Genetic and Clinical Profile of Retinopathies Due to Disease-Causing Variants in Leber Congenital Amaurosis (LCA)-Associated Genes in a Large German Cohort
Abstract
1. Introduction
2. Results
2.1. Genetic Findings
2.2. Clinical Findings
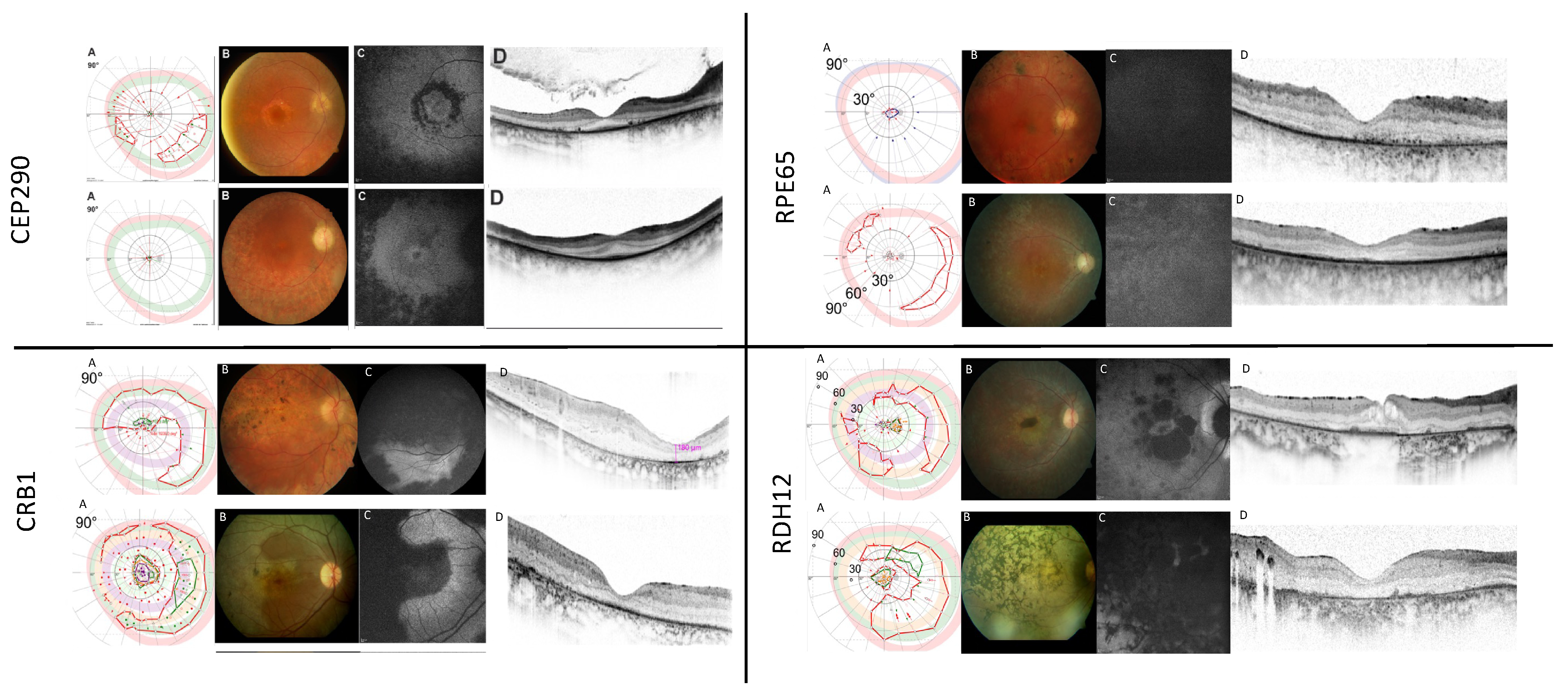
2.2.1. CEP290
2.2.2. CRB1
2.2.3. RPE65
2.2.4. RDH12
3. Discussion
4. Materials and Methods
4.1. Study Design and Study Population
4.2. Inclusion Criteria
4.3. Clinical Assessment
- -
- 1= Birth;
- -
- 2= Infancy (beginning 28 days to 12 months of age);
- -
- 3= Toddler (beginning 13 months -2 years of age);
- -
- 4= Early childhood (beginning 2 years to 5 years of age);
- -
- 5= Middle childhood (beginning 6 years to 11 years);
- -
- 6= Early adolescent (beginning 12 years to 18 years).
- -
- Counting fingers (CF) = 0.014;
- -
- Hand motion (HM) = 0.005;
- -
- Light perception (LP), no light perception (NLP) = 0.
4.4. Genetic Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leber, T.G. Ueber Retinitis pigmentosa und angeborene Amaurose. Graefes Archive of Clincal Experimental Ophthalmology. Arch. Für Ophthalmol. 1869, 15, 1–25. [Google Scholar]
- Kumaran, N.; Moore, A.T.; Weleber, R.G.; Michaelides, M. Leber congenital amaurosis/early-onset severe retinal dystrophy: Clinical features, molecular genetics and therapeutic interventions. Br. J. Ophthalmol. 2017, 101, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Daich Varela, M.; Cabral de Guimaraes, T.A.; Georgiou, M.; Michaelides, M. Leber congenital amaurosis/early-onset severe retinal dystrophy: Current management and clinical trials. Br. J. Ophthalmol. 2022, 106, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Retina, P. 2023. Available online: https://www.pro-retina.de (accessed on 1 March 2023).
- Sheck, L.; Davies, W.I.L.; Moradi, P.; Robson, A.G.; Kumaran, N.; Liasis, A.C.; Webster, A.R.; Moore, A.T.; Michaelides, M. Leber Congenital Amaurosis Associated with Mutations in CEP290, Clinical Phenotype, and Natural History in Preparation for Trials of Novel Therapies. Ophthalmology 2018, 125, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Hahn, L.C.; Georgiou, M.; Almushattat, H.; van Schooneveld, M.J.; de Carvalho, E.R.; Wesseling, N.L.; Ten Brink, J.B.; Florijn, R.J.; Lissenberg-Witte, B.I.; Strubbe, I.; et al. The Natural History of Leber Congenital Amaurosis and Cone-Rod Dystrophy Associated with Variants in the GUCY2D Gene. Ophthalmol. Retina 2022, 6, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, N.; Pennesi, M.E.; Yang, P.; Trzupek, K.M.; Schlechter, C.; Moore, A.T.; Weleber, R.G.; Michaelides, M. Leber Congenital Amaurosis/Early-Onset Severe Retinal Dystrophy Overview. In GeneReviews(®); Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Foxman, S.G.; Heckenlively, J.R.; Bateman, J.B.; Wirtschafter, J.D. Classification of congenital and early onset retinitis pigmentosa. Arch. Ophthalmol. 1985, 103, 1502–1506. [Google Scholar] [CrossRef] [PubMed]
- Cremers, F.P.; van den Hurk, J.A.; den Hollander, A.I. Molecular genetics of Leber congenital amaurosis. Hum. Mol. Genet. 2002, 11, 1169–1176. [Google Scholar] [CrossRef]
- den Hollander, A.I.; Black, A.; Bennett, J.; Cremers, F.P. Lighting a candle in the dark: Advances in genetics and gene therapy of recessive retinal dystrophies. J. Clin. Investig. 2010, 120, 3042–3053. [Google Scholar] [CrossRef]
- Weisschuh, N.; Feldhaus, B.; Khan, M.I.; Cremers, F.P.M.; Kohl, S.; Wissinger, B.; Zobor, D. Molecular and clinical analysis of 27 German patients with Leber congenital amaurosis. PLoS ONE 2018, 13, e0205380. [Google Scholar] [CrossRef]
- Retinal Information Network. 2023. Available online: https://web.sph.uth.edu/RetNet/ (accessed on 1 March 2023).
- Sergouniotis, P.I.; Maxime, E.; Leroux, D.; Olry, A.; Thompson, R.; Rath, A.; Robinson, P.N.; Dollfus, H. An ontological foundation for ocular phenotypes and rare eye diseases. Orphanet J. Rare Dis. 2019, 14, 8. [Google Scholar] [CrossRef]
- Cremers, F.P.M.; Lee, W.; Collin, R.W.J.; Allikmets, R. Clinical spectrum, genetic complexity and therapeutic approaches for retinal disease caused by ABCA4 mutations. Prog. Retin. Eye Res. 2020, 79, 100861. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, M.; Fujinami, K.; Vincent, A.; Nasser, F.; Khateb, S.; Vargas, M.E.; Thiadens, A.; de Carvalho, E.R.; Nguyen, X.T.; De Guimarães, T.A.C.; et al. KCNV2-Associated Retinopathy: Detailed Retinal Phenotype and Structural Endpoints-KCNV2 Study Group Report 2. Am. J. Ophthalmol. 2021, 230, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, M.; Robson, A.G.; Fujinami, K.; Leo, S.M.; Vincent, A.; Nasser, F.; Cabral De Guimarães, T.A.; Khateb, S.; Pontikos, N.; Fujinami-Yokokawa, Y.; et al. KCNV2-Associated Retinopathy: Genetics, Electrophysiology, and Clinical Course-KCNV2 Study Group Report 1. Am. J. Ophthalmol. 2021, 225, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, T.A.C.; Georgiou, M.; Robson, A.G.; Michaelides, M. KCNV2 retinopathy: Clinical features, molecular genetics and directions for future therapy. Ophthalmic. Genet. 2020, 41, 208–215. [Google Scholar] [CrossRef]
- Zobor, D.; Kohl, S.; Wissinger, B.; Zrenner, E.; Jägle, H. Rod and cone function in patients with KCNV2 retinopathy. PLoS ONE 2012, 7, e46762. [Google Scholar] [CrossRef]
- den Hollander, A.I.; Koenekoop, R.K.; Yzer, S.; Lopez, I.; Arends, M.L.; Voesenek, K.E.; Zonneveld, M.N.; Strom, T.M.; Meitinger, T.; Brunner, H.G.; et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006, 79, 556–561. [Google Scholar] [CrossRef]
- Perrault, I.; Delphin, N.; Hanein, S.; Gerber, S.; Dufier, J.L.; Roche, O.; Defoort-Dhellemmes, S.; Dollfus, H.; Fazzi, E.; Munnich, A.; et al. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum. Mutat. 2007, 28, 416. [Google Scholar] [CrossRef]
- Coppieters, F.; Casteels, I.; Meire, F.; De Jaegere, S.; Hooghe, S.; van Regemorter, N.; Van Esch, H.; Matuleviciene, A.; Nunes, L.; Meersschaut, V.; et al. Genetic screening of LCA in Belgium: Predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum. Mutat. 2010, 31, E1709–E1766. [Google Scholar] [CrossRef]
- Coppieters, F.; Lefever, S.; Leroy, B.P.; De Baere, E. CEP290, a gene with many faces: Mutation overview and presentation of CEP290base. Hum. Mutat. 2010, 31, 1097–1108. [Google Scholar] [CrossRef]
- Simonelli, F.; Ziviello, C.; Testa, F.; Rossi, S.; Fazzi, E.; Bianchi, P.E.; Fossarello, M.; Signorini, S.; Bertone, C.; Galantuomo, S.; et al. Clinical and molecular genetics of Leber’s congenital amaurosis: A multicenter study of Italian patients. Investig. Ophthalmol. Vis. Sci. 2007, 48, 4284–4290. [Google Scholar] [CrossRef]
- Vallespin, E.; Lopez-Martinez, M.A.; Cantalapiedra, D.; Riveiro-Alvarez, R.; Aguirre-Lamban, J.; Avila-Fernandez, A.; Villaverde, C.; Trujillo-Tiebas, M.J.; Ayuso, C. Frequency of CEP290 c.2991_1655A>G mutation in 175 Spanish families affected with Leber congenital amaurosis and early-onset retinitis pigmentosa. Mol. Vis. 2007, 13, 2160–2162. [Google Scholar]
- Seong, M.W.; Kim, S.Y.; Yu, Y.S.; Hwang, J.M.; Kim, J.Y.; Park, S.S. Molecular characterization of Leber congenital amaurosis in Koreans. Mol. Vis. 2008, 14, 1429–1436. [Google Scholar] [PubMed]
- Li, Y.; Wang, H.; Peng, J.; Gibbs, R.A.; Lewis, R.A.; Lupski, J.R.; Mardon, G.; Chen, R. Mutation survey of known LCA genes and loci in the Saudi Arabian population. Investig. Ophthalmol. Vis. Sci. 2009, 50, 1336–1343. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, P.; Vijayalakshmi, P.; Thompson, S.; Ko, A.C.; Fingert, J.H.; Stone, E.M. Mutations that are a common cause of Leber congenital amaurosis in northern America are rare in southern India. Mol. Vis. 2009, 15, 1781–1787. [Google Scholar] [PubMed]
- McKibbin, M.; Ali, M.; Mohamed, M.D.; Booth, A.P.; Bishop, F.; Pal, B.; Springell, K.; Raashid, Y.; Jafri, H.; Inglehearn, C.F. Genotype-phenotype correlation for leber congenital amaurosis in Northern Pakistan. Arch. Ophthalmol. 2010, 128, 107–113. [Google Scholar] [CrossRef]
- Astuti, G.D.; Bertelsen, M.; Preising, M.N.; Ajmal, M.; Lorenz, B.; Faradz, S.M.; Qamar, R.; Collin, R.W.; Rosenberg, T.; Cremers, F.P. Comprehensive genotyping reveals RPE65 as the most frequently mutated gene in Leber congenital amaurosis in Denmark. Eur. J. Hum. Genet. EJHG 2016, 24, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.H.; Sharma, T. Leber Congenital Amaurosis. Adv. Exp. Med. Biol. 2018, 1085, 131–137. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Available online: https://www.fda.gov/BiologicsBloodVaccines/CellularGeneTherapyProducts/ApprovedProducts/ucm589507.htm (accessed on 1 March 2023).
- Energy Market Authority (EMA). 2018. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/luxturna (accessed on 1 March 2023).
- Weisschuh, N.; Obermaier, C.D.; Battke, F.; Bernd, A.; Kuehlewein, L.; Nasser, F.; Zobor, D.; Zrenner, E.; Weber, E.; Wissinger, B.; et al. Genetic architecture of inherited retinal degeneration in Germany: A large cohort study from a single diagnostic center over a 9-year period. Hum. Mutat. 2020, 41, 1514–1527. [Google Scholar] [CrossRef]
- Feldhaus, B.; Weisschuh, N.; Nasser, F.; den Hollander, A.I.; Cremers, F.P.M.; Zrenner, E.; Kohl, S.; Zobor, D. CEP290 Mutation Spectrum and Delineation of the Associated Phenotype in a Large German Cohort: A Monocentric Study. Am. J. Ophthalmol. 2020, 211, 142–150. [Google Scholar] [CrossRef]
- Feldhaus, B.; Kohl, S.; Hörtnagel, K.; Weisschuh, N.; Zobor, D. Novel homozygous mutation in the SPATA7 gene causes autosomal recessive retinal degeneration in a consanguineous German family. Ophthalmic. Genet. 2018, 39, 131–134. [Google Scholar] [CrossRef]
- Perrault, I.; Estrada-Cuzcano, A.; Lopez, I.; Kohl, S.; Li, S.; Testa, F.; Zekveld-Vroon, R.; Wang, X.; Pomares, E.; Andorf, J.; et al. Union makes strength: A worldwide collaborative genetic and clinical study to provide a comprehensive survey of RD3 mutations and delineate the associated phenotype. PLoS ONE 2013, 8, e51622. [Google Scholar] [CrossRef]
- Siemiatkowska, A.M.; van den Born, L.I.; van Genderen, M.M.; Bertelsen, M.; Zobor, D.; Rohrschneider, K.; van Huet, R.A.; Nurohmah, S.; Klevering, B.J.; Kohl, S.; et al. Novel compound heterozygous NMNAT1 variants associated with Leber congenital amaurosis. Mol. Vis. 2014, 20, 753–759. [Google Scholar]
- Kaplan, J. Leber congenital amaurosis: From darkness to spotlight. Ophthalmic. Genet. 2008, 29, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Daich Varela, M.; Georgiou, M.; Alswaiti, Y.; Kabbani, J.; Fujinami, K.; Fujinami-Yokokawa, Y.; Khoda, S.; Mahroo, O.A.; Robson, A.G.; Webster, A.R.; et al. CRB1-Associated Retinal Dystrophies: Genetics, Clinical Characteristics, and Natural History. Am. J. Ophthalmol. 2023, 246, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Perrault, I.; Rozet, J.M.; Calvas, P.; Gerber, S.; Camuzat, A.; Dollfus, H.; Châtelin, S.; Souied, E.; Ghazi, I.; Leowski, C.; et al. Retinal-specific guanylate cyclase gene mutations in Leber’s congenital amaurosis. Nat. Genet. 1996, 14, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Skorczyk-Werner, A.; Sowińska-Seidler, A.; Wawrocka, A.; Walczak-Sztulpa, J.; Krawczyński, M.R. Molecular background of Leber congenital amaurosis in a Polish cohort of patients-novel variants discovered by NGS. J. Appl. Genet. 2023, 64, 89–104. [Google Scholar] [CrossRef]
- Di Iorio, V.; Karali, M.; Brunetti-Pierri, R.; Filippelli, M.; Di Fruscio, G.; Pizzo, M.; Mutarelli, M.; Nigro, V.; Testa, F.; Banfi, S.; et al. Clinical and Genetic Evaluation of a Cohort of Pediatric Patients with Severe Inherited Retinal Dystrophies. Genes 2017, 8, 280. [Google Scholar] [CrossRef]
- Sallum, J.M.F.; Motta, F.L.; Arno, G.; Porto, F.B.O.; Resende, R.G.; Belfort, R., Jr. Clinical and molecular findings in a cohort of 152 Brazilian severe early onset inherited retinal dystrophy patients. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 728–752. [Google Scholar] [CrossRef]
- Hull, S.; Kiray, G.; Chiang, J.P.; Vincent, A.L. Molecular and phenotypic investigation of a New Zealand cohort of childhood-onset retinal dystrophy. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 708–717. [Google Scholar] [CrossRef]
- Zhu, L.; Ouyang, W.; Zhang, M.; Wang, H.; Li, S.; Meng, X.; Yin, Z.Q. Molecular genetics with clinical characteristics of Leber congenital amaurosis in the Han population of western China. Ophthalmic. Genet. 2021, 42, 392–401. [Google Scholar] [CrossRef]
- Xu, K.; Xie, Y.; Sun, T.; Zhang, X.; Chen, C.; Li, Y. Genetic and clinical findings in a Chinese cohort with Leber congenital amaurosis and early onset severe retinal dystrophy. Br. J. Ophthalmol. 2020, 104, 932–937. [Google Scholar] [CrossRef]
- Hosono, K.; Nishina, S.; Yokoi, T.; Katagiri, S.; Saitsu, H.; Kurata, K.; Miyamichi, D.; Hikoya, A.; Mizobuchi, K.; Nakano, T.; et al. Molecular Diagnosis of 34 Japanese Families with Leber Congenital Amaurosis Using Targeted Next Generation Sequencing. Sci. Rep. 2018, 8, 8279. [Google Scholar] [CrossRef]
- Maguire, A.M.; High, K.A.; Auricchio, A.; Wright, J.F.; Pierce, E.A.; Testa, F.; Mingozzi, F.; Bennicelli, J.L.; Ying, G.S.; Rossi, S.; et al. Age-dependent effects of RPE65 gene therapy for Leber’s congenital amaurosis: A phase 1 dose-escalation trial. Lancet 2009, 374, 1597–1605. [Google Scholar] [CrossRef]
- Maguire, A.M.; Simonelli, F.; Pierce, E.A.; Pugh, E.N., Jr.; Mingozzi, F.; Bennicelli, J.; Banfi, S.; Marshall, K.A.; Testa, F.; Surace, E.M.; et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 2008, 358, 2240–2248. [Google Scholar] [CrossRef]
- Simonelli, F.; Maguire, A.M.; Testa, F.; Pierce, E.A.; Mingozzi, F.; Bennicelli, J.L.; Rossi, S.; Marshall, K.; Banfi, S.; Surace, E.M.; et al. Gene therapy for Leber’s congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther. 2010, 18, 643–650. [Google Scholar] [CrossRef]
- Acland, G.M.; Aguirre, G.D.; Bennett, J.; Aleman, T.S.; Cideciyan, A.V.; Bennicelli, J.; Dejneka, N.S.; Pearce-Kelling, S.E.; Maguire, A.M.; Palczewski, K.; et al. Long-term restoration of rod and cone vision by single dose rAAV-mediated gene transfer to the retina in a canine model of childhood blindness. Mol. Ther. 2005, 12, 1072–1082. [Google Scholar] [CrossRef]
- Acland, G.M.; Aguirre, G.D.; Ray, J.; Zhang, Q.; Aleman, T.S.; Cideciyan, A.V.; Pearce-Kelling, S.E.; Anand, V.; Zeng, Y.; Maguire, A.M.; et al. Gene therapy restores vision in a canine model of childhood blindness. Nat. Genet. 2001, 28, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.; Ashtari, M.; Wellman, J.; Marshall, K.A.; Cyckowski, L.L.; Chung, D.C.; McCague, S.; Pierce, E.A.; Chen, Y.; Bennicelli, J.L.; et al. AAV2 gene therapy readministration in three adults with congenital blindness. Sci. Transl. Med. 2012, 4, 120ra115. [Google Scholar] [CrossRef] [PubMed]
- Bennicelli, J.; Wright, J.F.; Komaromy, A.; Jacobs, J.B.; Hauck, B.; Zelenaia, O.; Mingozzi, F.; Hui, D.; Chung, D.; Rex, T.S.; et al. Reversal of blindness in animal models of leber congenital amaurosis using optimized AAV2-mediated gene transfer. Mol. Ther. 2008, 16, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, S.G.; Acland, G.M.; Aguirre, G.D.; Aleman, T.S.; Schwartz, S.B.; Cideciyan, A.V.; Zeiss, C.J.; Komaromy, A.M.; Kaushal, S.; Roman, A.J.; et al. Safety of recombinant adeno-associated virus type 2-RPE65 vector delivered by ocular subretinal injection. Mol. Ther. 2006, 13, 1074–1084. [Google Scholar] [CrossRef]
- Csaky, K.G.; Richman, E.A.; Ferris, F.L., 3rd. Report from the NEI/FDA Ophthalmic Clinical Trial Design and Endpoints Symposium. Investig. Ophthalmol. Vis. Sci. 2008, 49, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, M.; Fujinami, K.; Michaelides, M. Inherited retinal diseases: Therapeutics, clinical trials and end points-A review. Clin. Exp. Ophthalmol. 2021, 49, 270–288. [Google Scholar] [CrossRef] [PubMed]
- Talib, M.; Boon, C.J.F. Retinal Dystrophies and the Road to Treatment: Clinical Requirements and Considerations. Asia Pac. J. Ophthalmol. 2020, 9, 159–179. [Google Scholar] [CrossRef] [PubMed]
- Eunice Kennedy Shriver National Institute of Child Health and Human Development. 2016. Available online: https://evs.nci.nih.gov/ftp1/Pediatric_Terminologies/NICHD/ (accessed on 1 March 2023).
- Schulze-Bonsel, K.; Feltgen, N.; Burau, H.; Hansen, L.; Bach, M. Visual acuities “hand motion” and “counting fingers” can be quantified with the freiburg visual acuity test. Investig. Ophthalmol. Vis. Sci. 2006, 47, 1236–1240. [Google Scholar] [CrossRef]
- Lange, C.; Feltgen, N.; Junker, B.; Schulze-Bonsel, K.; Bach, M. Resolving the clinical acuity categories “hand motion” and “counting fingers” using the Freiburg Visual Acuity Test (FrACT). Graefes Arch. Clin. Exp. Ophthalmol. 2009, 247, 137–142. [Google Scholar] [CrossRef]
- Hoffmann, M.B.; Bach, M.; Kondo, M.; Li, S.; Walker, S.; Holopigian, K.; Viswanathan, S.; Robson, A.G. ISCEV standard for clinical multifocal electroretinography (mfERG) (2021 update). Doc. Ophthalmol. 2021, 142, 5–16. [Google Scholar] [CrossRef]
- Robson, A.G.; Frishman, L.J.; Grigg, J.; Hamilton, R.; Jeffrey, B.G.; Kondo, M.; Li, S.; McCulloch, D.L. ISCEV Standard for full-field clinical electroretinography (2022 update). Doc. Ophthalmol. 2022, 144, 165–177. [Google Scholar] [CrossRef]
- den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; McGowan-Jordan, J.; Roux, A.F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016, 37, 564–569. [Google Scholar] [CrossRef]
- Genoox, F.B. Available online: https://franklin.genoox.com (accessed on 1 March 2023).
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
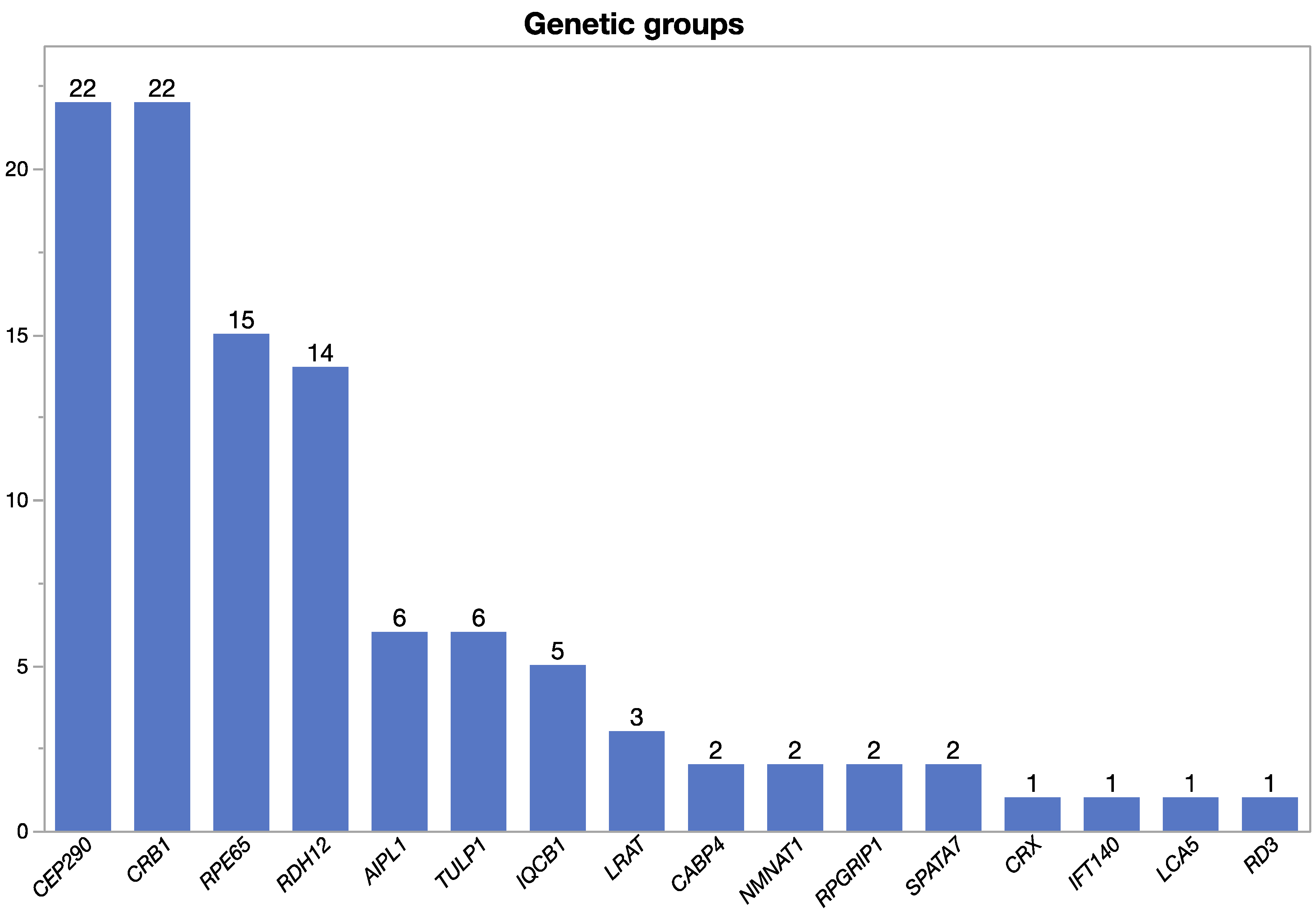
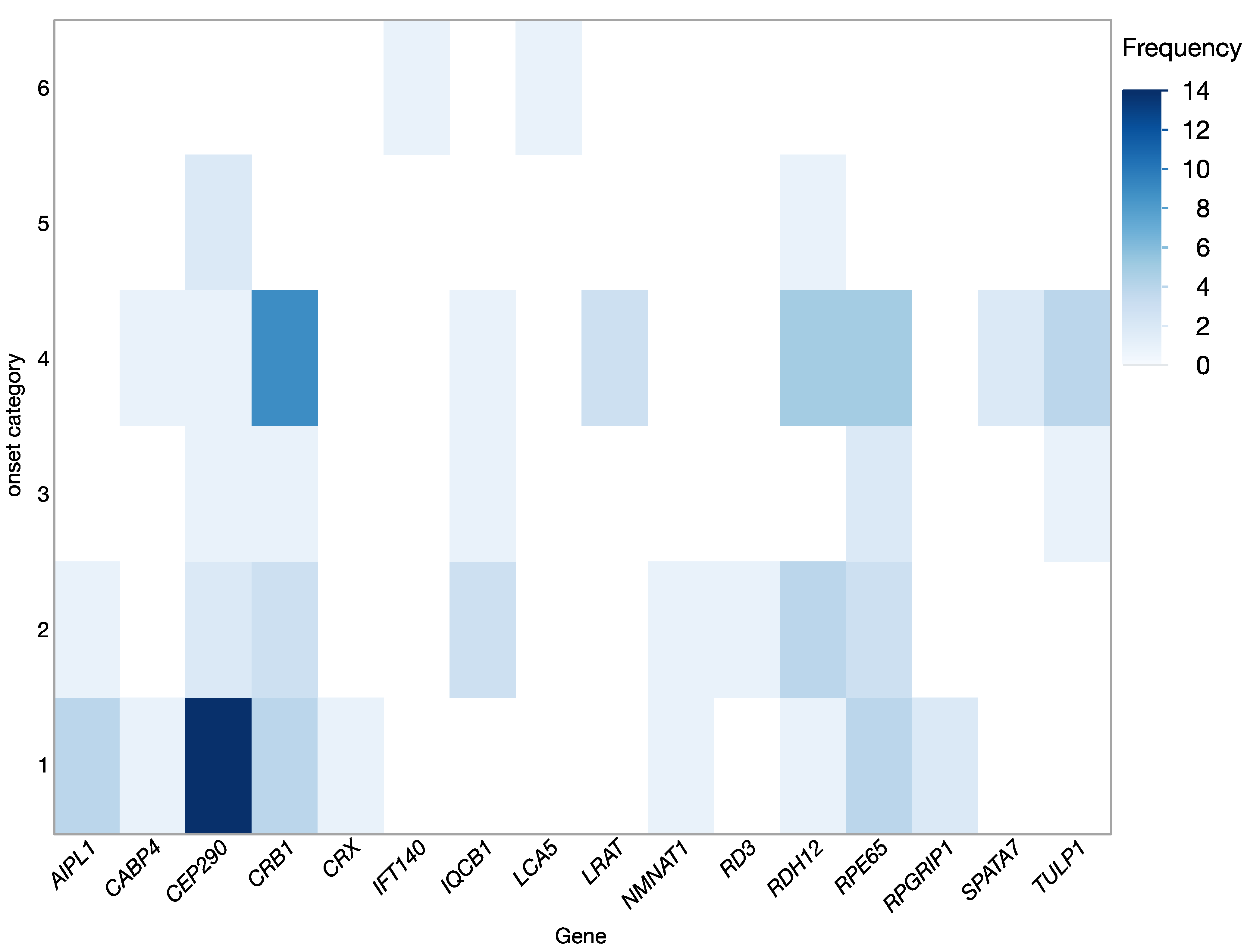
| Gene | N | Age (Years) M; Range | Eye | BCVA (Snellen) | VF III4e (deg2) | OCT CFT (µm) | |||
|---|---|---|---|---|---|---|---|---|---|
| N | M ± SD | N | M ± SD | N | M ± SD | ||||
| AIPL1 | 6 | 31; 10–49 | OD | 6 | 0.044 ± 0.092 | 2 | 5325 ± 5140 | 2 | 94 ± 0.7 |
| OS | 0.071 ± 0.146 | 3482 ± 3444 | 110 ± 57 | ||||||
| CABP4 | 2 | 28; 27–28 | OD | 2 | 0.365 ± 0.375 | 2 | 8847 ± 5048 | 2 | 204 ± 11 |
| OS | 0.365 ± 0.375 | 10370 ± 3896 | 214 ± 47 | ||||||
| CEP290 | 22 | 31; 4–72 | OD | 22 | 0.132 ± 0.246 | 14 | 804 ± 1741 | 8 | 184 ± 41 |
| OS | 0.184 ± 0.294 | 655 ± 1532 | 185 ± 37 | ||||||
| CRB1 | 22 | 32; 8–64 | OD | 22 | 0.101 ± 0.145 | 14 | 1476 ± 2474 | 13 | 151 ± 59 |
| OS | 0.113 ± 0.174 | 1250 ± 2437 | 9 | 161 ± 64 | |||||
| CRX | 1 | 46 | OD | 1 | * | 1 | * | 0 | * |
| OS | * | * | * | ||||||
| IFT140 | 1 | 76 | OD | 1 | * | 0 | * | 1 | * |
| OS | * | * | * | ||||||
| IQCB1 | 5 | 29; 10–61 | OD | 5 | 0.234 ± 0.194 | 5 | 36 ± 26 | 4 | 229 ± 8 |
| OS | 0.204 ± 0.173 | 31 ± 39 | 3 | 234 ± 3 | |||||
| LCA5 | 1 | 64 | OD | 1 | * | 1 | * | 1 | * |
| OS | * | * | * | ||||||
| LRAT | 3 | 39; 38–41 | OD | 3 | 0.028 ± 0.025 | 3 | * | 2 | 145 ± 4 |
| OS | 0.094 ± 0.136 | * | 169 ± 88 | ||||||
| NMNAT1 | 2 | 33; 10–55 | OD | 2 | 0 ± 0 | 2 | * | 0 | * |
| OS | 0 ± 0 | * | * | ||||||
| RD3 | 1 | 20 | OD | 1 | * | 1 | * | 1 | * |
| OS | * | * | * | ||||||
| RDH12 | 14 | 24; 3–51 | OD | 14 | 0.110 ± 0.183 | 8 | 1332 ± 2468 | 3 | 138 ± 72 |
| OS | 0.152 ± 0.215 | 1805 ± 638 | 145 ± 105 | ||||||
| RPE65 | 15 | 30; 9–51 | OD | 15 | 0.062 ± 0.084 | 15 | 257 ± 505 | 7 | 137 ± 47 |
| OS | 0.059 ± 0.100 | 329 ± 713 | 6 | 154 ± 27 | |||||
| RPGRIP1 | 2 | 29; 23–35 | OD | 2 | 0.04 ± 0.014 | 2 | 3083 ± 4168 | 2 | 194 ± 41 |
| OS | 0.045 ± 0.007 | 3190 ± 4345 | 1 | * | |||||
| SPATA7 | 2 | 50; 48–51 | OD | 2 | 0.163 ± 0.053 | 2 | 2137 ± 3019 | 2 | 187 ± 47 |
| OS | 0.143 ± 0.025 | 1646 ± 237 | 188 ± 63 | ||||||
| TULP1 | 6 | 21; 10–39 | OD | 6 | 0.25 ± 0.204 | 5 | 2469 ± 3521 | 5 | 135 ± 19 |
| OS | 0.272 ± 0.188 | 2278 ± 2449 | 134 ± 16 | ||||||
| Total | 105 | 31 ± 17 | OD | 108 | 0.124 ± 0.195 | 80 | 1403 ± 2702 | 53 | 165 ± 43 |
| OS | 0.147 ± 0.223 | 1277 ± 2544 | 49 | 183 ± 37 | |||||
| Gene | cDNA Position | Amino Acid Position | Number of Alleles |
|---|---|---|---|
| CEP290 | c.2991+1655A>G | p.(Cys998Ter) | 27 |
| AIPL1 | c.834G>A | p.(Trp278Ter) | 10 |
| RPE65 | c.1451G>T | p.(Gly484Val) | 10 |
| CRB1 | c.2843G>A | p.(Cys948Tyr) | 8 |
| IQCB1 | c.1558C>T | p.(Gln520Ter) | 6 |
| RDH12 | c.806_810del | p.(Ala269GlyfsTer2) | 6 |
| CRB1 | c.2234C>T | p.(Thr745Met) | 5 |
| IQCB1 | c.1558C>T | p.(Gln520Ter) | 5 |
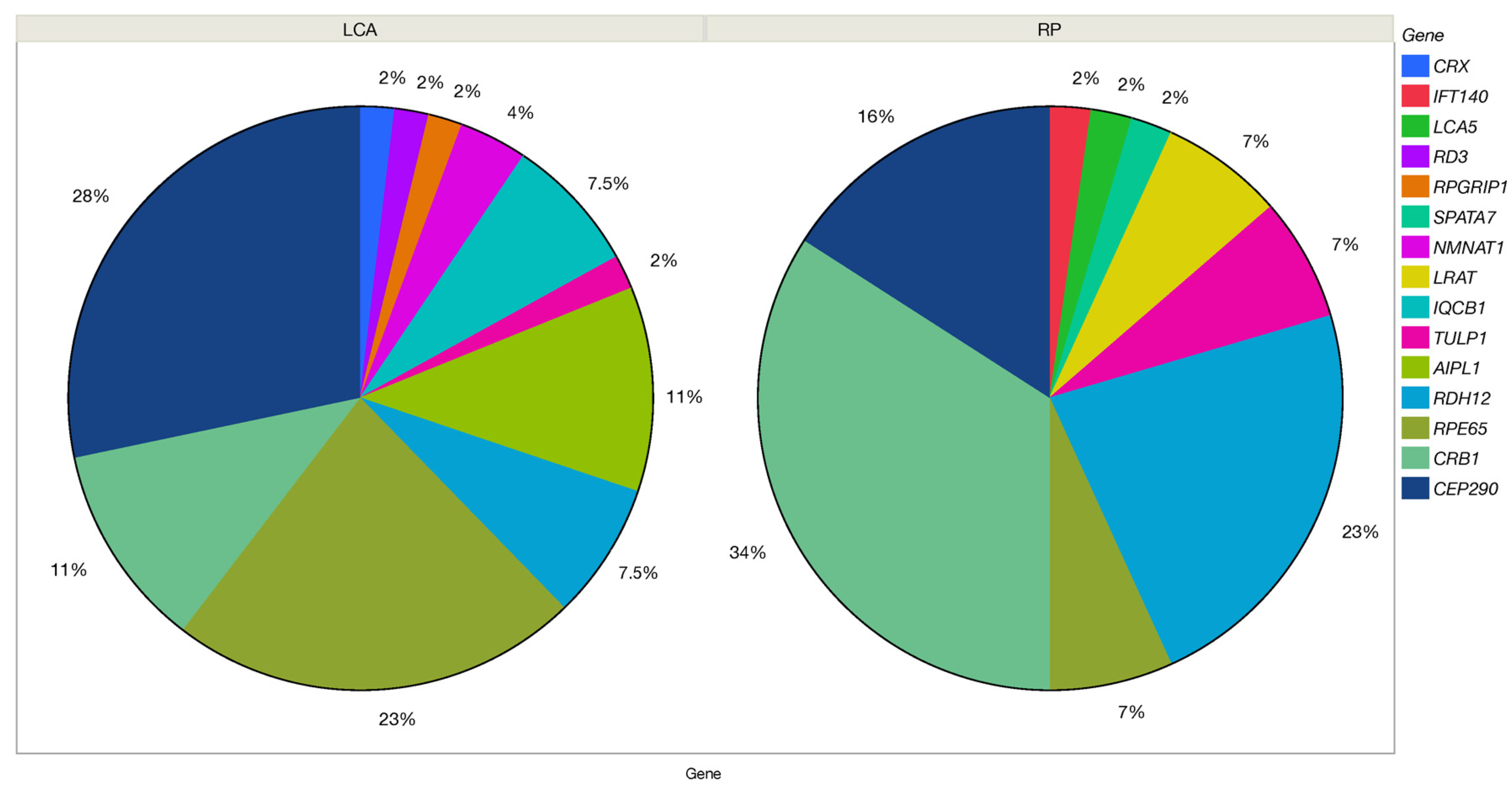
| Clinical Diagnosis | N (Patients) | Relative Frequency | Affected Genes |
| LCA/EOSRD | 56 | 53% | CEP290, RPE65, CRB1, AIPL1, IQCB1, RDH12, LRAT, NMNAT1, CRX, RD3, RPGRIP1 |
| RP | 42 | 40% | CRB1, RDH12, CEP290, TULP1, RPE65, AIPL1, LCA5, SPATA7, IFT140, LRAT |
| CRD | 5 | 5% | TULP1, CRB1, SPATA7, RPGRIP1 |
| CSNB | 2 | 2% | CABP4 |
| Gene | N | Nystagmus | Strabismus | Oculodigital Sign | Cataract | Keratoconus | Optic Nerve Head Drusen | Night Blindness | Photophobia | Color Vision Disturbance |
|---|---|---|---|---|---|---|---|---|---|---|
| N | ||||||||||
| AIPL1 | 6 | 6 | 2 | - | 2 | 1 | - | 2 | 4 | 4 |
| CABP4 | 2 | 1 | 2 | - | - | - | - | 1 | 1 | 2 |
| CEP290 | 22 | 17 | 9 | 4 | 11 | 4 | 3 | 13 | 13 | 12 |
| CRB1 | 22 | 11 | 9 | - | 10 | 1 | 3 | 18 | 15 | 18 |
| CRX | 1 | - | - | - | - | - | - | - | - | - |
| IFT140 | 1 | - | - | - | 1 | - | - | 1 | 1 | - |
| IQCB1 | 5 | 3 | 3 | - | 2 | - | - | 4 | 2 | 4 |
| LCA5 | 1 | - | - | - | - | - | - | 1 | 1 | - |
| LRAT | 3 | 3 | 1 | - | 1 | - | - | 1 | - | 1 |
| NMNAT1 | 2 | 1 | - | - | - | - | - | - | 1 | 1 |
| RD3 | 1 | 1 | 1 | - | - | - | - | - | 1 | 1 |
| RDH12 | 14 | 3 | 10 | - | 9 | - | - | 10 | 7 | 11 |
| RPE65 | 15 | 12 | 6 | 1 | 7 | - | - | 12 | 9 | 11 |
| RPGRIP1 | 2 | 2 | - | - | - | - | - | 1 | 2 | 2 |
| SPATA7 | 2 | 1 | 1 | - | 2 | - | - | 2 | 1 | 2 |
| TULP1 | 6 | 1 | - | - | 1 | - | - | 6 | 3 | 5 |
| General (N; frequency) | 105 | 62 59% | 47 44% | 5 5% | 54 51% | 6 6% | 6 6% | 72 68% | 63 60% | 74 * 70% * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zobor, D.; Brühwiler, B.; Zrenner, E.; Weisschuh, N.; Kohl, S. Genetic and Clinical Profile of Retinopathies Due to Disease-Causing Variants in Leber Congenital Amaurosis (LCA)-Associated Genes in a Large German Cohort. Int. J. Mol. Sci. 2023, 24, 8915. https://doi.org/10.3390/ijms24108915
Zobor D, Brühwiler B, Zrenner E, Weisschuh N, Kohl S. Genetic and Clinical Profile of Retinopathies Due to Disease-Causing Variants in Leber Congenital Amaurosis (LCA)-Associated Genes in a Large German Cohort. International Journal of Molecular Sciences. 2023; 24(10):8915. https://doi.org/10.3390/ijms24108915
Chicago/Turabian StyleZobor, Ditta, Britta Brühwiler, Eberhart Zrenner, Nicole Weisschuh, and Susanne Kohl. 2023. "Genetic and Clinical Profile of Retinopathies Due to Disease-Causing Variants in Leber Congenital Amaurosis (LCA)-Associated Genes in a Large German Cohort" International Journal of Molecular Sciences 24, no. 10: 8915. https://doi.org/10.3390/ijms24108915
APA StyleZobor, D., Brühwiler, B., Zrenner, E., Weisschuh, N., & Kohl, S. (2023). Genetic and Clinical Profile of Retinopathies Due to Disease-Causing Variants in Leber Congenital Amaurosis (LCA)-Associated Genes in a Large German Cohort. International Journal of Molecular Sciences, 24(10), 8915. https://doi.org/10.3390/ijms24108915






