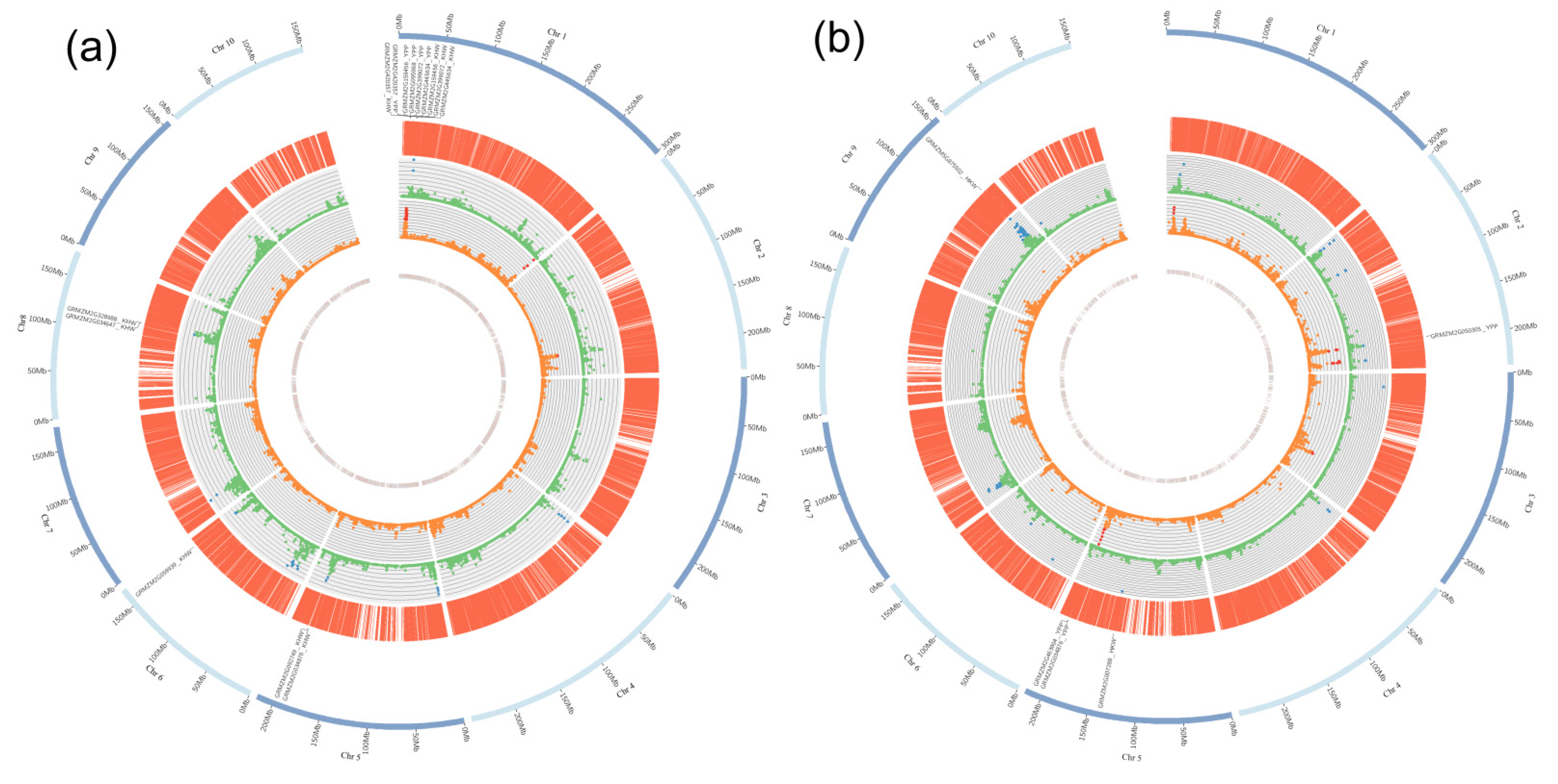
2.3. Genetic Features of the Significant SNPs
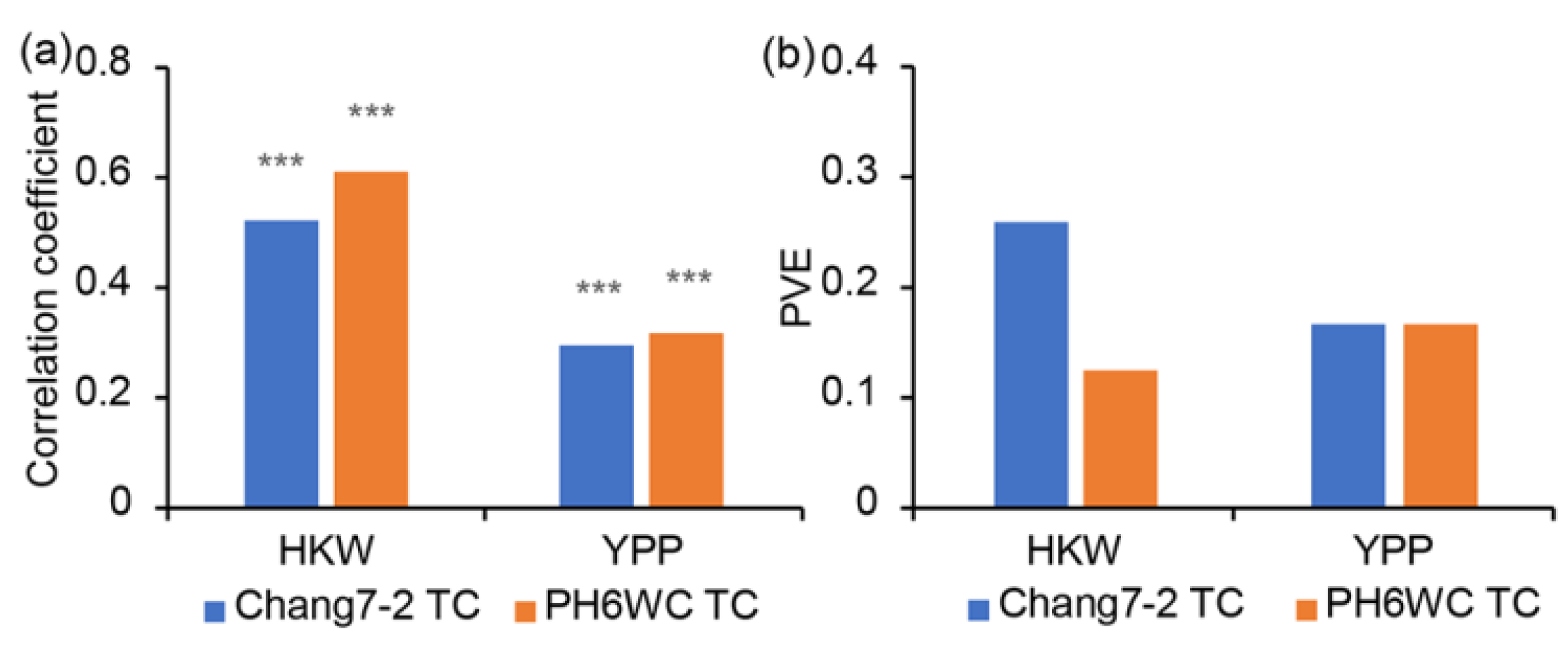
2.4. Identification of Common QTLs between LPS and TC Populations
2.5. RNA-seq Analysis Identified the Candidate Genes in the Surrounding Region of the Significant SNPs
3. Discussion
4. Materials and Methods
4.1. Population Construction, Phenotype Evaluation and Phenotypic Data Analysis
4.2. Genotype Processing
4.3. Marker–Trait Association Analysis and Calculation of PVE
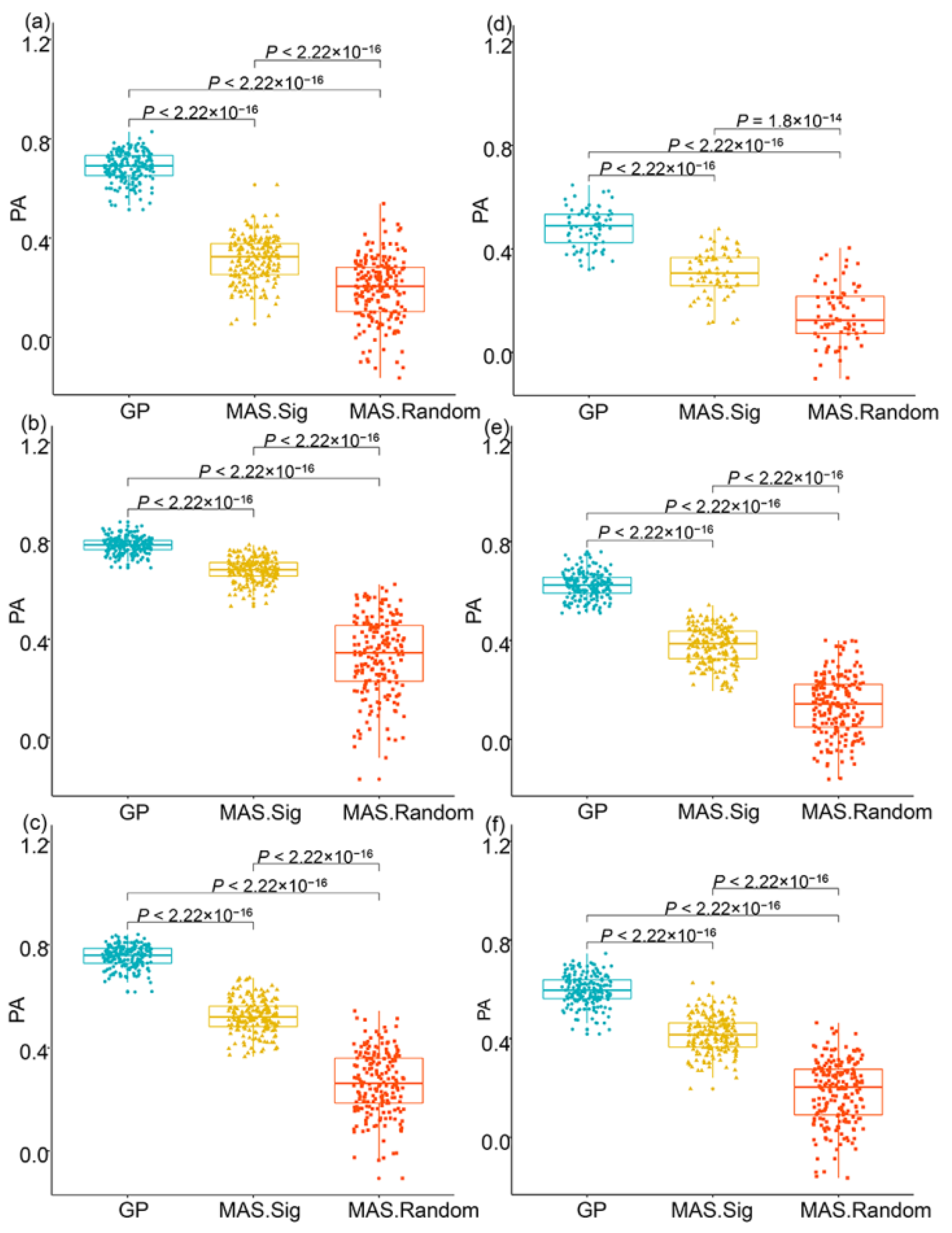
4.4. GP and MAS Analysis
4.5. Identification of Common QTLs among LPS and two TC Populations
4.6. RNA-seq Analysis and Identification of Differentially Expressed Genes around Significant SNPs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duvick, D.N. Biotechnology in the 1930s: The development of hybrid maize. Nat. Rev. Genet. 2001, 2, 69–74. [Google Scholar] [CrossRef]
- Bekavac, G.; Purar, B.; Jockovic, D. Relationships between line per se and testcross performance for agronomic traits in two broad-based populations of maize. Euphytica 2008, 162, 363–369. [Google Scholar] [CrossRef]
- Mihaljevic, R.; Schon, C.C.; Utz, H.F.; Melchinger, A.E. Correlations and QTL correspondence between line per se and testcross performance for agronomic traits in four populations of European maize. Crop Sci. 2005, 45, 114–122. [Google Scholar]
- Smith, O.S. Covariance between line per se and testcross performance. Crop Sci. 1986, 26, 540–543. [Google Scholar] [CrossRef]
- Schwegler, D.D.; Gowda, M.; Schulz, B.; Miedaner, T.; Liu, W.X.; Reif, J.C. Genotypic correlations and QTL correspondence between line per se and testcross performance in sugar beet (Beta vulgaris L.) for the three agronomic traits beet yield, potassium content, and sodium content. Mol. Breed. 2014, 34, 205–215. [Google Scholar] [CrossRef]
- Pace, J.; Gardner, C.; Romay, C.; Ganapathysubramanian, B.; Lubberstedt, T. Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genom. 2015, 16, 47. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, D.L.; Liu, S.S.; Ibrahim, R.; Blanco, M.; Lubberstedt, T. Genome-wide association studies of doubled haploid exotic introgression lines for root system architecture traits in maize (Zea mays L.). Plant Sci. 2018, 268, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Zaidi, P.H.; Seetharam, K.; Krishna, G.; Krishnamurthy, L.; Gajanan, S.; Babu, R.; Zerka, M.; Vinayan, M.T.; Vivek, B.S. Genomic regions associated with root traits under drought stress in tropical maize (Zea mays L.). PLoS ONE 2016, 11, e0164340. [Google Scholar] [CrossRef]
- Tibbs, C.L.; Zhang, Z.; Yu, J. Status and prospects of genome-wide association studies in plants. Plant Genome 2021, 14, e20077. [Google Scholar]
- Huang, X.; Yang, S.; Gong, J. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 2015, 6, 6258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Zhao, Y.; Beier, S. Exome association analysis sheds light onto leaf rust (Puccinia triticina) resistance genes currently used in wheat breeding (Triticum aestivum L.). Plant Biotechnol. J. 2020, 18, 1396–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.C.; Dai, X.B.; Wang, Q.S.; Xu, S.Z.; Zhao, P.X. PEPIS: A pipeline for estimating epistatic effects in quantitative trait locus mapping and genome-wide association studies. PLoS Comput. Biol. 2016, 12, e1004925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Yeh, C.T.; Tang, H.M.; Nettleton, D.; Schnable, P.S. Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE 2012, 7, e36406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Wang, X.; Pan, Q.; Li, P.; Liu, Y.; Lu, X.; Zhong, W.; Li, M.; Han, L.; Li, J.; et al. QTG-Seq accelerates QTL fine mapping through QTL partitioning and whole-genome sequencing of bulked segregant samples. Mol. Plant 2019, 12, 426–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Ma, Y.T.; Liu, Y.Y.; Lyle, D.; Li, D.D.; Wang, P.X.; Xu, J.L.; Zhen, S.H.; Lu, J.W.; Peng, Y.L.; et al. Dissecting the genetic basis of maize deep-sowing tolerance by combining association mapping and gene expression analysis. J. Integr. Agric. 2021, 21, 1266–1277. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Wang, H.; Guo, Z.; Xu, X.; Liu, J.; Wang, S.; Li, W.X.; Zou, C.; Prasanna, B.M. Factors affecting genomic selection revealed by empirical evidence in maize. Crop J. 2018, 6, 341–352. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, Y.; Ma, Y.; Fu, J.; Wang, G. Genetic and molecular control of grain yield in maize. Mol. Breed. 2021, 41, 18. [Google Scholar] [CrossRef]
- Andre, B.; Zheng, P.; Luck, S.; Shen, B.; Meyer, D.J.; Li, B.; Tingey, S.; Rafalskl, A. Whole genome scan detects an allelic variant of fad2 associated with increased oleic acid levels in maize. Mol. Genet. Genome 2008, 279, 1–10. [Google Scholar]
- Kump, K.L.; Bradbury, P.J.; Wisser, R.J. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping populating. Nat. Genet. 2011, 43, 163–168. [Google Scholar] [CrossRef]
- Li, L.; Hao, Z.; Li, X. An analysis of the poly morghisms in a gene for being involved in drought tolerance in maize. Genetics 2011, 139, 479–487. [Google Scholar]
- Tian, F.; Bradbury, P.J.; Brown, P.J.; Hung, H.; Sun, Q.; Flint-Garcia, S.; Rocheford, T.R.; Mcmullen, M.D.; Holland, J.B.; Buckler, E.S. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011, 43, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh, B.I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Zhao, Y.S.; Gowda, M.; Wurschum, T.; Longin, C.F.H.; Korzun, V.; Kollers, S.; Schachschneider, R. Dissecting the genetic architecture of frost tolerance in Central European winter wheat. J. Exp. Bot. 2013, 64, 4453–4460. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Han, L.Q.; Li, Q.; Wang, G.Y.; Zhang, H.W.; Li, L. Using interactome big data to crack genetic mysteries and enhance future crop breeding. Mol. Plant. 2021, 14, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.J.; Jiang, S.Q.; Cheng, Q.; Wang, X.Q.; Yan, J.; Zhang, R.Y.; Qiao, F.; Ma, C.; Luo, J.Y.; Li, W.Q.; et al. The genetic mechanism of heterosis utilization in maize improvement. Genome Biol. 2021, 22, 148. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.M.; Liu, H.J.; Yan, J.B.; Tian, F. Natural variation in crops, realized understanding continuing promise. Ann. Rev. Plant Biol. 2021, 72, 357–385. [Google Scholar] [CrossRef] [PubMed]
- Frascaroli, E.; Canè, M.A.; Pè, M.E. QTL detection in maize testcross progenies as affected by related and unrelated testers. Theor. Appl. Genet. 2009, 118, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhou, Z.; Lu, X. Genetic dissection of hybrid performance and heterosis for yield-related traits in maize. Front. Plant Sci. 2021, 12, 774478. [Google Scholar] [CrossRef]
- Jiang, L.; Ge, M.; Zhao, H.; Zhang, T. Analysis of heterosis and quantitative trait loci for kernel shape related traits using triple testcross population in maize. PLoS ONE 2015, 28, 10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Liu, Z.; Hu, Y. QTL analysis of kernel-related traits in maize using an immortalized F2 population. PLoS ONE 2014, 9, e89645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Xu, Z.; Gu, R.; Wang, P.; Lyle, D.; Xu, J.; Zhang, H.; Wang, G. Enhancing genomic selection by fitting large-effect SNPs as fixed effects and a genotypeby-environment effect using a maize BC1F3, 4 population. PLoS ONE 2019, 14, e0223898. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xu, Z.; Gu, R.; Wang, P.; Xu, J.; Du, D.; Fu, J.; Wang, J.; Zhang, H.; Wang, G. Genomic prediction across structured hybrid populations and environments in maize. Plants 2021, 10, 1174. [Google Scholar] [CrossRef]
- Liu, X.; Hu, X.; Li, K.; Liu, Z.; Wu, Y.; Wang, H.; Huang, C. Genetic mapping and genomic selection for maize stalk strength. BMC Plant Biol. 2020, 20, 196. [Google Scholar] [CrossRef]
- Tanaka, A.; Nakagawa, H.; Tomita, C.; Shimatani, Z.; Ohtake, M.; Nomura, T.; Jiang, C.J.; Dubouzet, J.G.; Kikuchi, S.; Sekimoto, H.; et al. BRASSINOSTEROID UPREGULATED1, encoding a Helix-Loop-Helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 2009, 151, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.S.; Feng, F.J.; Zhang, Y.; Elesawi, E.E.; Xu, K.; Li, T.F.; Mei, H.W.; Liu, H.Y.; Gao, N.N.; Chen, C.L.; et al. A novel rice grain size gene OsSNB was identified by genome-wide association study in natural population. PLoS Genet. 2019, 15, e1008191. [Google Scholar] [CrossRef] [PubMed]
- Hakata, M.; Kuroda, M.; Ohsumi, A.; Hirose, T.; Nakamura, H.; Muramatsu, M.; Ichikawa, H.; Yamakawa, H. Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci. Biotechnol. Biochem. 2012, 76, 2129–2134. [Google Scholar] [CrossRef] [Green Version]
- Che, R.H.; Tong, H.N.; Shi, B.H.; Liu, Y.Q.; Fang, S.R.; Liu, D.P.; Xiao, Y.H.; Hu, B.; Liu, L.C.; Wang, H.R.; et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2016, 2, 15195. [Google Scholar] [CrossRef]
- Hao, J.Q.; Wang, D.K.; Wu, Y.B.; Huang, K.; Duan, P.G.; Li, N.; Xu, R.; Zeng, D.L.; Dong, G.J.; Zhang, B.L.; et al. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol. Plant 2021, 14, 1266–1280. [Google Scholar] [CrossRef]
- Na, J.K.; Seo, M.H.; Moon, S.J.; Yoon, I.S.; Lee, Y.H.; Kim, J.K.; Lee, K.O.; Kim, D.Y. N-terminal region of rice polycomb group protein OsEZ1 is required for OsEZ1–OsFIE2 protein interaction. Plant Biotechnol. Rep. 2013, 7, 503–510. [Google Scholar] [CrossRef]
- Wang, A.H.; Garcia, D.; Zhang, H.Y.; Feng, K.; Chaudhury, A.; Berger, F.; Peacock, W.J.; Dennis, E.S.; Luo, M. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 2010, 63, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, J.; Huang, Y.; Liu, L.; Li, D.P.; Chen, L.B.; Luan, S. FERONIA receptor kinase controls seed size in Arabidopsis thaliana. Mol. Plant 2014, 7, 920–922. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Du, L.; Xu, R.; Cui, R.; Hao, J.; Sun, C.; Li, Y. Transcription factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana. Plant Cell 2015, 27, 620–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, D.Q.; Wang, X.C.; Yang, M.; Yang, L.; He, G.M.; Deng, X.W. A new regulator of seed size control in Arabidopsis identified by a genome-wide association study. N. Phytol. 2019, 222, 895–906. [Google Scholar] [CrossRef]
- Miller, C.; Wells, R.; Mckenzie, N.; Trick, M.; Ball, J.; Fatihi, A.; Du-Breucp, B.; Chardot, T.; Lepiniec, L.; Bevan, M. Variation in expression of the HECT E3 ligase UPL3 modulates LEC2 Levels, seed size, and crop yields in Brassica napus. Plant Cell 2019, 31, 1370–2385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.M.; Wang, J.; Dong, R.; Guan, H.Y.; Liu, T.S.; Liu, C.X.; Liu, Q.; Wang, L.M. Overexpression of an antisense RNA of maize receptor-like kinase gene ZmRLK7 enlarges the organ and seed size of transgenic Arabidopsis plants. Front. Plant Sci. 2020, 11, 1423. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.A.; Lee, H.S.; Kim, Y.S.; Paek, K.H.; Shin, J.S.; Bae, J.M. Down-regulation of the IbEXP1 gene enhanced storage root development in sweetpotato. J. Exp. Bot. 2013, 64, 129–142. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.M.; Yang, H.L.; Fang, Y.S.; Guo, W.; Chen, H.F.; Zhang, X.J.; Dai, W.J.; Chen, S.L.; Hao, Q.N.; Yuan, S.L.; et al. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 2020, 19, 702–716. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, D.F.; Cao, Y.Y.; Wang, L.F.; Li, J.J.; Lubberstedt, T.; Wang, T.Y.; Li, Y.; Li, H.Y. Heterosis-related genes under different planting densities in maize. J. Exp. Bot. 2018, 69, 5077–5087. [Google Scholar] [CrossRef]
- Song, W.; Shi, Z.; Xing, J.F.; Duan, M.X.; Su, A.G.; Li, C.H.; Zhang, R.Y.; Zhao, Y.X.; Luo, M.J.; Wang, J.D.; et al. Molecular mapping of quantitative trait loci for grain moisture at harvest in maize. Plant Breed. 2017, 136, 28–32. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.J.; Zhang, L.; Liu, X.J.; Wang, Z.H. Preliminary assessment of breeding potential of two exotic populations in improving Xianyu. Guizhou Agric. Sci. 2013, 41, 10–12. [Google Scholar]
- Zhang, X.G.; Ma, C.C.; Wang, X.Q.; Wu, M.B.; Shao, J.K.; Huang, L.; Yuan, L.; Fu, Z.Y.; Li, W.H.; Zhang, X.H.; et al. Global transcriptional profiling between inbred parents and hybrids provides comprehensive insights into ear-length heterosis of maize (Zea mays). BMC Plant Bio. 2021, 21, 118. [Google Scholar] [CrossRef] [PubMed]
- Hadasch, S.; Simko, I.; Hayes, R.J.; Ogutu, J.O.; Piepho, H.P. Comparing the predictive abilities of phenotypic and marker-assisted selection methods in a biparental lettuce population. Plant Genome 2016, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Mezmouk, S.; Baumgarten, A. Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLoS Genet. 2017, 13, e1007019. [Google Scholar] [CrossRef] [Green Version]
- Hallauer, A.R.; Carena, M.J.; Miranda Filho, J.B. Quantitative Genetics in Maize Breeding; Springer Science: New York, NY, USA, 2010. [Google Scholar]
- Bates, D.; Machler, M.; Bolker, B.M.; Walker, S.C. Fitting linear mixed-effects models using lme. J. Stat. Soft. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Senior, M.L.; Heun, M. Mapping maize microsatellites and polymerase chain reaction confirmation of the targeted repeats using a CT primer. Genome 1993, 36, 884–889. [Google Scholar] [CrossRef]
- Xu, C.; Ren, Y.H.; Jian, Y.Q.; Guo, Z.F.; Zhang, Y.; Xie, C.X.; Fu, J.J.; Wang, H.W.; Wang, G.Y.; Xu, Y.B.; et al. Development of a maize 55 K SNP array with improved genome coverage for molecular breeding. Mol. Breed. 2017, 37, 20. [Google Scholar] [CrossRef] [Green Version]
- Browning, B.L.; Browning, S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009, 84, 210–223. [Google Scholar] [CrossRef] [Green Version]
- Wimmer, V.; Albrecht, T.; Auinger, H.J.; Schon, C.C. Synbreed: A framework for the analysis of genomic prediction data using R. Bioinformatics 2012, 28, 2086–2087. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Li, R.; Li, G.; Zhang, F.; Zhu, T.; Zhang, Q.; Ali, J.; Li, Z.; Xu, S. Hybrid breeding of rice via genomic selection. Plant Biotechnol. J. 2019, 18, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Zhu, D.; Zhang, Q. Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proc. Natl. Acad. Sci. USA 2014, 111, 12456–12461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S. Mapping quantitative trait loci by controlling polygenic background effects. Genetics 2013, 12, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 2014, 4, 250–255. [Google Scholar] [CrossRef] [Green Version]
- Elisabetta, F.; Maria-Angela, C.; Pierangelo, L.; Giorgio, P.; Luca, G.; Marzio, V.; Michele, M.; Mario-Enrico, P. Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics 2007, 176, 625. [Google Scholar]
- Pan, Q.C.; Li, L.; Yang, X.H.; Tong, H.; Xu, S.T.; Li, Z.G.; Li, W.Y.; Muehlbauer, G.J.; Li, J.S.; Yan, J.B. Genome-wide recombination dynamics are associated with phenotypic variation in maize. N. Phytol. 2016, 210, 1083–1094. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Z.; Fu, J.; Wang, G.; Wang, J.; Liu, Y. Transcriptome analysis of maize immature embryos reveals the roles of cysteine in improving agrobacterium infection efficiency. Front. Plant Sci. 2017, 8, 1778. [Google Scholar] [CrossRef]
- Chen, L.; Bian, J.; Shi, S. Genetic analysis for the grain number heterosis of a super-hybrid rice WFYT025 combination using RNA-Seq. Rice 2018, 11, 37. [Google Scholar] [CrossRef] [Green Version]
- Howlader, J.; Robin, A.H.K.; Natarajan, S.; Biswas, M.K.; Sumi, K.R.; Song, C.Y.; Park, J.I.; Nou, I.S. Transcriptome analysis by RNA-Seq reveals genes related to plant height in two sets of parent-hybrid combinations in easter lily (Lilium longiflorum). Sci. Rep. 2020, 10, 9082. [Google Scholar]
- Ren, J.; Zhang, F.; Gao, F. Transcriptome and genome sequencing elucidates the molecular basis for the high yield and good quality of the hybrid rice variety Chuanyou. Sci. Rep. 2020, 10, 19935. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, K.; Zhang, X.; Guo, L. Comparative transcriptome analysis between inbred and hybrids reveals molecular insights into yield heterosis of upland cotton. BMC Plant Biol. 2020, 20, 239. [Google Scholar] [CrossRef] [PubMed]

| Trait | Population | Mean ± SD (g) | N | CV (%) | Range (g) | H2 (%) |
|---|---|---|---|---|---|---|
| HKW | LPS | 28.88 ± 2.45 | 481 | 8.49 | 21.73–35.52 | 82.47 |
| Chang7-2 | 31.94 ± 1.22 | 481 | 3.81 | 28.43–35.30 | 79.07 | |
| PH6WC | 36.96 ± 1.33 | 481 | 3.59 | 33.09–41.41 | 78.91 | |
| YPP | LPS | 101.50 ± 16.52 | 475 | 16.27 | 52.23–181.65 | 61.63 |
| Chang7-2 | 169.56 ± 9.13 | 469 | 5.38 | 145.28–202.65 | 55.09 | |
| PH6WC | 179.58 ± 9.14 | 475 | 5.09 | 154.86–204.93 | 58.27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Li, D.; Xu, Z.; Gu, R.; Wang, P.; Fu, J.; Wang, J.; Du, W.; Zhang, H. Dissection of the Genetic Basis of Yield Traits in Line per se and Testcross Populations and Identification of Candidate Genes for Hybrid Performance in Maize. Int. J. Mol. Sci. 2022, 23, 5074. https://doi.org/10.3390/ijms23095074
Ma Y, Li D, Xu Z, Gu R, Wang P, Fu J, Wang J, Du W, Zhang H. Dissection of the Genetic Basis of Yield Traits in Line per se and Testcross Populations and Identification of Candidate Genes for Hybrid Performance in Maize. International Journal of Molecular Sciences. 2022; 23(9):5074. https://doi.org/10.3390/ijms23095074
Chicago/Turabian StyleMa, Yuting, Dongdong Li, Zhenxiang Xu, Riliang Gu, Pingxi Wang, Junjie Fu, Jianhua Wang, Wanli Du, and Hongwei Zhang. 2022. "Dissection of the Genetic Basis of Yield Traits in Line per se and Testcross Populations and Identification of Candidate Genes for Hybrid Performance in Maize" International Journal of Molecular Sciences 23, no. 9: 5074. https://doi.org/10.3390/ijms23095074
APA StyleMa, Y., Li, D., Xu, Z., Gu, R., Wang, P., Fu, J., Wang, J., Du, W., & Zhang, H. (2022). Dissection of the Genetic Basis of Yield Traits in Line per se and Testcross Populations and Identification of Candidate Genes for Hybrid Performance in Maize. International Journal of Molecular Sciences, 23(9), 5074. https://doi.org/10.3390/ijms23095074








