Identification of lncRNAs Deregulated in Epithelial Ovarian Cancer Based on a Gene Expression Profiling Meta-Analysis
Abstract
1. Introduction
2. Results
2.1. Analysis of lncRNAs with Putative Diagnostic Value in EOC
2.2. Analysis of lncRNAs with Putative Prognostic Value in EOC
2.3. Analysis of lncRNAs Deregulated in EOC Metastasis
2.4. Looking for Outstanding lncRNAs Related in Common to Diagnosis, Prognosis, and Metastasis in EOC
2.5. Analysis of lncRNAs with Putative Influence in Resistance to Chemotherapy
2.6. Analysis of lncRNAs with a Putative Specific Value Associated with Histological EOC Subtypes
3. Discussion
4. Materials and Methods
4.1. Selection of Suitable Gene Expression Datasets for Meta-Analysis
4.2. Data Extraction and Processing
4.2.1. Microarray
4.2.2. RNA-Seq
4.3. Data Analysis
4.3.1. Differential Expression in Microarray
4.3.2. Differential Expression in RNA-Seq
4.3.3. Differential Survival in Microarray
4.3.4. Differential Survival in RNA-Seq (GEPIA2)
4.4. Pairwise lncRNA Analysis
4.5. ROC Analysis
4.6. Figures and Venn Diagrams
4.7. Nomenclature
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of Ovarian Cancer: A Review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [CrossRef] [PubMed]
- Ueland, F. A Perspective on Ovarian Cancer Biomarkers: Past, Present and Yet-To-Come. Diagnostics 2017, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, A.; Chen, L. LncRNAs in Ovarian Cancer. Clin. Chim. Acta 2019, 490, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Salamini-Montemurri, M.; Lamas-Maceiras, M.; Barreiro-Alonso, A.; Vizoso-Vázquez, Á.; Rodríguez-Belmonte, E.; Quindós-Varela, M.; Esperanza Cerdán, M. The Challenges and Opportunities of LncRNAs in Ovarian Cancer Research and Clinical Use. Cancers 2020, 12, 1020. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Doxtater, K.; Keramatnia, F.; Zacheaus, C.; Yallapu, M.M.; Jaggi, M.; Chauhan, S.C. Role of LncRNAs in Ovarian Cancer: Defining New Biomarkers for Therapeutic Purposes. Drug Discov. Today 2018, 23, 1635–1643. [Google Scholar] [CrossRef]
- Worku, T.; Bhattarai, D.; Ayers, D.; Wang, K.; Wang, C.; Rehman, Z.; Talpur, H.S.; Yang, L. Long Non-Coding RNAs: The New Horizon of Gene Regulation in Ovarian Cancer. Cell. Physiol. Biochem. 2017, 44, 948–966. [Google Scholar] [CrossRef]
- Negi, A.; Shukla, A.; Jaiswar, A.; Shrinet, J.; Jasrotia, R.S. Applications and Challenges of Microarray and RNA-Sequencing. In Bioinformatics; Elsevier: Amsterdam, The Netherlands, 2022; pp. 91–103. [Google Scholar]
- Pleasance, E.; Bohm, A.; Williamson, L.M.; Nelson, J.M.T.; Shen, Y.; Bonakdar, M.; Titmuss, E.; Csizmok, V.; Wee, K.; Hosseinzadeh, S.; et al. Whole-Genome and Transcriptome Analysis Enhances Precision Cancer Treatment Options. Ann. Oncol. 2022, 33, 939–949. [Google Scholar] [CrossRef]
- Ma, S.-Y.; Wei, P.; Qu, F. KCNMA1-AS1 Attenuates Apoptosis of Epithelial Ovarian Cancer Cells and Serves as a Risk Factor for Poor Prognosis of Epithelial Ovarian Cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4629–4641. [Google Scholar] [CrossRef]
- Shen, X.; Zhu, W. Long Non-coding RNA LINC01627 Is a Prognostic Risk Factor for Epithelial Ovarian Cancer. Oncol. Lett. 2019, 18, 2861–2868. [Google Scholar] [CrossRef]
- Chen, Y.; Bi, F.; An, Y.; Yang, Q. Identification of Pathological Grade and Prognosis-associated LncRNA for Ovarian Cancer. J. Cell. Biochem. 2019, 120, 14444–14454. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tian, Y.; Zheng, H.; Ding, Y.; Wang, X. An Integrated Analysis Reveals the Oncogenic Function of LncRNA LINC00511 in Human Ovarian Cancer. Cancer Med. 2019, 8, 3026–3035. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, Y.; Zhang, Z.; Zou, J.; Li, J.; Wei, R.; Guo, Q.; Zhu, X.; Chu, C.; Fu, X.; et al. Meta-Analysis Based Gene Expression Profiling Reveals Functional Genes in Ovarian Cancer. Biosci. Rep. 2020, 40, BSR20202911. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, Z.; Liang, B.; Chen, S.; Zhang, X.; Tong, X.; Lou, W.; Le, L.; Tang, X.; Fu, F. Identification of Core Genes in Ovarian Cancer by an Integrative Meta-Analysis. J. Ovarian Res. 2018, 11, 94. [Google Scholar] [CrossRef]
- Dong, H.; Hong, S.; Xu, X.; Xiao, Y.; Jin, L.; Xiong, M. Meta-Analysis and Network Analysis of Five Ovarian Cancer Gene Expression Dataset. In Proceedings of the 2010 Third International Joint Conference on Computational Science and Optimization, Huangshan, China, 28–31 May 2010; pp. 242–246. [Google Scholar]
- Fridley, B.L.; Dai, J.; Raghavan, R.; Li, Q.; Winham, S.J.; Hou, X.; Weroha, S.J.; Wang, C.; Kalli, K.R.; Cunningham, J.M.; et al. Transcriptomic Characterization of Endometrioid, Clear Cell, and High-Grade Serous Epithelial Ovarian Carcinoma. Cancer Epidemiol. Biomark. Prev. 2018, 27, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lei, Y.; Li, S.; Li, F.; Lei, J. LncRNA PART1 Stimulates the Development of Ovarian Cancer by Up-Regulating RACGAP1 and RRM2. Reprod. Sci. 2022, 29, 2224–2235. [Google Scholar] [CrossRef]
- Jiang, R.; Zhang, H.; Zhou, J.; Wang, J.; Xu, Y.; Zhang, H.; Gu, Y.; Fu, F.; Shen, Y.; Zhang, G.; et al. Inhibition of Long Non-Coding RNA XIST Upregulates MicroRNA-149-3p to Repress Ovarian Cancer Cell Progression. Cell Death Dis. 2021, 12, 145. [Google Scholar] [CrossRef]
- Wang, H.; Fu, Z.; Dai, C.; Cao, J.; Liu, X.; Xu, J.; Lv, M.; Gu, Y.; Zhang, J.; Hua, X.; et al. LncRNAs Expression Profiling in Normal Ovary, Benign Ovarian Cyst and Malignant Epithelial Ovarian Cancer. Sci. Rep. 2016, 6, 38983. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, T.; Xia, L.; Zhang, M. LncRNA WDFY3-AS2 Promotes Cisplatin Resistance and the Cancer Stem Cell in Ovarian Cancer by Regulating Hsa-MiR-139-5p/SDC4 Axis. Cancer Cell Int. 2021, 21, 284. [Google Scholar] [CrossRef]
- Mao, T.-L.; Fan, M.-H.; Dlamini, N.; Liu, C.-L. LncRNA MALAT1 Facilitates Ovarian Cancer Progression through Promoting Chemoresistance and Invasiveness in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 10201. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, J.; Yang, Y. Long Non-Coding RNA NRSN2-AS1 Facilitates Tumorigenesis and Progression of Ovarian Cancer via MiR-744-5p/PRKX Axis. Biol. Reprod. 2022, 106, 526–539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, G.; Qiu, J.; Zhang, N.; Ding, J.; Hua, K. E2F1-Regulated Long Non-Coding RNA RAD51-AS1 Promotes Cell Cycle Progression, Inhibits Apoptosis and Predicts Poor Prognosis in Epithelial Ovarian Cancer. Sci. Rep. 2017, 7, 4469. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Konno, Y.; Ihira, K.; Kobayashi, N.; Yue, J.; Watari, H. Long Non-Coding RNA DLEU2 Drives EMT and Glycolysis in Endometrial Cancer through HK2 by Competitively Binding with MiR-455 and by Modulating the EZH2/MiR-181a Pathway. J. Exp. Clin. Cancer Res. 2021, 40, 216. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xu, H.; Yang, L.; Zhan, M.; Shi, Y.; Zhang, C.; Gao, D.; Gu, M.; Chen, Y.; Wang, Z. E2F Transcription Factor 2-Activated DLEU2 Contributes to Prostate Tumorigenesis by Upregulating Serum and Glucocorticoid-Induced Protein Kinase 1. Cell Death Dis. 2022, 13, 77. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Zhang, Y.; Yang, X.; Li, X.; Zheng, Y.; Liu, Y.; Zhang, X. N6-Methyladenine- Induced LINC00667 Promoted Breast Cancer Progression through M6A/KIAA1429 Positive Feedback Loop. Bioengineered 2022, 13, 13462–13473. [Google Scholar] [CrossRef]
- Malek, E.; Kim, B.; Driscoll, J. Identification of Long Non-Coding RNAs Deregulated in Multiple Myeloma Cells Resistant to Proteasome Inhibitors. Genes 2016, 7, 84. [Google Scholar] [CrossRef]
- Gilks, C.B.; Prat, J. Ovarian Carcinoma Pathology and Genetics: Recent Advances. Hum. Pathol. 2009, 40, 1213–1223. [Google Scholar] [CrossRef]
- Lee, C.M.; Lo, H.-W.; Shao, R.-P.; Wang, S.-C.; Xia, W.; Gershenson, D.M.; Hung, M.-C. Selective Activation of Ceruloplasmin Promoter in Ovarian Tumors. Cancer Res. 2004, 64, 1788–1793. [Google Scholar] [CrossRef]
- Tarhriz, V.; Bandehpour, M.; Dastmalchi, S.; Ouladsahebmadarek, E.; Zarredar, H.; Eyvazi, S. Overview of CD24 as a New Molecular Marker in Ovarian Cancer. J. Cell. Physiol. 2019, 234, 2134–2142. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, M.; Cai, Y.; Rong, Z.; Wang, C.; Xu, Z.; Xu, H.; Song, W.; Hou, Y.; Lou, G. Platelet-derived Growth Factor-D Expression Mediates the Effect of Differentiated Degree on Prognosis in Epithelial Ovarian Cancer. J. Cell. Biochem. 2019, 120, 6920–6925. [Google Scholar] [CrossRef]
- Xu, P.; Xu, S.; Pan, H.; Dai, C.; Xu, Y.; Wang, L.; Cong, Y.; Zhang, H.; Cao, J.; Ge, L.; et al. Differential Effects of the LncRNA RNF157-AS1 on Epithelial Ovarian Cancer Cells through Suppression of DIRAS3- and ULK1-Mediated Autophagy. Cell Death Dis. 2023, 14, 140. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Spindler, T.J.; de Souza Fonseca, M.A.; Corona, R.I.; Seo, J.H.; Dezem, F.S.; Li, L.; Lee, J.M.; Long, H.W.; Sellers, T.A.; et al. Super-Enhancer-Associated LncRNA UCA1 Interacts Directly with AMOT to Activate YAP Target Genes in Epithelial Ovarian Cancer. iScience 2019, 17, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Chen, R.; Yang, Y.; Jiang, J. LncRNA BBOX1-AS1 Aggravates the Development of Ovarian Cancer by Sequestering MiR-361-3p to Augment PODXL Expression. Reprod. Sci. 2021, 28, 736–744. [Google Scholar] [CrossRef]
- Li, L.-M.; Hao, S.-J.; Ni, M.; Jin, S.; Tian, Y.-Q. DUXAP8 Promotes the Proliferation and Migration of Ovarian Cancer Cells via Down-Regulating MicroRNA-29a-3p Expression. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1837–1844. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhai, Y.; Chen, Y. GATA1-Induced Upregulation of LINC01503 Promotes Carboplatin Resistance in Ovarian Carcinoma by Upregulating PD-L1 via Sponging MiR-766-5p. J. Ovarian Res. 2021, 14, 108. [Google Scholar] [CrossRef]
- Zhang, Y.; Dun, Y.; Zhou, S.; Huang, X.-H. LncRNA HOXD-AS1 Promotes Epithelial Ovarian Cancer Cells Proliferation and Invasion by Targeting MiR-133a-3p and Activating Wnt/β-Catenin Signaling Pathway. Biomed. Pharmacother. 2017, 96, 1216–1221. [Google Scholar] [CrossRef]
- Xu, D.; Song, Q.; Liu, Y.; Chen, W.; LU, L.; Xu, M.; Fang, X.; Zhao, W.; Zhou, H. LINC00665 Promotes Ovarian Cancer Progression through Regulating the MiRNA-34a-5p/E2F3 Axis. J. Cancer 2021, 12, 1755–1763. [Google Scholar] [CrossRef]
- Zhu, L.; Mei, M. Interference of Long Non-coding RNA HAGLROS Inhibits the Proliferation and Promotes the Apoptosis of Ovarian Cancer Cells by Targeting MiR-26b-5p. Exp. Ther. Med. 2021, 22, 879. [Google Scholar] [CrossRef]
- Gokulnath, P.; de Cristofaro, T.; Manipur, I.; Di Palma, T.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long Non-Coding RNA MAGI2-AS3 Is a New Player with a Tumor Suppressive Role in High Grade Serous Ovarian Carcinoma. Cancers 2019, 11, 2008. [Google Scholar] [CrossRef]
- Gokulnath, P.; de Cristofaro, T.; Manipur, I.; Di Palma, T.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long Non-Coding RNA HAND2-AS1 Acts as a Tumor Suppressor in High-Grade Serous Ovarian Carcinoma. Int. J. Mol. Sci. 2020, 21, 4059. [Google Scholar] [CrossRef]
- Gloss, B.; Moran-Jones, K.; Lin, V.; Gonzalez, M.; Scurry, J.; Hacker, N.F.; Sutherland, R.L.; Clark, S.J.; Samimi, G. ZNF300P1 Encodes a LincRNA That Regulates Cell Polarity and Is Epigenetically Silenced in Type II Epithelial Ovarian Cancer. Mol. Cancer 2014, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Kamikihara, T.; Arima, T.; Kato, K.; Matsuda, T.; Kato, H.; Douchi, T.; Nagata, Y.; Nakao, M.; Wake, N. Epigenetic Silencing of the Imprinted GeneZAC by DNA Methylation Is an Early Event in the Progression of Human Ovarian Cancer. Int. J. Cancer 2005, 115, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Profumo, V.; Forte, B.; Percio, S.; Rotundo, F.; Doldi, V.; Ferrari, E.; Fenderico, N.; Dugo, M.; Romagnoli, D.; Benelli, M.; et al. LEADeR Role of MiR-205 Host Gene as Long Noncoding RNA in Prostate Basal Cell Differentiation. Nat. Commun. 2019, 10, 307. [Google Scholar] [CrossRef] [PubMed]
- Di Agostino, S.; Valenti, F.; Sacconi, A.; Fontemaggi, G.; Pallocca, M.; Pulito, C.; Ganci, F.; Muti, P.; Strano, S.; Blandino, G. Long Non-Coding MIR205HG Depletes Hsa-MiR-590-3p Leading to Unrestrained Proliferation in Head and Neck Squamous Cell Carcinoma. Theranostics 2018, 8, 1850–1868. [Google Scholar] [CrossRef]
- Du, Q.; Hoover, A.R.; Dozmorov, I.; Raj, P.; Khan, S.; Molina, E.; Chang, T.-C.; de la Morena, M.T.; Cleaver, O.B.; Mendell, J.T.; et al. MIR205HG Is a Long Noncoding RNA That Regulates Growth Hormone and Prolactin Production in the Anterior Pituitary. Dev. Cell 2019, 49, 618–631.e5. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; Zhang, R.; Li, C.; Xiong, J.; Wei, Y. MIR205HG Acts as a CeRNA to Expedite Cell Proliferation and Progression in Lung Squamous Cell Carcinoma via Targeting MiR-299-3p/MAP3K2 Axis. BMC Pulm. Med. 2020, 20, 163. [Google Scholar] [CrossRef]
- Wang, X.; Yu, X.; Long, X.; Pu, Q. MIR205 Host Gene (MIR205HG) Drives Osteosarcoma Metastasis via Regulating the MicroRNA 2114-3p (MiR-2114-3p)/Twist Family BHLH Transcription Factor 2 (TWIST2) Axis. Bioengineered 2021, 12, 1576–1586. [Google Scholar] [CrossRef]
- Guo, J.; Gan, Q.; Gan, C.; Zhang, X.; Ma, X.; Dong, M. LncRNA MIR205HG Regulates Melanomagenesis via the MiR-299-3p/VEGFA Axis. Aging 2021, 13, 5297–5311. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, Y.; Zheng, L. Analysis of Differentially Expressed Long Non-coding RNAs Revealed a Pro-tumor Role of MIR205HG in Cervical Cancer. Mol. Med. Rep. 2021, 25, 42. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Huang, H. Long Non-Coding RNA MIR205HG Function as a CeRNA to Accelerate Tumor Growth and Progression via Sponging MiR-122–5p in Cervical Cancer. Biochem. Biophys. Res. Commun. 2019, 514, 78–85. [Google Scholar] [CrossRef]
- Hongle, L.; Jia, J.; Yang, L.; Chu, J.; Sheng, J.; Wang, C.; Meng, W.; Jia, Z.; Yin, H.; Wan, J.; et al. LncRNA MIR205HG Drives Esophageal Squamous Cell Carcinoma Progression by Regulating MiR-214/SOX4 Axis. OncoTargets Ther. 2020, 13, 13097–13109. [Google Scholar]
- Dong, X.; Chen, X.; Lu, D.; Diao, D.; Liu, X.; Mai, S.; Feng, S.; Xiong, G. LncRNA MiR205HG Hinders HNRNPA0 Translation: Anti-oncogenic Effects in Esophageal Carcinoma. Mol. Oncol. 2022, 16, 795–812. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Tieu, A.H.; Cheng, Y.; Ma, K.; Akshintala, V.S.; Simsek, C.; Prasath, V.; Shin, E.J.; Ngamruengphong, S.; Khashab, M.A.; et al. Novel Long Noncoding RNA MiR205HG Functions as an Esophageal Tumor-Suppressive Hedgehog Inhibitor. Cancers 2021, 13, 1707. [Google Scholar] [CrossRef] [PubMed]
- Bezzecchi, E.; Pagani, G.; Forte, B.; Percio, S.; Zaffaroni, N.; Dolfini, D.; Gandellini, P. MIR205HG/LEADR Long Noncoding RNA Binds to Primed Proximal Regulatory Regions in Prostate Basal Cells Through a Triplex- and Alu-Mediated Mechanism. Front. Cell Dev. Biol. 2022, 10, 1293. [Google Scholar] [CrossRef]
- Hao, C.; Lin, S.; Liu, P.; Liang, W.; Li, Z.; Li, Y. Potential Serum Metabolites and Long-chain Noncoding RNA Biomarkers for Endometrial Cancer Tissue. J. Obstet. Gynaecol. Res. 2023, 49, 725–743. [Google Scholar] [CrossRef]
- Zhang, T.; Xia, W.; Song, X.; Mao, Q.; Huang, X.; Chen, B.; Liang, Y.; Wang, H.; Chen, Y.; Yu, X.; et al. Super-Enhancer Hijacking LINC01977 Promotes Malignancy of Early-Stage Lung Adenocarcinoma Addicted to the Canonical TGF-β/SMAD3 Pathway. J. Hematol. Oncol. 2022, 15, 114. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Wang, X.; Liang, Y.; Luo, D.; Han, D.; Li, C.; Chen, T.; Zhang, H.; Liu, Y.; et al. LINC01977 Promotes Breast Cancer Progression and Chemoresistance to Doxorubicin by Targeting MiR-212-3p/GOLM1 Axis. Front. Oncol. 2021, 11, 657094. [Google Scholar] [CrossRef]
- Van Grembergen, O.; Bizet, M.; de Bony, E.J.; Calonne, E.; Putmans, P.; Brohée, S.; Olsen, C.; Guo, M.; Bontempi, G.; Sotiriou, C.; et al. Portraying Breast Cancers with Long Noncoding RNAs. Sci. Adv. 2016, 2, e1600220. [Google Scholar] [CrossRef]
- Hu, J.; Wang, T.; Chen, Q. Competitive Endogenous RNA Network Identifies Four Long Non-Coding RNA Signature as a Candidate Prognostic Biomarker for Lung Adenocarcinoma. Transl. Cancer Res. 2019, 8, 1046–1064. [Google Scholar] [CrossRef]
- Seitz, A.K.; Christensen, L.L.; Christensen, E.; Faarkrog, K.; Ostenfeld, M.S.; Hedegaard, J.; Nordentoft, I.; Nielsen, M.M.; Palmfeldt, J.; Thomson, M.; et al. Profiling of Long Non-Coding RNAs Identifies LINC00958 and LINC01296 as Candidate Oncogenes in Bladder Cancer. Sci. Rep. 2017, 7, 395. [Google Scholar] [CrossRef]
- Li, L.; Peng, Q.; Gong, M.; Ling, L.; Xu, Y.; Liu, Q. Using LncRNA Sequencing to Reveal a Putative LncRNA-MRNA Correlation Network and the Potential Role of PCBP1-AS1 in the Pathogenesis of Cervical Cancer. Front. Oncol. 2021, 11, 634732. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Guo, H.; Tang, J. Long Non-Coding RNA TFAP2A-AS1 Inhibits Cell Proliferation and Invasion in Breast Cancer via MiR-933/SMAD2. Med. Sci. Monit. 2019, 25, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, L.; Wu, J.; You, J.; Hong, Q.; Ye, F. Transcription Factor KLF15 Inhibits the Proliferation and Migration of Gastric Cancer Cells via Regulating the TFAP2A-AS1/NISCH Axis. Biol. Direct 2021, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Hui, B.; Lu, C.; Wang, J.; Xu, Y.; Yang, Y.; Ji, H.; Li, X.; Xu, L.; Wang, J.; Tang, W.; et al. Engineered Exosomes for Co-delivery of PGM5-AS1 and Oxaliplatin to Reverse Drug Resistance in Colon Cancer. J. Cell. Physiol. 2022, 237, 911–933. [Google Scholar] [CrossRef]
- Liu, W.; Liu, P.; Gao, H.; Wang, X.; Yan, M. Long Non-coding RNA PGM5-AS1 Promotes Epithelial-mesenchymal Transition, Invasion and Metastasis of Osteosarcoma Cells by Impairing MiR-140-5p-mediated FBN1 Inhibition. Mol. Oncol. 2020, 14, 2660–2677. [Google Scholar] [CrossRef]
- Liu, J.-Q.; Deng, M.; Xue, N.-N.; Li, T.-X.; Guo, Y.-X.; Gao, L.; Zhao, D.; Fan, R.-T. LncRNA KLF3-AS1 Suppresses Cell Migration and Invasion in ESCC by Impairing MiR-185-5p-Targeted KLF3 Inhibition. Mol. Ther.-Nucleic Acids 2020, 20, 231–241. [Google Scholar] [CrossRef]
- Jiang, H.; Hu, K.; Xia, Y.; Liang, L.; Zhu, X. Long Noncoding RNA KLF3-AS1 Acts as an Endogenous RNA of MiR-223 to Attenuate Gastric Cancer Progression and Chemoresistance. Front. Oncol. 2021, 11, 704339. [Google Scholar] [CrossRef]
- Chen, C.; Liu, L. Silencing of LncRNA KLF3-AS1 Represses Cell Growth in Osteosarcoma via MiR-338-3p/MEF2C Axis. J. Clin. Lab. Anal. 2022, 36, e24698. [Google Scholar] [CrossRef]
- Jin, L.; Chen, C.; Huang, L.; Sun, Q.; Bu, L. Long Noncoding RNA NR2F1-AS1 Stimulates the Tumorigenic Behavior of Non-Small Cell Lung Cancer Cells by Sponging MiR-363-3p to Increase SOX4. Open Med. 2021, 17, 87–95. [Google Scholar] [CrossRef]
- Guo, F.; Fu, Q.; Wang, Y.; Sui, G. Long Non-coding RNA NR2F1-AS1 Promoted Proliferation and Migration yet Suppressed Apoptosis of Thyroid Cancer Cells through Regulating MiRNA-338-3p/CCND1 Axis. J. Cell. Mol. Med. 2019, 23, 5907–5919. [Google Scholar] [CrossRef]
- Luo, D.; Liu, Y.; Li, Z.; Zhu, H.; Yu, X. NR2F1-AS1 Promotes Pancreatic Ductal Adenocarcinoma Progression Through Competing Endogenous RNA Regulatory Network Constructed by Sponging MiRNA-146a-5p/MiRNA-877-5p. Front. Cell Dev. Biol. 2021, 9, 736980. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chen, J.; Ding, C.; Jin, X.; Jia, Z.; Peng, J. LncRNA NR2F1-AS1 Regulates Hepatocellular Carcinoma Oxaliplatin Resistance by Targeting ABCC1 via MiR-363. J. Cell. Mol. Med. 2018, 22, 3238–3245. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, S.; Cai, K.; Zheng, D.; Zhu, C.; Li, L.; Wang, F.; He, Z.; Yu, C.; Sun, C. Hypoxia-Induced Long Noncoding RNA NR2F1-AS1 Maintains Pancreatic Cancer Proliferation, Migration, and Invasion by Activating the NR2F1/AKT/MTOR Axis. Cell Death Dis. 2022, 13, 232. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, P.; Wu, Q.; Fang, H.; Wang, Y.; Xiao, Y.; Cong, M.; Wang, T.; He, Y.; Ma, C.; et al. Long Non-Coding RNA NR2F1-AS1 Induces Breast Cancer Lung Metastatic Dormancy by Regulating NR2F1 and ΔNp63. Nat. Commun. 2021, 12, 5232. [Google Scholar] [CrossRef]
- Yu, Q.; Li, X.; Feng, T. GLIDR Promotes the Progression of Glioma by Regulating the MiR-4677-3p/MAGI2 Axis. Exp. Cell Res. 2021, 406, 112726. [Google Scholar] [CrossRef] [PubMed]
- Tai, G.; Fu, H.; Bai, H.; Liu, H.; Li, L.; Song, T. Long Non-Coding RNA GLIDR Accelerates the Tumorigenesis of Lung Adenocarcinoma by MiR-1270/TCF12 Axis. Cell Cycle 2021, 20, 1653–1662. [Google Scholar] [CrossRef]
- Li, S.; Wang, Y.; Cao, Q.; Li, H.; Zhao, Z.; Wei, B.; Yuan, H.; Chen, Z.; Yang, S. GLIDR Promotes the Aggressiveness Progression of Prostate Cancer Cells by Sponging MiR-128–3p. Pathol.-Res. Pract. 2023, 242, 154343. [Google Scholar] [CrossRef]
- Xie, H.; Dai, L.; Ye, B.; Chen, R.; Wang, B.; Zhang, N.; Miao, H.; Liang, W. Long Non-Coding RNA ERVK13-1 Aggravates Osteosarcoma through the Involvement of MicroRNA-873-5p/KLF5 Axis. Acta Biochim. Pol. 2022, 69, 703–710. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, Z.; Wang, X.; Zhang, Q.; Lyu, L.; Tang, B. The Predictive Competing Endogenous RNA Regulatory Networks and Potential Prognostic and Immunological Roles of Cyclin A2 in Pan-Cancer Analysis. Front. Mol. Biosci. 2022, 9, 809509. [Google Scholar] [CrossRef]
- Cao, W.; Liu, J.; Liu, Z.; Wang, X.; Han, Z.-G.; Ji, T.; Chen, W.; Zou, X. A Three-LncRNA Signature Derived from the Atlas of NcRNA in Cancer (TANRIC) Database Predicts the Survival of Patients with Head and Neck Squamous Cell Carcinoma. Oral Oncol. 2017, 65, 94–101. [Google Scholar] [CrossRef]
- Cai, J.; Xie, H.; Yan, Y.; Huang, Z.; Tang, P.; Cao, X.; Wang, Z.; Yang, C.; Wen, J.; Tan, M.; et al. A Novel Cuproptosis-Related LncRNA Signature Predicts Prognosis and Therapeutic Response in Bladder Cancer. Front. Genet. 2023, 13, 1082691. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Liao, X.; Zhang, Y.; Xu, B.; Song, Y.; Bian, G.; Fu, X. Anti-Tumor Role of CAMK2B in Remodeling the Stromal Microenvironment and Inhibiting Proliferation in Papillary Renal Cell Carcinoma. Front. Oncol. 2022, 12, 29. [Google Scholar] [CrossRef]
- Lyu, H.; Zhang, J.; Wei, Q.; Huang, Y.; Zhang, R.; Xiao, S.; Guo, D.; Chen, X.-Z.; Zhou, C.; Tang, J. Identification of Wnt/β-Catenin- and Autophagy-Related LncRNA Signature for Predicting Immune Efficacy in Pancreatic Adenocarcinoma. Biology 2023, 12, 319. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, X.; Yang, B.; Xia, Z.; Chen, Q. MiR-924 as a Tumor Suppressor Inhibits Non-Small Cell Lung Cancer by Inhibiting RHBDD1/Wnt/β-Catenin Signaling Pathway. Cancer Cell Int. 2020, 20, 491. [Google Scholar] [CrossRef]
- Fan, H.; Lv, P.; Mu, T.; Zhao, X.; Liu, Y.; Feng, Y.; Lv, J.; Liu, M.; Tang, H. LncRNA N335586/MiR-924/CKMT1A Axis Contributes to Cell Migration and Invasion in Hepatocellular Carcinoma Cells. Cancer Lett. 2018, 429, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-H.; Shi, Y.-F.; Cheng, Q.; Deng, L. Expression and Localization of Aquaporin-5 in the Epithelial Ovarian Tumors. Gynecol. Oncol. 2006, 100, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Zhu, Y.; Zhang, X.; Chen, X.; Zheng, W.; Yang, J. Down-Regulated Aquaporin 5 Inhibits Proliferation and Migration of Human Epithelial Ovarian Cancer 3AO Cells. J. Ovarian Res. 2014, 7, 78. [Google Scholar] [CrossRef]
- Chetry, M.; Li, S.; Liu, H.; Hu, X.; Zhu, X. Prognostic Values of Aquaporins MRNA Expression in Human Ovarian Cancer. Biosci. Rep. 2018, 38, BSR20180108. [Google Scholar] [CrossRef]
- Wei, W.; Xu, T.; Zhang, Y.; Huang, Y.; Wang, X. Upregulation of Long Noncoding RNA Linc02544 and Its Association with Overall Survival Rate and the Influence on Cell Proliferation and Migration in Lung Squamous Cell Carcinoma. Discov. Oncol. 2022, 13, 41. [Google Scholar] [CrossRef]
- Guo, Z.-H.; Yao, L.-T.; Guo, A.-Y. Clinical and Biological Impact of LINC02544 Expression in Breast Cancer after Neoadjuvant Chemotherapy. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10573–10585. [Google Scholar]
- Zhang, C.; Liang, Y.; Zhang, C.-D.; Pei, J.-P.; Wu, K.-Z.; Li, Y.-Z.; Dai, D.-Q. The Novel Role and Function of LINC01235 in Metastasis of Gastric Cancer Cells by Inducing Epithelial-Mesenchymal Transition. Genomics 2021, 113, 1504–1513. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.-E.; Xing, Y.; Ran, B.-L.; Zhang, C.; Pan, S.-W.; An, W.; Chen, Q.-C.; Xu, H.-M. LINC01235-TWIST2 Feedback Loop Facilitates Epithelial–Mesenchymal Transition in Gastric Cancer by Inhibiting THBS2. Aging 2020, 12, 25060–25075. [Google Scholar] [CrossRef]
- Tu, C.; Ren, X.; He, J.; Li, S.; Qi, L.; Duan, Z.; Wang, W.; Li, Z. The Predictive Value of LncRNA MIR31HG Expression on Clinical Outcomes in Patients with Solid Malignant Tumors. Cancer Cell Int. 2020, 20, 115. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Hu, D.; Yang, P.; Li, Y.; Bashir, S.; Nai, A.; Ma, F.; Jia, G.; Xu, M. A Novel Cuproptosis-Related Prognostic LncRNA Signature and LncRNA MIR31HG/MiR-193a-3p/TNFRSF21 Regulatory Axis in Lung Adenocarcinoma. Front. Oncol. 2022, 12, 927706. [Google Scholar] [CrossRef]
- Wang, R.; Ma, Z.; Feng, L.; Yang, Y.; Tan, C.; Shi, Q.; Lian, M.; He, S.; Ma, H.; Fang, J. LncRNA MIR31HG Targets HIF1A and P21 to Facilitate Head and Neck Cancer Cell Proliferation and Tumorigenesis by Promoting Cell-Cycle Progression. Mol. Cancer 2018, 17, 162. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.-W.; Hung, W.-W.; Chou, C.-H.; Tu, H.-F.; Chang, S.-R.; Liu, Y.-C.; Liu, C.-J.; Lin, S.-C. LncRNA MIR31HG Drives Oncogenicity by Inhibiting the Limb-Bud and Heart Development Gene (LBH) during Oral Carcinoma. Int. J. Mol. Sci. 2021, 22, 8383. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, B.; Cao, J.; Zhao, L.; Wang, G. MIR31HG Expression Predicts Poor Prognosis and Promotes Colorectal Cancer Progression. Cancer Manag. Res. 2022, 14, 1973–1986. [Google Scholar] [CrossRef]
- Zheng, S.; Zhang, X.; Wang, X.; Li, J. MIR31HG Promotes Cell Proliferation and Invasion by Activating the Wnt/Β-catenin Signaling Pathway in Non-small Cell Lung Cancer. Oncol. Lett. 2018, 17, 221–229. [Google Scholar] [CrossRef]
- Ghorbanzadeh, S.; Poor-Ghassem, N.; Afsa, M.; Nikbakht, M.; Malekzadeh, K. Long Non-Coding RNA NR2F2-AS1: Its Expanding Oncogenic Roles in Tumor Progression. Hum. Cell 2022, 35, 1355–1363. [Google Scholar] [CrossRef]
- Hawkins, S.M.; Loomans, H.A.; Wan, Y.-W.; Ghosh-Choudhury, T.; Coffey, D.; Xiao, W.; Liu, Z.; Sangi-Haghpeykar, H.; Anderson, M.L. Expression and Functional Pathway Analysis of Nuclear Receptor NR2F2 in Ovarian Cancer. J. Clin. Endocrinol. Metab. 2013, 98, E1152–E1162. [Google Scholar] [CrossRef]
- Zheng, J.; Qin, W.; Jiao, D.; Ren, J.; Wei, M.; Shi, S.; Xi, W.; Wang, H.; Yang, A.-G.; Huan, Y.; et al. Knockdown of COUP-TFII Inhibits Cell Proliferation and Induces Apoptosis through Upregulating BRCA1 in Renal Cell Carcinoma Cells. Int. J. Cancer 2016, 139, 1574–1585. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Nomura, N.; Mori, H.; Wake, N. Poor Correlation with Loss of Heterozygosity on Chromosome 17p and P53 Mutations in Ovarian Cancers. Gynecol. Oncol. 1996, 63, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Phillips, N.J.; Ziegler, M.R.; Radford, D.M.; Fair, K.L.; Steinbrueck, T.; Xynos, F.P.; Donis-Keller, H. Allelic Deletion on Chromosome 17p13.3 in Early Ovarian Cancer. Cancer Res. 1996, 56, 606–611. [Google Scholar] [PubMed]
- Wei, M.; Liu, L.; Wang, Z. Long Non-coding RNA Heart and Neural Crest Derivatives Expressed 2-antisense RNA 1 Overexpression Inhibits the Proliferation of Cancer Cells by Reducing RUNX2 Expression in Triple-negative Breast Cancer. Oncol. Lett. 2019, 18, 6775–6780. [Google Scholar] [CrossRef]
- Mortlock, S.; Corona, R.I.; Kho, P.F.; Pharoah, P.; Seo, J.-H.; Freedman, M.L.; Gayther, S.A.; Siedhoff, M.T.; Rogers, P.A.W.; Leuchter, R.; et al. A Multi-Level Investigation of the Genetic Relationship between Endometriosis and Ovarian Cancer Histotypes. Cell Rep. Med. 2022, 3, 100542. [Google Scholar] [CrossRef]
- Li, Z.; Niu, H.; Qin, Q.; Yang, S.; Wang, Q.; Yu, C.; Wei, Z.; Jin, Z.; Wang, X.; Yang, A.; et al. LncRNA UCA1 Mediates Resistance to Cisplatin by Regulating the MiR-143/FOSL2-Signaling Pathway in Ovarian Cancer. Mol. Ther.-Nucleic Acids 2019, 17, 92–101. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Z.; Pan, D.; Xin, Z.; Bu, F.; Zhang, Y.; Tian, Q.; Feng, X. Circulating LncRNA UCA1 and LncRNA PGM5-AS1 Act as Potential Diagnostic Biomarkers for Early-Stage Colorectal Cancer. Biosci. Rep. 2021, 41, BSR20211115. [Google Scholar] [CrossRef]
- Filippov-Levy, N.; Cohen-Schussheim, H.; Tropé, C.G.; Hetland Falkenthal, T.E.; Smith, Y.; Davidson, B.; Reich, R. Expression and Clinical Role of Long Non-Coding RNA in High-Grade Serous Carcinoma. Gynecol. Oncol. 2018, 148, 559–566. [Google Scholar] [CrossRef]
- Zheng, R.; Liang, J.; Lu, J.; Li, S.; Zhang, G.; Wang, X.; Liu, M.; Wang, W.; Chu, H.; Tao, G.; et al. Genome-Wide Long Non-Coding RNAs Identified a Panel of Novel Plasma Biomarkers for Gastric Cancer Diagnosis. Gastric Cancer 2019, 22, 731–741. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Mudunuri, U.; Che, A.; Yi, M.; Stephens, R.M. BioDBnet: The Biological Database Network. Bioinformatics 2009, 25, 555–556. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-Generation Characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Goksuluk, D.; Korkmaz, S.; Zararsiz, G.; Karaagaoglu, A.E. EasyROC: An Interactive Web-Tool for ROC Curve Analysis Using R Language Environment. R J. 2016, 8, 213. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. GENCODE 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- Tone, A.A.; Begley, H.; Sharma, M.; Murphy, J.; Rosen, B.; Brown, T.J.; Shaw, P.A. Gene Expression Profiles of Luteal Phase Fallopian Tube Epithelium from BRCA Mutation Carriers Resemble High-Grade Serous Carcinoma. Clin. Cancer Res. 2008, 14, 4067–4078. [Google Scholar] [CrossRef]
- Tung, C.S.; Mok, S.C.; Tsang, Y.T.M.; Zu, Z.; Song, H.; Liu, J.; Deavers, M.T.; Malpica, A.; Wolf, J.K.; Lu, K.H.; et al. PAX2 Expression in Low Malignant Potential Ovarian Tumors and Low-Grade Ovarian Serous Carcinomas. Mod. Pathol. 2009, 22, 1243–1250. [Google Scholar] [CrossRef]
- Bowen, N.J.; Walker, L.D.; Matyunina, L.V.; Logani, S.; Totten, K.A.; Benigno, B.B.; McDonald, J.F. Gene Expression Profiling Supports the Hypothesis That Human Ovarian Surface Epithelia Are Multipotent and Capable of Serving as Ovarian Cancer Initiating Cells. BMC Med. Genom. 2009, 2, 71. [Google Scholar] [CrossRef]
- Mok, S.C.; Bonome, T.; Vathipadiekal, V.; Bell, A.; Johnson, M.E.; Wong, K.; Park, D.-C.; Hao, K.; Yip, D.K.P.; Donninger, H.; et al. A Gene Signature Predictive for Outcome in Advanced Ovarian Cancer Identifies a Survival Factor: Microfibril-Associated Glycoprotein 2. Cancer Cell 2009, 16, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Bonome, T.; Levine, D.A.; Shih, J.; Randonovich, M.; Pise-Masison, C.A.; Bogomolniy, F.; Ozbun, L.; Brady, J.; Barrett, J.C.; Boyd, J.; et al. A Gene Signature Predicting for Survival in Suboptimally Debulked Patients with Ovarian Cancer. Cancer Res. 2008, 68, 5478–5486. [Google Scholar] [CrossRef] [PubMed]
- Stany, M.P.; Vathipadiekal, V.; Ozbun, L.; Stone, R.L.; Mok, S.C.; Xue, H.; Kagami, T.; Wang, Y.; McAlpine, J.N.; Bowtell, D.; et al. Identification of Novel Therapeutic Targets in Microdissected Clear Cell Ovarian Cancers. PLoS ONE 2011, 6, e21121. [Google Scholar] [CrossRef] [PubMed]
- Lili, L.N.; Matyunina, L.V.; Walker, L.D.; Benigno, B.B.; McDonald, J.F. Molecular Profiling Predicts the Existence of Two Functionally Distinct Classes of Ovarian Cancer Stroma. Biomed Res. Int. 2013, 2013, 846387. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.-L.; Leung, C.S.; Wong, K.-K.; Samimi, G.; Thompson, M.S.; Liu, J.; Zaid, T.M.; Ghosh, S.; Birrer, M.J.; Mok, S.C. TGF-β Modulates Ovarian Cancer Invasion by Upregulating CAF-Derived Versican in the Tumor Microenvironment. Cancer Res. 2013, 73, 5016–5028. [Google Scholar] [CrossRef]
- Hill, C.G.; Matyunina, L.V.; Walker, D.; Benigno, B.B.; McDonald, J.F. Transcriptional Override: A Regulatory Network Model of Indirect Responses to Modulations in MicroRNA Expression. BMC Syst. Biol. 2014, 8, 36. [Google Scholar] [CrossRef]
- Yeung, T.-L.; Leung, C.S.; Wong, K.-K.; Gutierrez-Hartmann, A.; Kwong, J.; Gershenson, D.M.; Mok, S.C. ELF3 Is a Negative Regulator of Epithelial-Mesenchymal Transition in Ovarian Cancer Cells. Oncotarget 2017, 8, 16951–16963. [Google Scholar] [CrossRef]
- Dong, S.; Wang, R.; Wang, H.; Ding, Q.; Zhou, X.; Wang, J.; Zhang, K.; Long, Y.; Lu, S.; Hong, T.; et al. HOXD-AS1 Promotes the Epithelial to Mesenchymal Transition of Ovarian Cancer Cells by Regulating MiR-186-5p and PIK3R3. J. Exp. Clin. Cancer Res. 2019, 38, 110. [Google Scholar] [CrossRef]
- Mitra, S.; Tiwari, K.; Podicheti, R.; Pandhiri, T.; Rusch, D.B.; Bonetto, A.; Zhang, C.; Mitra, A.K. Transcriptome Profiling Reveals Matrisome Alteration as a Key Feature of Ovarian Cancer Progression. Cancers 2019, 11, 1513. [Google Scholar] [CrossRef]
- Shahab, S.W.; Matyunina, L.V.; Mezencev, R.; Walker, L.D.; Bowen, N.J.; Benigno, B.B.; McDonald, J.F. Evidence for the Complexity of MicroRNA-Mediated Regulation in Ovarian Cancer: A Systems Approach. PLoS ONE 2011, 6, e22508. [Google Scholar] [CrossRef]
- King, E.R.; Tung, C.S.; Tsang, Y.T.M.; Zu, Z.; Lok, G.T.M.; Deavers, M.T.; Malpica, A.; Wolf, J.K.; Lu, K.H.; Birrer, M.J.; et al. The Anterior Gradient Homolog 3 (AGR3) Gene Is Associated With Differentiation and Survival in Ovarian Cancer. Am. J. Surg. Pathol. 2011, 35, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Lee, J.; Haemmerle, M.; Ling, H.; Previs, R.A.; Pradeep, S.; Wu, S.Y.; Ivan, C.; Ferracin, M.; Dennison, J.B.; et al. Long Noncoding RNA Ceruloplasmin Promotes Cancer Growth by Altering Glycolysis. Cell Rep. 2015, 13, 2395–2402. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Hendrix-Lucas, N.; Kuick, R.; Zhai, Y.; Schwartz, D.R.; Akyol, A.; Hanash, S.; Misek, D.E.; Katabuchi, H.; Williams, B.O.; et al. Mouse Model of Human Ovarian Endometrioid Adenocarcinoma Based on Somatic Defects in the Wnt/β-Catenin and PI3K/Pten Signaling Pathways. Cancer Cell 2007, 11, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Kim, J.H.; Zhou, H.; Kim, B.W.; Wong, D.T. Salivary Transcriptomic Biomarkers for Detection of Ovarian Cancer: For Serous Papillary Adenocarcinoma. J. Mol. Med. 2012, 90, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Merritt, M.A.; Parsons, P.G.; Newton, T.R.; Martyn, A.C.; Webb, P.M.; Green, A.C.; Papadimos, D.J.; Boyle, G.M. Expression Profiling Identifies Genes Involved in Neoplastic Transformation of Serous Ovarian Cancer. BMC Cancer 2009, 9, 378. [Google Scholar] [CrossRef]
- Dou, Z.; Qiu, C.; Zhang, X.; Yao, S.; Zhao, C.; Wang, Z.; Chu, R.; Chen, J.; Chen, Z.; Li, R.; et al. HJURP Promotes Malignant Progression and Mediates Sensitivity to Cisplatin and WEE1-Inhibitor in Serous Ovarian Cancer. Int. J. Biol. Sci. 2022, 18, 1188–1210. [Google Scholar] [CrossRef]
- Zhao, J.; Song, X.; Xu, T.; Yang, Q.; Liu, J.; Jiang, B.; Wu, J. Identification of Potential Prognostic Competing Triplets in High-Grade Serous Ovarian Cancer. Front. Genet. 2021, 11, 1586. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, C. LncRNA DLX6-AS1 Aggravates the Development of Ovarian Cancer via Modulating FHL2 by Sponging MiR-195-5p. Cancer Cell Int. 2020, 20, 370. [Google Scholar] [CrossRef]
- Xie, M.; Fu, Q.; Wang, P.; Cui, Y. STAT1-Induced Upregulation LncRNA LINC00958 Accelerates the Epithelial Ovarian Cancer Tumorigenesis by Regulating Wnt/β-Catenin Signaling. Dis. Mrk. 2021, 2021, 1405045. [Google Scholar] [CrossRef]
- Cremaschi, P.; Carriero, R.; Astrologo, S.; Colì, C.; Lisa, A.; Parolo, S.; Bione, S. An Association Rule Mining Approach to Discover LncRNAs Expression Patterns in Cancer Datasets. Biomed Res. Int. 2015, 2015, 146250. [Google Scholar] [CrossRef]
- Jing, L.; Gong, M.; Lu, X.; Jiang, Y.; Li, H.; Cheng, W. LINC01127 Promotes the Development of Ovarian Tumors by Regulating the Cell Cycle. Am. J. Transl. Res. 2019, 11, 406–417. [Google Scholar] [PubMed]
- Wang, W.; Song, F.; Feng, X.; Chu, X.; Dai, H.; Tian, J.; Fang, X.; Song, F.; Liu, B.; Li, L.; et al. Functional Interrogation of Enhancer Connectome Prioritizes Candidate Target Genes at Ovarian Cancer Susceptibility Loci. Front. Genet. 2021, 12, 646179. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, T.; Ji, J.; Zhao, F.; Li, C.; Han, X. RHPN1-AS1 Promotes Cell Proliferation and Migration via MiR-665/Akt3 in Ovarian Cancer. Cancer Gene Ther. 2021, 28, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Zhou, J.-Q.; Liao, Y.-S. Autophagy-Related Long Non-Coding RNA Signature for Ovarian Cancer. J. Int. Med. Res. 2020, 48, 030006052097076. [Google Scholar] [CrossRef]
- Chang, H.; Li, B.; Zhang, X.; Meng, X. NCK1-AS1 Promotes NCK1 Expression to Facilitate Tumorigenesis and Chemo-Resistance in Ovarian Cancer. Biochem. Biophys. Res. Commun. 2020, 522, 292–299. [Google Scholar] [CrossRef]
- Wang, L.-L.; Sun, K.-X.; Wu, D.-D.; Xiu, Y.-L.; Chen, X.; Chen, S.; Zong, Z.-H.; Sang, X.-B.; Liu, Y.; Zhao, Y. DLEU1 Contributes to Ovarian Carcinoma Tumourigenesis and Development by Interacting with MiR-490-3p and Altering CDK1 Expression. J. Cell. Mol. Med. 2017, 21, 3055–3065. [Google Scholar] [CrossRef]
- Hu, X.; Li, Y.; Kong, D.; Hu, L.; Liu, D.; Wu, J. Long Noncoding RNA CASC9 Promotes LIN7A Expression via MiR-758-3p to Facilitate the Malignancy of Ovarian Cancer. J. Cell. Physiol. 2019, 234, 10800–10808. [Google Scholar] [CrossRef]
- Manichaikul, A.; Peres, L.C.; Wang, X.; Barnard, M.E.; Chyn, D.; Sheng, X.; Du, Z.; Tyrer, J.; Dennis, J.; Schwartz, A.G.; et al. Identification of Novel Epithelial Ovarian Cancer Loci in Women of African Ancestry. Int. J. Cancer 2020, 146, 2987–2998. [Google Scholar] [CrossRef]
- Xue, H.; Wu, Z.; Rao, D.; Zhuo, B.; Chen, Q. Long Non-Coding RNA LINC00858 Aggravates the Oncogenic Phenotypes of Ovarian Cancer Cells through MiR-134-5p/RAD18 Signaling. Arch. Gynecol. Obstet. 2020, 302, 1243–1254. [Google Scholar] [CrossRef]
- Ding, Y.; Tan, X.; Abasi, A.; Dai, Y.; Wu, R.; Zhang, T.; Li, K.; Yan, M.; Huang, X. LncRNA TRPM2-AS Promotes Ovarian Cancer Progression and Cisplatin Resistance by Sponging MiR-138-5p to Release SDC3 MRNA. Aging 2021, 13, 6832–6848. [Google Scholar] [CrossRef]
- Feng, S.; Yin, H.; Zhang, K.; Shan, M.; Ji, X.; Luo, S.; Shen, Y. Integrated Clinical Characteristics and Omics Analysis Identifies a Ferroptosis and Iron-Metabolism-Related LncRNA Signature for Predicting Prognosis and Therapeutic Responses in Ovarian Cancer. J. Ovarian Res. 2022, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.R.; Lv, Y.H.; Yao, H.M.; Zhang, H.Y.; Zhou, Y.; Liu, S.E. LncRNA PCAT6 Promotes Occurrence and Development of Ovarian Cancer by Inhibiting PTEN. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8230–8238. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhai, J.; Fu, Y. LncRNA CDKN2B-AS1 Promotes the Progression of Ovarian Cancer by MiR-143-3p/SMAD3 Axis and Predicts a Poor Prognosis. Neoplasma 2020, 67, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wang, H.; Liu, G.; Tang, R.; Ding, Y.; Xu, P.; Wang, H.; Miao, J.; Gu, X.; Han, S. LBX2-AS1 Promotes Ovarian Cancer Progression by Facilitating E2F2 Gene Expression via MiR-455-5p and MiR-491-5p Sponging. J. Cell. Mol. Med. 2021, 25, 1178–1189. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tan, S.; Bai, X.; Ma, S.; Chen, X. Long Non-Coding RNA LINC01215 Promotes Epithelial-Mesenchymal Transition and Lymph Node Metastasis in Epithelial Ovarian Cancer through RUNX3 Promoter Methylation. Transl. Oncol. 2021, 14, 101135. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Lin, Y.-B.; Li, L.; Liu, J. LncRNA TLR8-AS1 Promotes Metastasis and Chemoresistance of Ovarian Cancer through Enhancing TLR8 MRNA Stability. Biochem. Biophys. Res. Commun. 2020, 526, 857–864. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, M.; Wang, Z.; Guo, W.; Yang, D. MYC-binding LncRNA EPIC1 Promotes AKT-mTORC1 Signaling and Rapamycin Resistance in Breast and Ovarian Cancer. Mol. Carcinog. 2020, 59, 1188–1198. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Zhang, Y.; Luo, C.; Zhu, T.; Zhang, R.; Yao, R. LINC01342 Promotes the Progression of Ovarian Cancer by Absorbing MicroRNA-30c-2-3p to Upregulate HIF3A. J. Cell. Physiol. 2020, 235, 3939–3949. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, S.; Lv, Z.X.; Zhao, X.J. Promoting Action of Long Non-Coding RNA Small Nucleolar RNA Host Gene 4 in Ovarian Cancer. Acta Biochim. Pol. 2023, 70, 59–68. [Google Scholar] [CrossRef]
- Chu, Z.-P.; Dai, J.; Jia, L.-G.; Li, J.; Zhang, Z.-Y.; Yan, P. Increased Expression of Long Noncoding RNA HMMR-AS1 in Epithelial Ovarian Cancer: An Independent Prognostic Factor. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8145–8150. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, H.; Huang, Q.; Wu, J.; Zhang, M. A Prognostic Model Based on Immune-Related Long Noncoding RNAs for Patients with Epithelial Ovarian Cancer. J. Ovarian Res. 2022, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Hua, T.; Tian, Y.; Wang, R.; Zhao, C.; Kong, Y.; Tian, R.; Wang, W.; Ma, L. FOXP4-AS1 Is a Favorable Prognostic-Related Enhancer RNA in Ovarian Cancer. Biosci. Rep. 2021, 41, BSR20204008. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gong, M.; Zhao, M.; Wang, X.; Cheng, W.; Xia, Y. LncRNAs KB-1836B5, LINC00566 and FAM27L Are Associated with the Survival Time of Patients with Ovarian Cancer. Oncol. Lett. 2018, 16, 3735–3745. [Google Scholar] [CrossRef] [PubMed]
- Jie, Y.; Ye, L.; Chen, H.; Yu, X.; Cai, L.; He, W.; Fu, Y. ELFN1-AS1 Accelerates Cell Proliferation, Invasion and Migration via Regulating MiR-497-3p/CLDN4 Axis in Ovarian Cancer. Bioengineered 2020, 11, 872–882. [Google Scholar] [CrossRef]
- Wen, A.; Luo, L.; Du, C.; Luo, X. Long Non-coding RNA MiR155HG Silencing Restrains Ovarian Cancer Progression by Targeting the MicroRNA-155-5p/Tyrosinase-related Protein 1 Axis. Exp. Ther. Med. 2021, 22, 1237. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhan, X. Identification of Clinical Trait–Related LncRNA and MRNA Biomarkers with Weighted Gene Co-Expression Network Analysis as Useful Tool for Personalized Medicine in Ovarian Cancer. EPMA J. 2019, 10, 273–290. [Google Scholar] [CrossRef]
- Zhou, M.; Sun, Y.; Sun, Y.; Xu, W.; Zhang, Z.; Zhao, H.; Zhong, Z.; Sun, J. Comprehensive Analysis of LncRNA Expression Profiles Reveals a Novel LncRNA Signature to Discriminate Nonequivalent Outcomes in Patients with Ovarian Cancer. Oncotarget 2016, 7, 32433–32448. [Google Scholar] [CrossRef]
- Tao, F.; Tian, X.; Lu, M.; Zhang, Z. A Novel LncRNA, Lnc-OC1, Promotes Ovarian Cancer Cell Proliferation and Migration by Sponging MiR-34a and MiR-34c. J. Genet. Genom. 2018, 45, 137–145. [Google Scholar] [CrossRef]
- Wang, H.; Liu, J.; Yang, J.; Wang, Z.; Zhang, Z.; Peng, J.; Wang, Y.; Hong, L. A Novel Tumor Mutational Burden-Based Risk Model Predicts Prognosis and Correlates with Immune Infiltration in Ovarian Cancer. Front. Immunol. 2022, 13, 943389. [Google Scholar] [CrossRef]
- Li, S.; Shen, S.; Ge, W.; Cen, Y.; Zhang, S.; Cheng, X.; Wang, X.; Xie, X.; Lu, W. Long Non-Coding RNA SLC25A21-AS1 Inhibits the Development of Epithelial Ovarian Cancer by Specifically Inducing PTBP3 Degradation. Biomark. Res. 2023, 11, 12. [Google Scholar] [CrossRef]
- Xu, H.; Wang, X.; Zhang, Y.; Zheng, W.; Zhang, H. GATA6-AS1 Inhibits Ovarian Cancer Cell Proliferation and Migratory and Invasive Abilities by Sponging MiR-19a-5p and Upregulating TET2. Oncol. Lett. 2021, 22, 718. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, G.; Yang, F.; He, L.; Xie, X.; Li, L.; Yang, L.; Ma, Y.; Zhang, Q.; Chen, J.; et al. Elevated LINC00909 Promotes Tumor Progression of Ovarian Cancer via Regulating the MiR-23b-3p/MRC2 Axis. Oxid. Med. Cell. Longev. 2021, 2021, 5574130. [Google Scholar] [CrossRef] [PubMed]
- Yao, N.; Yu, L.; Zhu, B.; Gan, H.Y.; Guo, B.Q. LncRNA GIHCG Promotes Development of Ovarian Cancer by Regulating MicroRNA-429. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8127–8134. [Google Scholar] [CrossRef] [PubMed]
- Miao, W.; Lu, T.; Liu, X.; Yin, W.; Zhang, H. LncRNA SNHG8 Induces Ovarian Carcinoma Cells Cellular Process and Stemness through Wnt/β-Catenin Pathway. Cancer Biomark. 2020, 28, 459–471. [Google Scholar] [CrossRef]
- Dai, C.; Xu, P.; Liu, S.; Xu, S.; Xu, J.; Fu, Z.; Cao, J.; Lv, M.; Zhou, J.; Liu, G.; et al. Long Noncoding RNA ZEB1-AS1 Affects Paclitaxel and Cisplatin Resistance by Regulating MMP19 in Epithelial Ovarian Cancer Cells. Arch. Gynecol. Obstet. 2021, 303, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Wang, W.; Wu, L.; Qi, C.; Yan, W.; Lu, W.; Tian, J.; Shang, A. LINC00936/MicroRNA-221-3p Regulates Tumor Progression in Ovarian Cancer by Interacting with LAMA3. Recent Pat. Anticancer. Drug Discov. 2023, 18, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, L.-X.; Zhang, C.-Y.; Bai, N.; Feng, C.; Zhang, Z.-M.; Wang, L.; Gao, Z.-Z. LncRNA CRNDE Promotes Cell Proliferation, Migration and Invasion of Ovarian Cancer via MiR-423-5p/FSCN1 Axis. Mol. Cell. Biochem. 2022, 477, 1477–1488. [Google Scholar] [CrossRef]
- Wang, A.; Jin, C.; Li, H.; Qin, Q.; Li, L. LncRNA ADAMTS9-AS2 Regulates Ovarian Cancer Progression by Targeting MiR-182-5p/FOXF2 Signaling Pathway. Int. J. Biol. Macromol. 2018, 120, 1705–1713. [Google Scholar] [CrossRef]
- Aichen, Z.; Kun, W.; Xiaochun, S.; Lingling, T. LncRNA FGD5-AS1 Promotes the Malignant Phenotypes of Ovarian Cancer Cells via Targeting MiR-142-5p. Apoptosis 2021, 26, 348–360. [Google Scholar] [CrossRef]
- Zheng, J.; Guo, J.; Zhang, H.; Cao, B.; Xu, G.; Zhang, Z.; Tong, J. Four Prognosis-Associated LncRNAs Serve as Biomarkers in Ovarian Cancer. Front. Genet. 2021, 12, 672674. [Google Scholar] [CrossRef]
- Han, S.; Li, D.; Xiao, M. LncRNA ZFAS1 Serves as a Prognostic Biomarker to Predict the Survival of Patients with Ovarian Cancer. Exp. Ther. Med. 2019, 18, 4673–4681. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yuan, D.; Zhu, D.; Xu, T.; Huang, A.; Jiang, L.; Liu, C.; Qian, H.; Bu, X. LncRNA-MSC-AS1 Inhibits the Ovarian Cancer Progression by Targeting MiR-425-5p. J. Ovarian Res. 2021, 14, 109. [Google Scholar] [CrossRef]
- Han, Z.; Li, D.; Yang, Y.; Zhang, H. LINC-DUBR Suppresses Malignant Progression of Ovarian Cancer by Downregulating MiR-107 to Induce SMAC Expression. J. Healthc. Eng. 2022, 2022, 4535655. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Shen, L.; Lin, Q.; Dong, C.; Maswela, B.; Illahi, G.S.; Wu, X. SNHG5 Enhances Paclitaxel Sensitivity of Ovarian Cancer Cells through Sponging MiR-23a. Biomed. Pharmacother. 2020, 123, 109711. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Reid, B.M.; Sellers, T.A.; Karreth, F.A. Abstract B34: LINC00886, a Risk Locus-Associated Long Noncoding RNA, Promotes Ovarian Cancer Progression. Clin. Cancer Res. 2020, 26, B34. [Google Scholar] [CrossRef]
- Li, W.; Ma, S.; Bai, X.; Pan, W.; Ai, L.; Tan, W. Long Noncoding RNA WDFY3-AS2 Suppresses Tumor Progression by Acting as a Competing Endogenous RNA of MicroRNA-18a in Ovarian Cancer. J. Cell. Physiol. 2020, 235, 1141–1154. [Google Scholar] [CrossRef]
- Liu, X.; Liu, C.; Zhang, A.; Wang, Q.; Ge, J.; Li, Q.; Xiao, J. Long Non-Coding RNA SDCBP2-AS1 Delays the Progression of Ovarian Cancer via MicroRNA-100-5p-Targeted EPDR1. World J. Surg. Oncol. 2021, 19, 199. [Google Scholar] [CrossRef]
- Feng, H.; Gu, Z.-Y.; Li, Q.; Liu, Q.-H.; Yang, X.-Y.; Zhang, J.-J. Identification of Significant Genes with Poor Prognosis in Ovarian Cancer via Bioinformatical Analysis. J. Ovarian Res. 2019, 12, 35. [Google Scholar] [CrossRef]
- Cai, L.; Hu, X.; Ye, L.; Bai, P.; Jie, Y.; Shu, K. Long Non-Coding RNA ADAMTS9-AS1 Attenuates Ferroptosis by Targeting MicroRNA-587/Solute Carrier Family 7 Member 11 Axis in Epithelial Ovarian Cancer. Bioengineered 2022, 13, 8226–8239. [Google Scholar] [CrossRef]
- Yang, M.; Zhai, Z.; Guo, S.; Li, X.; Zhu, Y.; Wang, Y. Long Non-Coding RNA FLJ33360 Participates in Ovarian Cancer Progression by Sponging MiR-30b-3p. OncoTargets. Ther. 2019, 12, 4469–4480. [Google Scholar] [CrossRef]
- Lei, J.; He, Z.; Wang, J.; Hu, M.; Zhou, P.; Lian, C.; Hua, L.; Wu, S.; Zhou, J. Identification of MEG8/MiR-378d/SOBP Axis as a Novel Regulatory Network and Associated with Immune Infiltrates in Ovarian Carcinoma by Integrated Bioinformatics Analysis. Cancer Med. 2021, 10, 2924–2939. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Wang, F.; Gao, C.; Cao, Y.; Wang, J. Development and Verification of an Autophagy-Related LncRNA Signature to Predict Clinical Outcomes and Therapeutic Responses in Ovarian Cancer. Front. Med. 2021, 8, 666973. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, H.; Xi, M.; Lu, X. Long Noncoding RNA C17orf91 Is a Potential Prognostic Marker and Functions as an Oncogene in Ovarian Cancer. J. Ovarian Res. 2016, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Gao, H.; Li, X.; Zhu, Y.; Peng, S.; Yu, J.; Zhan, G.; Wang, J.; Liu, N.; Guo, X. LncRNA TPT1-AS1 Promotes Tumorigenesis and Metastasis in Epithelial Ovarian Cancer by Inducing TPT1 Expression. Cancer Sci. 2019, 110, 1587–1598. [Google Scholar] [CrossRef]
- Xu, R.; Peng, H.; Yang, N.; Liu, Z.; Lu, W. Nuclear LncRNA CERNA1 Enhances the Cisplatin-Induced Cell Apoptosis and Overcomes Chemoresistance via Epigenetic Activation of BCL2L10 in Ovarian Cancer. Genes Dis. 2023, 10, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Konstantinopoulos, P.A.; Spentzos, D.; Karlan, B.Y.; Taniguchi, T.; Fountzilas, E.; Francoeur, N.; Levine, D.A.; Cannistra, S.A. Gene Expression Profile of BRCA Ness That Correlates With Responsiveness to Chemotherapy and With Outcome in Patients With Epithelial Ovarian Cancer. J. Clin. Oncol. 2010, 28, 3555–3561. [Google Scholar] [CrossRef]
- Mateescu, B.; Batista, L.; Cardon, M.; Gruosso, T.; de Feraudy, Y.; Mariani, O.; Nicolas, A.; Meyniel, J.-P.; Cottu, P.; Sastre-Garau, X.; et al. MiR-141 and MiR-200a Act on Ovarian Tumorigenesis by Controlling Oxidative Stress Response. Nat. Med. 2011, 17, 1627–1635. [Google Scholar] [CrossRef]
- Lisowska, K.M.; Olbryt, M.; Student, S.; Kujawa, K.A.; Cortez, A.J.; Simek, K.; Dansonka-Mieszkowska, A.; Rzepecka, I.K.; Tudrej, P.; Kupryjańczyk, J. Unsupervised Analysis Reveals Two Molecular Subgroups of Serous Ovarian Cancer with Distinct Gene Expression Profiles and Survival. J. Cancer Res. Clin. Oncol. 2016, 142, 1239–1252. [Google Scholar] [CrossRef]
- Ferriss, J.S.; Kim, Y.; Duska, L.; Birrer, M.; Levine, D.A.; Moskaluk, C.; Theodorescu, D.; Lee, J.K. Multi-Gene Expression Predictors of Single Drug Responses to Adjuvant Chemotherapy in Ovarian Carcinoma: Predicting Platinum Resistance. PLoS ONE 2012, 7, e30550. [Google Scholar] [CrossRef]
- Pils, D.; Hager, G.; Tong, D.; Aust, S.; Heinze, G.; Kohl, M.; Schuster, E.; Wolf, A.; Sehouli, J.; Braicu, I.; et al. Validating the Impact of a Molecular Subtype in Ovarian Cancer on Outcomes: A Study of the OVCAD Consortium. Cancer Sci. 2012, 103, 1334–1341. [Google Scholar] [CrossRef]
- Yoshihara, K.; Tajima, A.; Yahata, T.; Kodama, S.; Fujiwara, H.; Suzuki, M.; Onishi, Y.; Hatae, M.; Sueyoshi, K.; Fujiwara, H.; et al. Gene Expression Profile for Predicting Survival in Advanced-Stage Serous Ovarian Cancer Across Two Independent Datasets. PLoS ONE 2010, 5, e9615. [Google Scholar] [CrossRef] [PubMed]
- Spentzos, D.; Levine, D.A.; Kolia, S.; Otu, H.; Boyd, J.; Libermann, T.A.; Cannistra, S.A. Unique Gene Expression Profile Based on Pathologic Response in Epithelial Ovarian Cancer. J. Clin. Oncol. 2005, 23, 7911–7918. [Google Scholar] [CrossRef]
- Yoshihara, K.; Tsunoda, T.; Shigemizu, D.; Fujiwara, H.; Hatae, M.; Fujiwara, H.; Masuzaki, H.; Katabuchi, H.; Kawakami, Y.; Okamoto, A.; et al. High-Risk Ovarian Cancer Based on 126-Gene Expression Signature Is Uniquely Characterized by Downregulation of Antigen Presentation Pathway. Clin. Cancer Res. 2012, 18, 1374–1385. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Shen, J.; He, Q.; Xu, S. Identification of a Novel Immune-Related LncRNA CTD-2288O8.1 Regulating Cisplatin Resistance in Ovarian Cancer Based on Integrated Analysis. Front. Genet. 2022, 13, 814291. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, Z.; Yan, Y. Role of a Pyroptosis-Related LncRNA Signature in Risk Stratification and Immunotherapy of Ovarian Cancer. Front. Med. 2022, 8, 2842. [Google Scholar] [CrossRef]
- Cao, X.; Zhang, Q.; Zhu, Y.; Huo, X.; Bao, J.; Su, M. Derivation, Comprehensive Analysis, and Assay Validation of a Pyroptosis-Related LncRNA Prognostic Signature in Patients With Ovarian Cancer. Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef]
- Li, M.; Zhang, L.; Feng, M.; Huang, X. M6A-Related LncRNA Signature Is Involved in Immunosuppression and Predicts the Patient Prognosis of the Age-Associated Ovarian Cancer. J. Immunol. Res. 2022, 2022, 3258400. [Google Scholar] [CrossRef]
- Liang, H.; Bai, Y.; Wang, H.; Yang, X. Identification of LncRNA Prognostic Markers for Ovarian Cancer by Integration of Co-Expression and CeRNA Network. Front. Genet. 2021, 11, 566497. [Google Scholar] [CrossRef]
- Guo, Q.; Cheng, Y.; Liang, T.; He, Y.; Ren, C.; Sun, L.; Zhang, G. Comprehensive Analysis of LncRNA-MRNA Co-Expression Patterns Identifies Immune-Associated LncRNA Biomarkers in Ovarian Cancer Malignant Progression. Sci. Rep. 2016, 5, 17683. [Google Scholar] [CrossRef]
- Lin, N.; Lin, J.; Tanaka, Y.; Sun, P.; Zhou, X. Identification and Validation of a Five-LncRNA Signature for Predicting Survival with Targeted Drug Candidates in Ovarian Cancer. Bioengineered 2021, 12, 3263–3274. [Google Scholar] [CrossRef]
- Wang, L.; He, M.; Fu, L.; Jin, Y. Role of LncRNAHCP5/MicroRNA-525–5p/PRC1 Crosstalk in the Malignant Behaviors of Ovarian Cancer Cells. Exp. Cell Res. 2020, 394, 112129. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Tan, J. N6-Methyladenosine-Related LncRNAs Is a Potential Marker for Predicting Prognosis and Immunotherapy in Ovarian Cancer. Hereditas 2022, 159, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, H.; Zhang, M.; Xing, L.; Yang, C.; Xia, B.; Lou, G. Integrated Analysis of a Competing Endogenous RNA Network Reveals an 11-LncRNA Prognostic Signature in Ovarian Cancer. Aging 2020, 12, 25153–25171. [Google Scholar] [CrossRef]
- Newtson, A.; Reyes, H.; Devor, E.J.; Goodheart, M.J.; Bosquet, J.G. Identification of Novel Fusion Transcripts in High Grade Serous Ovarian Cancer. Int. J. Mol. Sci. 2021, 22, 4791. [Google Scholar] [CrossRef] [PubMed]
- Hyter, S.; Hirst, J.; Pathak, H.; Pessetto, Z.Y.; Koestler, D.C.; Raghavan, R.; Pei, D.; Godwin, A.K. Developing a Genetic Signature to Predict Drug Response in Ovarian Cancer. Oncotarget 2018, 9, 14828–14848. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Jia, Y.; Wang, J.; Duan, Y.; Wang, X.; Liu, T.; Hao, S.; Liu, L. Long Non-Coding RNA SNHG10 Upregulates BIN1 to Suppress the Tumorigenesis and Epithelial–Mesenchymal Transition of Epithelial Ovarian Cancer via Sponging MiR-200a-3p. Cell Death Discov. 2022, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Xia, Y. LncRNA HLA-F-AS1 Attenuates the Ovarian Cancer Development by Targeting MiR-21-3p/PEG3 Axis. Anticancer. Drugs 2022, 33, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Hou, Y.; Li, A.; Li, Z.; Wang, W.; Xie, H.; Rong, Z.; Lou, G.; Li, K. Identification of a Six-LncRNA Signature Associated with Recurrence of Ovarian Cancer. Sci. Rep. 2017, 7, 752. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.-G.; Zhang, K.-Q. LINC00452 Promotes Ovarian Carcinogenesis through Increasing ROCK1 by Sponging MiR-501-3p and Suppressing Ubiquitin-Mediated Degradation. Aging 2020, 12, 21129–21146. [Google Scholar] [CrossRef]
- Fu, Y.; Biglia, N.; Wang, Z.; Shen, Y.; Risch, H.A.; Lu, L.; Canuto, E.M.; Jia, W.; Katsaros, D.; Yu, H. Long Non-Coding RNAs, ASAP1-IT1, FAM215A, and LINC00472, in Epithelial Ovarian Cancer. Gynecol. Oncol. 2016, 143, 642–649. [Google Scholar] [CrossRef]
- Gong, M.; Luo, C.; Meng, H.; Li, S.; Nie, S.; Jiang, Y.; Wan, Y.; Li, H.; Cheng, W. Upregulated LINC00565 Accelerates Ovarian Cancer Progression By Targeting GAS6. Onco. Targets. Ther. 2019, 12, 10011–10022. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Huang, S.; Han, F. LINC-PINT Suppresses Tumour Cell Proliferation, Migration and Invasion through Targeting MiR-374a-5p in Ovarian Cancer. Cell Biochem. Funct. 2020, 38, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Yang, D.-Z.; Zhai, Y.-N.; Xue, K.; Xu, F.; Gu, X.-Y.; Wang, S.-M. Microarray Expression Profiling of Long Non-Coding RNAs in Epithelial Ovarian Cancer. Oncol. Lett. 2017, 14, 2523–2530. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.; Zhang, L.; Li, H.; Duan, H.; Wang, D.; Zhao, X.; Xie, Y. The Enhancer RNA ADCY10P1 Is Associated with the Progression of Ovarian Cancer. J. Ovarian Res. 2022, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Wang, F.; Wang, L. Hypoxia-Related LncRNA Prognostic Model of Ovarian Cancer Based on Big Data Analysis. J. Oncol. 2023, 2023, 6037121. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhan, X. Anti-Parasite Drug Ivermectin Can Suppress Ovarian Cancer by Regulating LncRNA-EIF4A3-MRNA Axes. EPMA J. 2020, 11, 289–309. [Google Scholar] [CrossRef]
- Zhang, W.; Fei, J.; Yu, S.; Shen, J.; Zhu, X.; Sadhukhan, A.; Lu, W.; Zhou, J. LINC01088 Inhibits Tumorigenesis of Ovarian Epithelial Cells by Targeting MiR-24-1-5p. Sci. Rep. 2018, 8, 2876. [Google Scholar] [CrossRef]
- Shao, S.; Tian, J.; Zhang, H.; Wang, S. LncRNA Myocardial Infarction-Associated Transcript Promotes Cell Proliferation and Inhibits Cell Apoptosis by Targeting MiR-330-5p in Epithelial Ovarian Cancer Cells. Arch. Med. Sci. 2018, 14, 1263–1270. [Google Scholar] [CrossRef]
- Liu, S.; Lai, W.; Shi, Y.; Liu, N.; Ouyang, L.; Zhang, Z.; Chen, L.; Wang, X.; Qian, B.; Xiao, D.; et al. Annotation and Cluster Analysis of Long Noncoding RNA Linked to Male Sex and Estrogen in Cancers. NPJ Precis. Oncol. 2020, 4, 5. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, H.; Fu, Y. Effects of Autophagy-Related Genes on the Prognosis and Immune Microenvironment of Ovarian Cancer. Biomed Res. Int. 2022, 2022, 6609195. [Google Scholar] [CrossRef]
- Luan, A.; Hou, L.; Zhang, F. Silencing of SBF2-AS1 Inhibits Cell Growth and Invasion by Sponging MicroRNA-338-3p in Serous Ovarian Carcinoma. Kaohsiung J. Med. Sci. 2022, 38, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Gao, L.; Zhang, M.; Ning, N.; Wang, Y.; Wu, D.; Li, X. Identification and Analysis of An Epigenetically Regulated Five-LncRNA Signature Associated With Outcome and Chemotherapy Response in Ovarian Cancer. Front. Cell Dev. Biol. 2021, 9, 644940. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, A.S.; Fischer, A.; Miller, D.H.; Vang, S.; MacLaughlan, S.; Wu, H.-T.; Yu, J.; Steinhoff, M.; Collins, C.; Smith, P.J.S.; et al. Expression Profiling of Primary and Metastatic Ovarian Tumors Reveals Differences Indicative of Aggressive Disease. PLoS ONE 2014, 9, e94476. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yang, Z.; Xu, S.; Li, X.; Yang, X.; Jin, P.; Liu, Y.; Zhou, X.; Zhang, T.; Gong, C.; et al. Heterotypic CAF-Tumor Spheroids Promote Early Peritoneal Metastasis of Ovarian Cancer. J. Exp. Med. 2019, 216, 688–703. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Taylor-Harding, B.; Raz, Y.; Haro, M.; Recouvreux, M.S.; Taylan, E.; Lester, J.; Millstein, J.; Walts, A.E.; Karlan, B.Y.; et al. Are Epithelial Ovarian Cancers of the Mesenchymal Subtype Actually Intraperitoneal Metastases to the Ovary? Front. Cell Dev. Biol. 2020, 8, 647. [Google Scholar] [CrossRef]
- Sallinen, H.; Janhonen, S.; Pölönen, P.; Niskanen, H.; Liu, O.H.; Kivelä, A.; Hartikainen, J.M.; Anttila, M.; Heinäniemi, M.; Ylä-Herttuala, S.; et al. Comparative Transcriptome Analysis of Matched Primary and Distant Metastatic Ovarian Carcinoma. BMC Cancer 2019, 19, 1121. [Google Scholar] [CrossRef]
- Wang, L.; Ren, C.; Xu, Y.; Yang, L.; Chen, Y.; Zhu, Y. The LINC00922 Aggravates Ovarian Cancer Progression via Sponging MiR-361-3p. J. Ovarian Res. 2021, 14, 77. [Google Scholar] [CrossRef]
- Eoh, K.; Lee, D.; Nam, E.; Kim, J.; Moon, H.; Kim, S.; Kim, Y. HOXA-AS3 Induces Tumor Progression through the Epithelial-mesenchymal Transition Pathway in Epithelial Ovarian Cancer. Oncol. Rep. 2023, 49, 64. [Google Scholar] [CrossRef]
- Yao, N.; Sun, J.-Q.; Yu, L.; Ma, L.; Guo, B.-Q. LINC00968 Accelerates the Progression of Epithelial Ovarian Cancer via Mediating the Cell Cycle Progression. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4642–4649. [Google Scholar]
- Liu, M.; Shen, C.; Wang, C. Long Noncoding RNA LINC01133 Confers Tumor-Suppressive Functions in Ovarian Cancer by Regulating Leucine-Rich Repeat Kinase 2 as an MiR-205 Sponge. Am. J. Pathol. 2019, 189, 2323–2339. [Google Scholar] [CrossRef]
- Wang, K.; Sun, Y.; Wang, Y.; Liu, L. An Integration Analysis of MRNAs and MiRNAs Microarray Data to Identify Key Regulators for Ovarian Endometriosis Based on Competing Endogenous RNAs. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 252, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Guo, J.; Zhu, L.; Zhou, Y.; Tong, J. Comprehensive Analyses of Glycolysis-Related LncRNAs for Ovarian Cancer Patients. J. Ovarian Res. 2021, 14, 124. [Google Scholar] [CrossRef] [PubMed]
- Guan, N.; Zheng, H.; Wu, X.; Xie, L.; Tong, X. SP1-Regulated Non-Coding RNA SNHG22 Promotes Ovarian Cancer Growth and Glycolysis. Cancer Manag. Res. 2021, 13, 7299–7309. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Yu, Y.; Yin, W.; Qian, S. Development and Verification of a 7-LncRNA Prognostic Model Based on Tumor Immunity for Patients with Ovarian Cancer. J. Ovarian Res. 2023, 16, 31. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Qu, H. Identification and Validation of a Seven M6A-Related LncRNAs Signature Predicting Prognosis of Ovarian Cancer. BMC Cancer 2022, 22, 633. [Google Scholar] [CrossRef]
- Li, J.; Feng, L.; Tian, C.; Tang, Y.L.; Tang, Y.; Hu, F.Q. Long Noncoding RNA-JPX Predicts the Poor Prognosis of Ovarian Cancer Patients and Promotes Tumor Cell Proliferation, Invasion and Migration by the PI3K/Akt/MTOR Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8135–8144. [Google Scholar] [CrossRef]
- Lin, H.; Xu, X.; Chen, K.; Fu, Z.; Wang, S.; Chen, Y.; Zhang, H.; Niu, Y.; Chen, H.; Yu, H.; et al. LncRNA CASC15, MiR-23b Cluster and SMAD3 Form a Novel Positive Feedback Loop to Promote Epithelial-Mesenchymal Transition and Metastasis in Ovarian Cancer. Int. J. Biol. Sci. 2022, 18, 1989–2002. [Google Scholar] [CrossRef]
- Wang, L.; Ye, T.Y.; Wu, H.; Chen, S.Y.; Weng, J.R.; Xi, X.W. LINC00702 Accelerates the Progression of Ovarian Cancer through Interacting with EZH2 to Inhibit the Transcription of KLF2. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 201–208. [Google Scholar] [CrossRef]
- Tian, J.; Yang, L.; Wang, Z.; Yan, H. MIR503HG Impeded Ovarian Cancer Progression by Interacting with SPI1 and Preventing TMEFF1 Transcription. Aging 2022, 14, 5390–5405. [Google Scholar] [CrossRef]
- Pan, X.; Guo, Z.; Chen, Y.; Zheng, S.; Peng, M.; Yang, Y.; Wang, Z. STAT3-Induced LncRNA SNHG17 Exerts Oncogenic Effects on Ovarian Cancer through Regulating CDK6. Mol. Ther.-Nucleic Acids 2020, 22, 38–49. [Google Scholar] [CrossRef]
- Zheng, M.; Hu, Y.; Gou, R.; Nie, X.; Li, X.; Liu, J.; Lin, B. Identification Three LncRNA Prognostic Signature of Ovarian Cancer Based on Genome-Wide Copy Number Variation. Biomed. Pharmacother. 2020, 124, 109810. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, G. DNM3OS Facilitates Ovarian Cancer Progression by Regulating MiR-193a-3p/MAP3K3 Axis. Yonsei Med. J. 2021, 62, 535. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ji, G.; Le, X.; Wang, C.; Xu, L.; Feng, M.; Zhang, Y.; Yang, H.; Xuan, Y.; Yang, Y.; et al. Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer. Cancer Res. 2017, 77, 1369–1382. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, B.; Zhan, X. Comprehensive Analysis of Tumor Microenvironment Identified Prognostic Immune-Related Gene Signature in Ovarian Cancer. Front. Genet. 2021, 12, 616073. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Shi, X.-Y.; Li, Z.-L.; Li, M.; Zhang, M.-M.; Yan, S.-J.; Wei, Z.-L. Downregulation of LINC01508 Contributes to Cisplatin Resistance in Ovarian Cancer via the Regulation of the Hippo-YAP Pathway. J. Gynecol. Oncol. 2021, 32, e77. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Xiao, X.; Chen, J. Silencing of the Long Noncoding RNA LINC01132 Alleviates the Oncogenicity of Epithelial Ovarian Cancer by Regulating the MicroRNA-431-5p/SOX9 Axis. Int. J. Mol. Med. 2021, 48, 151. [Google Scholar] [CrossRef]
- Liu, S.; Chen, X.; Huang, K.; Xiong, X.; Shi, Y.; Wang, X.; Pan, X.; Cong, Y.; Sun, Y.; Ge, L.; et al. Long Noncoding RNA RFPL1S-202 Inhibits Ovarian Cancer Progression by Downregulating the IFN-β/STAT1 Signaling. Exp. Cell Res. 2023, 422, 113438. [Google Scholar] [CrossRef]
- Kaneuchi, M.; Sasaki, M.; Tanaka, Y.; Shiina, H.; Yamada, H.; Yamamoto, R.; Sakuragi, N.; Enokida, H.; Verma, M.; Dahiya, R. WT1 and WT1-AS Genes Are Inactivated by Promoter Methylation in Ovarian Clear Cell Adenocarcinoma. Cancer 2005, 104, 1924–1930. [Google Scholar] [CrossRef]
- Liu, B.; Yan, L.; Chi, Y.; Sun, Y.; Yang, X. Long Non-Coding RNA AFAP1-AS1 Facilitates Ovarian Cancer Progression by Regulating the MiR-107/PDK4 Axis. J. Ovarian Res. 2021, 14, 60. [Google Scholar] [CrossRef]
- Taniguchi-Ponciano, K.; Huerta-Padilla, V.; Baeza-Xochihua, V.; Ponce-Navarrete, G.; Salcedo, E.; Gomez-Apo, E.; Chavez-Macias, L.; Aviles-Duran, J.; Ruiz-Sanchez, H.; Valdivia, A.; et al. Revisiting the Genomic and Transcriptomic Landscapes from Female Malignancies Could Provide Molecular Markers and Targets for Precision Medicine. Arch. Med. Res. 2019, 50, 428–436. [Google Scholar] [CrossRef]
- Chen, X.; Yang, Y.; Sun, J.; Hu, C.; Ge, X.; Li, R. LncRNA HCG11 Represses Ovarian Cancer Cell Growth via AKT Signaling Pathway. J. Obstet. Gynaecol. Res. 2022, 48, 796–805. [Google Scholar] [CrossRef]
- Xue, Z.; Zhu, X.; Teng, Y. Long Non-coding RNA CASC2 Inhibits Progression and Predicts Favorable Prognosis in Epithelial Ovarian Cancer. Mol. Med. Rep. 2018, 18, 5173–5181. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhan, X.; Zhan, X. The LncRNA SNHG3 Regulates Energy Metabolism of Ovarian Cancer by an Analysis of Mitochondrial Proteomes. Gynecol. Oncol. 2018, 150, 343–354. [Google Scholar] [CrossRef]
- Liu, H.; Liu, G.; Pang, W.; Zhang, H.; Zeng, Z.; Wang, H. LncRNA LUCAT1 Promotes Proliferation of Ovarian Cancer Cells by Regulating MiR-199a-5p Expression. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1682–1687. [Google Scholar] [PubMed]
- Fang, Y.-N.; Huang, Z.-L.; Li, H.; Tan, W.-B.; Zhang, Q.-G.; Wang, L.; Wu, J.-L. LINC01116 Promotes the Progression of Epithelial Ovarian Cancer via Regulating Cell Apoptosis. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5127–5133. [Google Scholar] [PubMed]
- Guo, R.; Qin, Y. LEMD1-AS1 Suppresses Ovarian Cancer Progression Through Regulating MiR-183-5p/TP53 Axis. Onco. Targets. Ther. 2020, 13, 7387–7398. [Google Scholar] [CrossRef]
- Ruan, Z.; Zhao, D. Long Intergenic Noncoding RNA LINC00284 Knockdown Reduces Angiogenesis in Ovarian Cancer Cells via Up-regulation of MEST through NF-κB1. FASEB J. 2019, 33, 12047–12059. [Google Scholar] [CrossRef]
- Wu, X.; Qiu, L.; Feng, H.; Zhang, H.; Yu, H.; Du, Y.; Wu, H.; Zhu, S.; Ruan, Y.; Jiang, H. KHDRBS3 Promotes Paclitaxel Resistance and Induces Glycolysis through Modulated MIR17HG/CLDN6 Signaling in Epithelial Ovarian Cancer. Life Sci. 2022, 293, 120328. [Google Scholar] [CrossRef]
- Yang, H.; Han, C. Abstract 2139: A Seven LncRNA Based Risk Score System for Predicting the Recurrence and Prognosis of Ovarian Cancer Patients. Cancer Res. 2020, 80, 2139. [Google Scholar] [CrossRef]
- Geng, L.; Wang, Z.; Tian, Y. Down-Regulation of ZNF252P-AS1 Alleviates Ovarian Cancer Progression by Binding MiR-324-3p to Downregulate LY6K. J. Ovarian Res. 2022, 15, 1. [Google Scholar] [CrossRef]
- Wang, K.; Mei, S.; Cai, M.; Zhai, D.; Zhang, D.; Yu, J.; Ni, Z.; Yu, C. Ferroptosis-Related Long Noncoding RNAs as Prognostic Biomarkers for Ovarian Cancer. Front. Oncol. 2022, 12, 888699. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Huang, H.; Qi, X.; Bian, C.; Cheng, M.; Liu, L.; Xue, L.; Zhao, X.; Yi, T.; Quan, Y. Hypoxia-Induced LncRNA-MIR210HG Promotes Cancer Progression By Inhibiting HIF-1α Degradation in Ovarian Cancer. Front. Oncol. 2021, 11, 4971. [Google Scholar] [CrossRef] [PubMed]
- Buttarelli, M.; De Donato, M.; Raspaglio, G.; Babini, G.; Ciucci, A.; Martinelli, E.; Baccaro, P.; Pasciuto, T.; Fagotti, A.; Scambia, G.; et al. Clinical Value of LncRNA MEG3 in High-Grade Serous Ovarian Cancer. Cancers 2020, 12, 966. [Google Scholar] [CrossRef]
- Abildgaard, C.; do Canto, L.M.; Rainho, C.A.; Marchi, F.A.; Calanca, N.; Waldstrøm, M.; Steffensen, K.D.; Rogatto, S.R. The Long Non-Coding RNA SNHG12 as a Mediator of Carboplatin Resistance in Ovarian Cancer via Epigenetic Mechanisms. Cancers 2022, 14, 1664. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Jin, H.; Duan, X.; Liu, H.; Zhao, X.; Fan, S.; Wang, Y.; Yao, T. LncRNA PSMA3-AS1 Promotes Cell Proliferation, Migration, and Invasion in Ovarian Cancer by Activating the PI3K/Akt Pathway via the MiR-378a-3p/GALNT3 Axis. Environ. Toxicol. 2021, 36, 2562–2577. [Google Scholar] [CrossRef]
- Koti, M.; Gooding, R.J.; Nuin, P.; Haslehurst, A.; Crane, C.; Weberpals, J.; Childs, T.; Bryson, P.; Dharsee, M.; Evans, K.; et al. Identification of the IGF1/PI3K/NF ΚB/ERK Gene Signalling Networks Associated with Chemotherapy Resistance and Treatment Response in High-Grade Serous Epithelial Ovarian Cancer. BMC Cancer 2013, 13, 549. [Google Scholar] [CrossRef]
- Xu, S.; Jia, G.; Zhang, H.; Wang, L.; Cong, Y.; Lv, M.; Xu, J.; Ruan, H.; Jia, X.; Xu, P.; et al. LncRNA HOXB-AS3 Promotes Growth, Invasion and Migration of Epithelial Ovarian Cancer by Altering Glycolysis. Life Sci. 2021, 264, 118636. [Google Scholar] [CrossRef]
- Ramakrishna, M.; Williams, L.H.; Boyle, S.E.; Bearfoot, J.L.; Sridhar, A.; Speed, T.P.; Gorringe, K.L.; Campbell, I.G. Identification of Candidate Growth Promoting Genes in Ovarian Cancer through Integrated Copy Number and Expression Analysis. PLoS ONE 2010, 5, e9983. [Google Scholar] [CrossRef]
- Akasu-Nagayoshi, Y.; Hayashi, T.; Kawabata, A.; Shimizu, N.; Yamada, A.; Yokota, N.; Nakato, R.; Shirahige, K.; Okamoto, A.; Akiyama, T. PHOSPHATE Exporter XPR1/SLC53A1 Is Required for the Tumorigenicity of Epithelial Ovarian Cancer. Cancer Sci. 2022, 113, 2034–2043. [Google Scholar] [CrossRef]
- Zhao, Q.; Fan, C. A Novel Risk Score System for Assessment of Ovarian Cancer Based on Co-Expression Network Analysis and Expression Level of Five LncRNAs. BMC Med. Genet. 2019, 20, 103. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Z.; Huang, Y.; Wang, K.; Xie, Y.; Yang, N. LncRNA GAS5 Inhibits the Proliferation and Invasion of Ovarian Clear Cell Carcinoma via the MiR-31-5p/ARID1A Axis. Kaohsiung J. Med. Sci. 2021, 37, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhang, Q.; Huang, X. Abnormal 5-Methylcytosine LncRNA Methylome Is Involved in Human High-Grade Serous Ovarian Cancer. Am. J. Transl. Res. 2021, 13, 13625–13639. [Google Scholar] [PubMed]
- Duan, M.; Fang, M.; Wang, C.; Wang, H.; Li, M. LncRNA EMX2OS Induces Proliferation, Invasion and Sphere Formation of Ovarian Cancer Cells via Regulating the MiR-654-3p/AKT3/PD-L1 Axis. Cancer Manag. Res. 2020, 12, 2141–2154. [Google Scholar] [CrossRef]
- Chen, X.; Wu, W.; Cao, X.; Zhao, X.; Li, W.; Deng, C.; Huang, Z. LncRNA Mortal Obligate RNA Transcript Was Downregulated in Ovarian Carcinoma and Inhibits Cancer Cell Proliferation by Downregulating MiRNA-21. J. Cell. Biochem. 2019, 120, 11949–11954. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, Y.; Li, X.; Wang, D. Ovarian Cancer-specific Dysregulated Genes with Prognostic Significance: ScRNA-Seq with Bulk RNA-Seq Data and Experimental Validation. Ann. N. Y. Acad. Sci. 2022, 1512, 154–173. [Google Scholar] [CrossRef]
- Gao, L.; Li, X.; Nie, X.; Guo, Q.; Liu, Q.; Qi, Y.; Liu, J.; Lin, B. Construction of Novel MRNA-MiRNA-LncRNA Regulatory Networks Associated with Prognosis of Ovarian Cancer. J. Cancer 2020, 11, 7057–7072. [Google Scholar] [CrossRef]
- Wang, D.; Xu, Y.; Feng, L.; Yin, P.; Song, S.; Wu, F.; Yan, P.; Liang, Z. RGS5 Decreases the Proliferation of Human Ovarian Carcinoma-derived Primary Endothelial Cells through the MAPK/ERK Signaling Pathway in Hypoxia. Oncol. Rep. 2018, 41, 165–177. [Google Scholar] [CrossRef]
- Gao, J.; Liu, F.; Zhao, X.; Zhang, P. Long Non-coding RNA FOXD2-AS1 Promotes Proliferation, Migration and Invasion of Ovarian Cancer Cells via Regulating the Expression of MiR-4492. Exp. Ther. Med. 2021, 21, 307. [Google Scholar] [CrossRef]
- Liu, Q.-Y.; Jiang, X.-X.; Tian, H.-N.; Guo, H.-L.; Guo, H.; Guo, Y. Long Non-Coding RNA OIP5-AS1 Plays an Oncogenic Role in Ovarian Cancer through Targeting MiR-324-3p/NFIB Axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7266–7275. [Google Scholar]
- Zheng, Q.; Zhang, J.; Liu, Y.; Dong, W.; Dai, X.; Du, X.; Gu, D. LINC01119 Encapsulated by Cancer-Associated Adipocytes-Derived Exosomes Promotes M2 Polarization of Macrophages to Induce Immune Escape in Ovarian Cancer in a 3D Co-Culture Cell-Based Model. Clin. Transl. Oncol. 2023. [Google Scholar] [CrossRef]
- Naghsh-Nilchi, A.; Ebrahimi Ghahnavieh, L.; Dehghanian, F. Construction of MiRNA-lncRNA-mRNA Co-expression Network Affecting EMT- Mediated Cisplatin Resistance in Ovarian Cancer. J. Cell. Mol. Med. 2022, 26, 4530–4547. [Google Scholar] [CrossRef]
- Liu, W.; Yang, Y.-J.; An, Q. LINC00963 Promotes Ovarian Cancer Proliferation, Migration and EMT via the MiR-378g/CHI3L1 Axis. Cancer Manag. Res. 2020, 12, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Burdennyy, A.M.; Filippova, E.A.; Ivanova, N.A.; Lukina, S.S.; Pronina, I.V.; Loginov, V.I.; Fridman, M.V.; Kazubskaya, T.P.; Utkin, D.O.; Braga, E.A.; et al. Hypermethylation of Genes in New Long Noncoding RNA in Ovarian Tumors and Metastases: A Dual Effect. Bull. Exp. Biol. Med. 2021, 171, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Kerslake, R.; Sisu, C.; Panfilov, S.; Hall, M.; Khan, N.; Jeyaneethi, J.; Randeva, H.; Kyrou, I.; Karteris, E. Differential Regulation of Genes by the Glucogenic Hormone Asprosin in Ovarian Cancer. J. Clin. Med. 2022, 11, 5942. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Jiang, Y.; Yang, J. Long Noncoding RNA LINC01554 as a Novel Biomarker for Diagnosis and Prognosis Prediction of Epithelial Ovarian Cancer. Dis. Mrk. 2021, 2021, 1244612. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Hao, Y.; Rao, B.; Zhang, Z. A Ferroptosis-Related LncRNA Signature Predicts Prognosis in Ovarian Cancer Patients. Transl. Cancer Res. 2021, 10, 4802–4816. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, B.; Liu, M.; Qiao, H.; Zhang, S.; Qiu, J.; Ying, X. Long Non-Coding RNA SNHG25 Promotes Epithelial Ovarian Cancer Progression by up-Regulating COMP. J. Cancer 2021, 12, 1660–1668. [Google Scholar] [CrossRef]
- Chen, H.; Tian, X.; Luan, Y.; Lu, H. Downregulated Long Noncoding RNA DGCR5 Acts as a New Promising Biomarker for the Diagnosis and Prognosis of Ovarian Cancer. Technol. Cancer Res. Treat. 2019, 18, 153303381989680. [Google Scholar] [CrossRef]
- Arab, K.; Park, Y.J.; Lindroth, A.M.; Schäfer, A.; Oakes, C.; Weichenhan, D.; Lukanova, A.; Lundin, E.; Risch, A.; Meister, M.; et al. Long Noncoding RNA TARID Directs Demethylation and Activation of the Tumor Suppressor TCF21 via GADD45A. Mol. Cell 2014, 55, 604–614. [Google Scholar] [CrossRef]
- Gao, C.; Zhao, D.; Zhao, Q.; Dong, D.; Mu, L.; Zhao, X.; Guo, M.; Xu, A.; Fang, L.; Liu, Q.; et al. Microarray Profiling and Co-Expression Network Analysis of LncRNAs and MRNAs in Ovarian Cancer. Cell Death Discov. 2019, 5, 93. [Google Scholar] [CrossRef]
- Cardillo, N.; Russo, D.; Newtson, A.; Reyes, H.; Lyons, Y.; Devor, E.; Bender, D.; Goodheart, M.J.; Gonzalez-Bosquet, J. Identification of Novel LncRNAs in Ovarian Cancer and Their Impact on Overall Survival. Int. J. Mol. Sci. 2021, 22, 1079. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; You, J.; Han, Y.; Liu, Y.; Huang, M.; Lu, X.; Chen, J.; Zheng, Y. LINC00184 Promotes Ovarian Cancer Cells Proliferation and Cisplatin Resistance by Elevating CNTN1 Expression via Sponging MiR-1305. Onco. Targets. Ther. 2021, 14, 2711–2726. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Luo, B.-H.; Wu, Q.-H.; Li, Q.-L.; Yang, K.-D. LncRNA HCG18 Upregulates TRAF4/TRAF5 to Facilitate Proliferation, Migration and EMT of Epithelial Ovarian Cancer by Targeting MiR-29a/B. Mol. Med. 2022, 28, 2. [Google Scholar] [CrossRef]
- Yang, Q.; Dong, Y.-J. LncRNA SNHG20 Promotes Migration and Invasion of Ovarian Cancer via Modulating the MicroRNA-148a/ROCK1 Axis. J. Ovarian Res. 2021, 14, 168. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Su, H.; Tan, Y. UNC5B-AS1 Promoted Ovarian Cancer Progression by Regulating the H3K27me on NDRG2 via EZH2. Cell Biol. Int. 2020, 44, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fang, L.; Hu, D.; Chen, S.; Shen, S.; Chen, K.; Mu, J.; Li, J.; Zhang, H.; Yong-lin, L.; et al. Necroptosis-Associated Long Noncoding RNAs Can Predict Prognosis and Differentiate between Cold and Hot Tumors in Ovarian Cancer. Front. Oncol. 2022, 12, 967207. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Shi, J.; Xu, Y. Long Non-coding RNA NNT-AS1 Contributes to Cell Proliferation, Metastasis and Apoptosis in Human Ovarian Cancer. Oncol. Lett. 2018, 15, 9264–9270. [Google Scholar] [CrossRef]
- Qi, B.; Liu, S.; Liu, D.; Yao, H.; Yan, R. Comprehensive Analysis of CRIP1 in Patients with Ovarian Cancer, Including CeRNA Network, Immune-Infiltration Pattern, and Clinical Benefit. Dis. Mrk. 2022, 2022, 2687867. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, Y.; Guo, Q.; Zhu, J.; Cao, L.; Guo, X.; Xu, F.; Weng, W.; Ju, X.; Wu, X. Long Non-coding RNA SNHG6 Promotes Cell Proliferation and Migration through Sponging MiR-4465 in Ovarian Clear Cell Carcinoma. J. Cell. Mol. Med. 2019, 23, 5025–5036. [Google Scholar] [CrossRef]


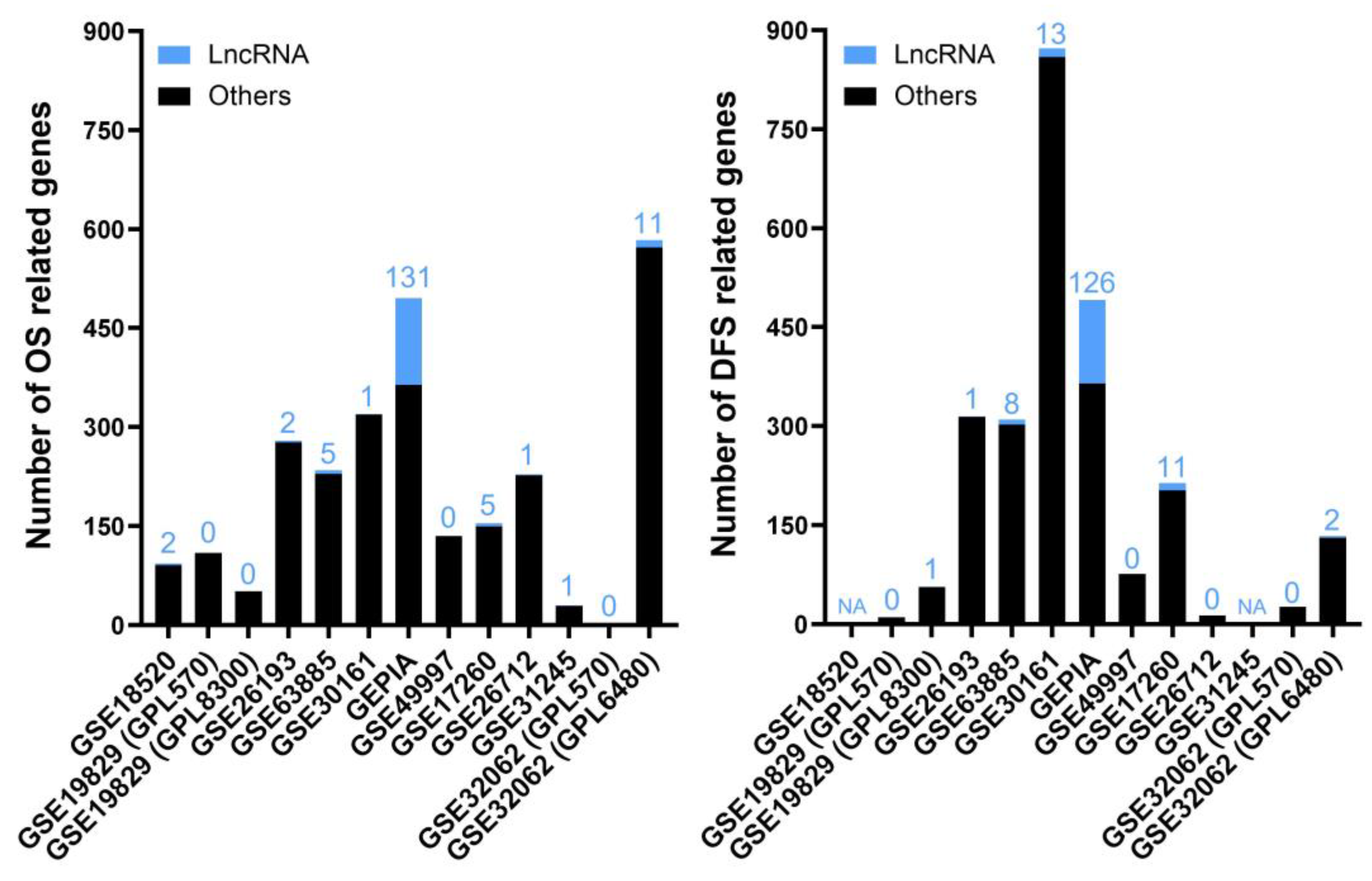

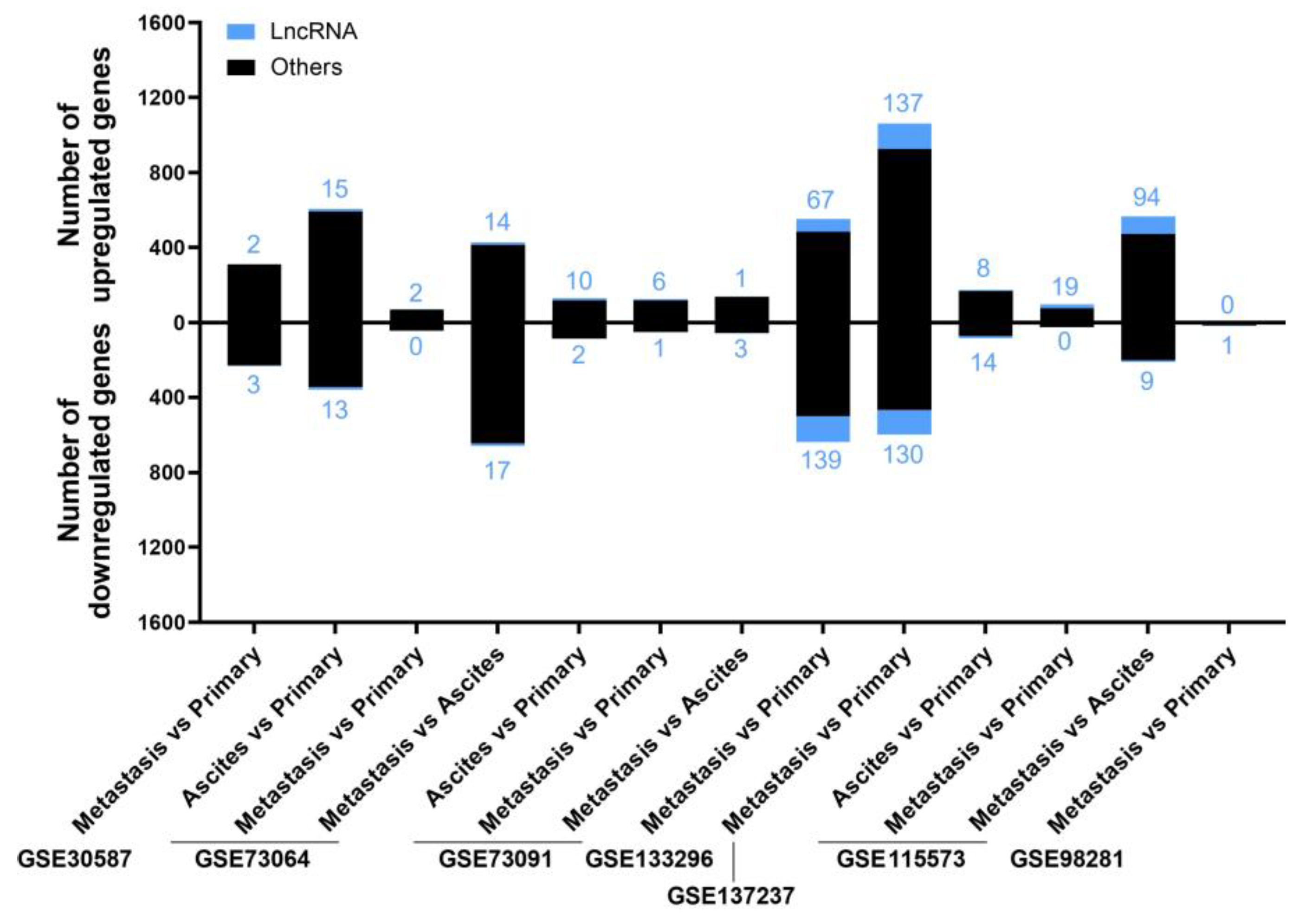







| Number of Studies | Gene Count | Gene Name |
|---|---|---|
| 12 | 2 | RNF157-AS1, BBOX1-AS1 |
| 11 | 1 | DUXAP8 |
| 10 | 6 | ATP2A1-AS1, ENSG00000187951, FOXP4-AS1, MIR205HG, UCA1, ZNF232-AS1 |
| 9 | 2 | LINC00664, LINC01503 |
| 8 | 10 | ENSG00000285756, ESRG, HAGLR, LINC00665, LINC01297, LINC01770, LINC01977, PRKCQ-AS1, TFAP2A-AS1, TIAM1-AS1 |
| 7 | 8 | ASH1L-AS1, HAGLROS, LINC00839, LINC01135, LINC01558, MRPL20-AS1, SPINT1-AS1, VPS13B-DT |
| 6 | 11 | C10orf95-AS1, DLX6-AS1, ENSG00000286546, FZD10-AS1, KLHDC7B-DT, LINC03014, NCK1-DT, PCAT6, PP7080, RNASEH1-AS1, TDRKH-AS1 |
| 5 | 25 | CCDC140, CDKN2B-AS1, DLEU1, DPP3-DT, ELFN1-AS1, WDR35-DT, PPP1R3B-DT, ENSG00000256802, KDM7A-DT, KIAA1614-AS1, KIF25-AS1, LBX2-AS1, LINC00853, LINC00884, LINC01142, LINC01215, LINC01532, LINC02159, LINC02387, LYRM4-AS1, MIR155HG, MYCNOS, OBSCN-AS1, TLR8-AS1, TNFRSF10A-DT |
| 4 | 46 | ATP11A-AS1, CLMAT3, DSP-AS1, ENSG00000231119, ENSG00000260418, ENSG00000267665, EPHA1-AS1, EPIC1, EXOSC10-AS1, GAPLINC, ITGB2-AS1, LAMA5-AS1, LINC00943, LINC00944, LINC00958, LINC01224, LINC01271, LINC01315, LINC01333, LINC01342, LINC01356, LINC01362, LINC01410, LINC01545, LINC01547, LINC02041, LINC02043, LINC02492, LINC03011, LINGO1-AS1, MINCR, MIR1915HG, MIR200CHG, NAV2-AS5, PCAT4, PPP1R14B-AS1, PPP1R26-AS1, PRRT3-AS1, PRRX2-AS1, PSORS1C3, RALY-AS1, SLC2A1-DT, SNHG4, ZNF503-AS1, ZNF687-AS1, ENSG00000215022 |
| 3 | 136 | ANKRD44-IT1, BISPR, BRWD1-AS2, C6orf99, C8orf31, CA3-AS1, CARD8-AS1, CASC9, CSMD2-AS1, CT69, CTB-178M22.2, DHCR24-DT, DHX35-DT, EGFLAM-AS4, ENSG00000224504, ENSG00000226527, ENSG00000259540, ENSG00000260912, ENSG00000261095, ENSG00000261924, ENSG00000273523, ENSG00000289161, ENSG00000290993, ENSG00000291232, FAM151B-DT, FZD4-DT, GNAS-AS1, GPRC5D-AS1, HMMR-AS1, IQCF5-AS1, KANSL1-AS1, KCNMB2-AS1, LCAL1, LCMT1-AS2, LINC00308, LINC00354, LINC00461, LINC00513, LINC00589, LINC00620, LINC00656, LINC00668, LINC00858, LINC00868, LINC00896, LINC00907, LINC01049, LINC01111, LINC01123, LINC01127, LINC01144, LINC01512, LINC01737, LINC01812, LINC01833, LINC01904, LINC02064, LINC02152, LINC02223, LINC02240, LINC02257, LINC02517, LINC02580, LINC02712, LINC02837, LNCOC1, LZTS1-AS1, MAP3K9-DT, MAPT-AS1, MIR3681HG, MIR3945HG, MIR4458HG, MRPL20-DT, NDUFB2-AS1, NEBL-AS1, NINJ2-AS1, NUP50-DT, NUTM2A-AS1, OGFRP1, PARTICL, PIK3CD-AS2, POU2F2-AS1, PSLNR, PTPRN2-AS1, RAPGEF4-AS1, RFX5-AS1, RHPN1-AS1, RNASE11-AS1, SAMD12-AS1, SCIRT, SLC8A1-AS1, STAU2-AS1, TIPARP-AS1, TRPM2-AS, TYMSOS, UBAC2-AS1, UBR5-DT, ENSG00000236529, ENSG00000261211, ENSG00000268204, ENSG00000272275, AQP5-AS1, ENSG00000272763, ENSG00000261068, ENSG00000261135, ENSG00000261061, ENSG00000272872, ENSG00000224272, ENSG00000275216, ENSG00000225335, ENSG00000236924, ENSG00000250899, ENSG00000269155, ENSG00000245719, ENSG00000256234, ENSG00000259163, ENSG00000225489, ENSG00000260920, ENSG00000255114, ENSG00000270460, ENSG00000278238, ENSG00000267698, ENSG00000233461, ENSG00000270174, ENSG00000253174, ENSG00000269968, ENSG00000231295, ENSG00000232386, ENSG00000269399, ENSG00000226334, ENSG00000265415, ENSG00000234694, ENSG00000243224, ENSG00000267512, ENSG00000244184, ENSG00000261071 |
| Number of Studies | Gene Count | Gene Name |
|---|---|---|
| 15 | 1 | MAGI2-AS3 |
| 13 | 2 | ADAMTS9-AS2, HAND2-AS1 |
| 12 | 4 | CRNDE, GAS1RR, MIR22HG, PGM5-AS1 |
| 11 | 4 | ADAMTS9-AS1, ENSG00000267058, EPM2A-DT, NR2F1-AS1 |
| 10 | 4 | GATA6-AS1, GIHCG, KLF3-AS1, WDFY3-AS2 |
| 9 | 6 | ATP2B1-AS1, ERVK13-1, GLIDR, HHIP-AS1, HYMAI, SNHG8 |
| 8 | 9 | C2orf27A, CLN8-AS1, DPP10-AS1, ENSG00000272447, EPB41L4A-AS1, FGF14-AS2, MIR100HG, RASSF8-AS1, SP2-AS1 |
| 7 | 6 | ID2-AS1, LINC00667, LINC00842, LINC02941, LINC03013, ZFAS1 |
| 6 | 12 | CKMT2-AS1, DUBR, EGOT, FGD5-AS1, GABPB1-AS1, LINC00924, LINC01616, PAN3-AS1, SDCBP2-AS1, SLC25A21-AS1, SNHG18, ZEB1-AS1 |
| 5 | 13 | CERNA1, CPVL-AS2, ENSG00000204814, FRMD6-AS2, LINC00342, LINC00526, LINC00683, LINC02754, MEF2C-AS1, NIFK-AS1, PAXBP1-AS1, PWRN1, VLDLR-AS1 |
| 4 | 44 | ADD3-AS1, ANXA2R-AS1, ARHGEF26-AS1, CRIM1-DT, DPH1-AS1, ENSG00000145075, ENSG00000204666, ENSG00000230393, ENSG00000244953, ENSG00000245025, ENSG00000248115, ENSG00000253123, ENSG00000255495, ENSG00000258181, ENSG00000259976, ENSG00000261305, ENSG00000261671, ENSG00000270589, FGF13-AS1, FLRT2-AS1, GASK1B-AS1, KLRK1-AS1, LETR1, LINC00324, LINC00926, LINC00997, LINC01018, LINC01391, LINC02360, LNCRNA-IUR, MIR3936HG, MRGPRF-AS1, NPEPPSP1, NR2F2-AS1, NR4A1AS, OR2A1-AS1, PCAT19, PTPRD-AS1, RPH3AL-AS1, SNHG26, TMEM220-AS1, TPT1-AS1, TRHDE-AS1, WARS2-IT1 |
| 3 | 138 | CAVIN2-AS1, CEROX1, CNN3-DT, DHDDS-AS1, DNAJC27-AS1, ELOA-AS1, ENSG00000226622, ENSG00000226862, ENSG00000227733, ENSG00000231609, ENSG00000232934, ENSG00000233178, ENSG00000233760, ENSG00000233894, ENSG00000234394, ENSG00000234699, ENSG00000235159, ENSG00000235563, ENSG00000238018, ENSG00000243004, ENSG00000246250, ENSG00000249679, ENSG00000249849, ENSG00000250286, ENSG00000250326, ENSG00000254141, ENSG00000254855, ENSG00000256139, ENSG00000256973, ENSG00000257894, ENSG00000259275, ENSG00000259359, ENSG00000259367, ENSG00000259969, ENSG00000260269, ENSG00000260277, ENSG00000260563, ENSG00000260583, ENSG00000260645, ENSG00000260686, ENSG00000260693, ENSG00000260859, ENSG00000261167, ENSG00000261269, ENSG00000261292, ENSG00000266677, ENSG00000267042, ENSG00000267082, ENSG00000267636, ENSG00000267774, ENSG00000268047, ENSG00000268912, ENSG00000269068, ENSG00000269194, ENSG00000269210, ENSG00000270096, ENSG00000270140, ENSG00000270640, ENSG00000271730, ENSG00000271930, ENSG00000272158, ENSG00000272159, ENSG00000272823, ENSG00000272831, ENSG00000273650, ENSG00000274719, ENSG00000275120, ENSG00000277351, ENSG00000277954, ENTPD1-AS1, FAM167A-AS1, FAM182A, FAM53B-AS1, FAM66A, FAM85B, FOXO6-AS1, GARS1-DT, GCC2-AS1, HOXA-AS2, HSD11B1-AS1, INE2, IRS4-AS1, JAZF1-AS1, LINC00602, LINC00844, LINC00847, LINC00886, LINC00891, LINC00921, LINC01229, LINC01402, LINC01474, LINC01560, LINC01619, LINC01625, LINC01852, LINC02126, LINC02145, LINC02202, LINC02308, LINC02345, LINC02613, LINC02691, LINC02731, LINC03007, LRRC8C-DT, LRRK2-DT, MAFTRR, MAP3K4-AS1, MEG8, MIMT1, MIOS-DT, MIR223HG, MIR497HG, MRPS30-DT, MSC-AS1, NALT1, PACERR, PGM5P3-AS1, PGM5P4-AS1, PRR34-AS1, PRSS23-AS1, PSMG3-AS1, RASGRF2-AS1, RBPMS-AS1, RORA-AS1, SAP30-DT, SEMA6A-AS1, SMC5-DT, SNHG5, TICAM2-AS1, TMC3-AS1, TSPEAR-AS1, TTC3-AS1, XPC-AS1, ZEB2-AS1, ZNF300P1, ZNF594-DT |
| LncRNAs Correlating with Longer OS | LncRNAs Correlating with Shorter OS |
|---|---|
| RNF157-AS1, AQP5-AS1, AKAP1-DT, ANKRD44-AS1, ARHGAP28-AS1, BMPR1B-DT, CD2AP-DT, CEACAM16-AS1, DCTN6-DT, DNAJC9-AS1, DNM1P35, DTD1-AS1, EBLN3P, FAM27C, FOXP4-AS1, FZD4-DT, GPRC5D-AS1, HAGLROS, HCG14, HCP5, HLA-L, IFNG-AS1, IL6R-AS1, JARID2-DT, LINC00467, LINC00488, LINC00592, LINC00664, LINC01431, LINC01635, LINC01695, LINC01829, LINC01970, LINC02006, LINC02073, LINC02091, LINC02167, LINC02321, LINC02346, LINC02629, LINC02754, LINC02777, LUNAR1, MIR762HG, MYCNOS, MYL12-AS1, NUTM2A-AS1, PCAT18, POLH-AS1, RAB11B-AS1, RFX5-AS1, RGMB-AS1, RMST, RNASEH1-AS1, SCIRT, SNHG10, SRD5A3-AS1, TRBV11-2, TWSG1-DT, USP30-AS1, ENSG00000228863, ENSG00000259834, ENSG00000235021, ENSG00000273221, ENSG00000232739, ENSG00000270087, ENSG00000255135, ENSG00000256101, ENSG00000256948, SLC38A4-AS1, ENSG00000275769, ENSG00000276727, ENSG00000277863, ENSG00000258521, ENSG00000278022, ENSG00000259772, ENSG00000276408, ENSG00000263063, ENSG00000263427, ENSG00000265840, ENSG00000274213, ENSG00000277597, ENSG00000266149, ENSG00000267627, ENSG00000273368, ENSG00000268555, ENSG00000268650, ENSG00000230432, ENSG00000235319, ENSG00000273063, ENSG00000276517, ENSG00000281920, ENSG00000277901, ENSG00000273210, ENSG00000272858, ENSG00000250039, ENSG00000272626, ENSG00000272986, ENSG00000272203, ENSG00000272417, ENSG00000261071, ENSG00000272209, ENSG00000228506, ENSG00000261189, ENSG00000272009, ENSG00000272243, ENSG00000272379, ENSG00000231794, ENSG00000214870, ENSG00000237773, ENSG00000260997, ENSG00000273391, ENSG00000253982, PPP1R3B-DT, ENSG00000253369, ENSG00000254812, ENSG00000260484, ENSG00000253400, ENSG00000232412, ENSG00000290574, ENSG00000290689, ENSG00000290796 | CRNDE, ZFAS1, C2orf27A, DUXAP8, FLG-AS1, GUSBP11, LINC00484, LINC01127, LINC02881, MAGI2-AS3, MIR600HG, MIR924HG, NRSN2-AS1, PAXBP1-AS1, PCAT4, PIK3R5-DT, RP9P, ENSG00000260917, ENSG00000257545, ENSG00000259341, ENSG00000261069, ENSG00000271725, ENSG00000270020, ENSG00000275438, ENSG00000266709, LINC02958, ENSG00000213963, ENSG00000229839, ENSG00000270696, ENSG00000240057, ENSG00000230074, ENSG00000290659 |
| LncRNAs Correlating with Longer DFS | LncRNAs Correlating with Shorter DFS |
|---|---|
| ADCY10P1, ANO1-AS1, ATXN2-AS, B4GALT1-AS1, C1orf21-DT, C6orf223, CRTC3-AS1, CT62, CXXC5-AS1, DIAPH2-AS1, DLGAP1-AS1, DNM1P35, EIF1B-AS1, EML2-AS1, FAM111A-DT, FAM27C, FOXP4-AS1, FZD4-DT, HCG15, IFNG-AS1, JARID2-DT, LINC00221, LINC00243, LINC00582, LINC00592, LINC00927, LINC01003, LINC01144, LINC01825, LINC02298, LINC02397, LINC02541, LINC02601, LINC02875, LL22NC03-63E9.3, LUNAR1, MAN2A1-DT, MIAT, MIR3142HG, NCKAP5-AS2, NDUFV2-AS1, NIFK-AS1, NR2F2-AS1, NSMCE1-DT, OLMALINC, PKP4-AS1, POLG-DT, PP2672, PPP1R21-DT, PPP1R26-AS1, RNF157-AS1, RORA-AS1, RPH3AL-AS1, SBF2-AS1, SLC25A5-AS1, SUGCT-AS1, TAX1BP1-AS1, THOC7-AS1, TRBV11-2, XIAP-AS1, ZSCAN5A-AS1, ENSG00000224152, ENSG00000226334, ENSG00000226889, ENSG00000228021, ENSG00000228084, ENSG00000228863, ENSG00000229311, ENSG00000230912, ENSG00000231081, ENSG00000231098, ENSG00000231519, ENSG00000231794, ENSG00000232533, ENSG00000233230, ENSG00000233242, ENSG00000234134, ENSG00000235586, ENSG00000236140, ENSG00000254290, ENSG00000254343, ENSG00000255440, ENSG00000257042, ENSG00000257327, AQP5-AS1, ENSG00000258534, ENSG00000258572, ENSG00000258904, ENSG00000259802, ENSG00000259834, ENSG00000260038, ENSG00000260352, ENSG00000260369, ENSG00000261071, ENSG00000261204, ENSG00000261320, ENSG00000261655, ENSG00000262115, ENSG00000263063, ENSG00000266830, ENSG00000267255, ENSG00000267666, ENSG00000269951, ENSG00000270115, ENSG00000270265, ENSG00000271367, ENSG00000272172, ENSG00000272209, ENSG00000272456, ENSG00000272672, ENSG00000272831, ENSG00000273004, ENSG00000273104, ENSG00000273162, ENSG00000273210, ENSG00000273456, ENSG00000273989, ENSG00000275155, ENSG00000275202, ENSG00000275297, ENSG00000275484, ENSG00000276408, ENSG00000277901, ENSG00000278022, ENSG00000291006, ENSG00000291136 | GUSBP11, MALAT1, AQP4-AS1, DSG2-AS1, HLA-F-AS1, HOTAIRM1, LINC00452, LINC00472, LINC00565, LINC01140, LINC02328, LINC02432, LINC-PINT, LNCOC1, MIR924HG, PHACTR2-AS1, RRP7BP, TMEM161B-AS1, ZNF295-AS1, ZNF503-AS2, ENSG00000233834, ENSG00000243004, ENSG00000248268, ENSG00000249476, ENSG00000258603, ENSG00000259065, ENSG00000260412, ENSG00000261292, ENSG00000265975, ENSG00000266283, ENSG00000270074, ENSG00000272632, ENSG00000278668, ENSG00000280604 |
| Number of Studies | Number of Comparisons | Gene Count | Gene Name |
|---|---|---|---|
| 3 | 4 | 2 | LINC02544, LINC01235 |
| 2 | 3 | 2 | HECW2-AS1, MIR31HG |
| 2 | 2 | 20 | ENSG00000282057, UNC5C-AS1, ENSG00000261327, ENSG00000259807, MEG9, LINC01561, SNHG18, HOTAIRM1, LINC00922, LINC01619, FAM225A, HOXA-AS3, LINC00968, ENSG00000271811, ENSG00000272755, ENSG00000233682, ENSG00000248540, ENSG00000259663, LINC02593, HOXA-AS2 |
| 1 + cell line | 1 | 6 | TENM3-AS1, LINC01678, ENSG00000250410, ENSG00000261604, ENSG0000022749, LNCOG |
| Number of Studies | Number of Comparisons | Gene Count | Gene Name |
|---|---|---|---|
| 1 + cell line | 1 | 8 | ENSG00000260604, ENSG00000267284, LINC01508, LINC01138, FAM160A1-DT, ENSG00000255118, ENSG00000249049, ENSG00000225649 |
| Status in Ascites | Gene Count | Gene Name |
|---|---|---|
| Up | 13 | TP53TG1, ENSG00000253982, MIR210HG, LINC00667, ATP2B1-AS1, ENSG00000275210, LINC00847, ENSG00000259153, ENSG00000243655, FCGR1BP, LINC00888, ENSG00000291107, ENSG00000291230 |
| Down | 13 | MEG3, PCBP1-AS1, ENSG00000223774, FLJ16779, ENSG00000263065, SNHG12, IGFBP7-AS1, IDI2-AS1, PSMA3-AS1, NUTM2B-AS1, SNHG3, MALAT1, SMG1P5 |
| HGSOC | |
|---|---|
| Upregulated lncRNA Genes | Downregulated lncRNA Genes |
| WT1-AS *, PART1, ENSG00000255135 | FAM155A-IT1, DLX6-AS1 |
| CCOC | |
|---|---|
| Upregulated lncRNA Genes | Downregulated lncRNA Genes |
| SNHG12, LINC00472, C8orf31 *, RBPMS-AS1, CPNE8-AS1, FAM155A-IT1, LINC01137, LINC02765, LINC01637, PPP1R3B-DT, PCCA-DT, LINC00598, LINC02435, ENSG00000271992, ENSG00000259052, ENSG00000218416, ENSG00000253307, ENSG00000223786, ENSG00000224583, ENSG00000264596, ENSG00000274718, ENSG00000187185, ENSG00000253666, ENSG00000257681, ENSG00000257831, ENSG00000236283, ENSG00000260317, ENSG00000265625, ENSG00000286546, ENSG00000249125 | PART1, ZNF667-AS1, WT1-AS, HCP5, LINC01139, TARID, PAXIP1-DT PRKCQ-AS1, EMX2OS, ENSG00000227733, ENSG00000235560, ENSG00000255135 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salamini-Montemurri, M.; Lamas-Maceiras, M.; Lorenzo-Catoira, L.; Vizoso-Vázquez, Á.; Barreiro-Alonso, A.; Rodríguez-Belmonte, E.; Quindós-Varela, M.; Cerdán, M.E. Identification of lncRNAs Deregulated in Epithelial Ovarian Cancer Based on a Gene Expression Profiling Meta-Analysis. Int. J. Mol. Sci. 2023, 24, 10798. https://doi.org/10.3390/ijms241310798
Salamini-Montemurri M, Lamas-Maceiras M, Lorenzo-Catoira L, Vizoso-Vázquez Á, Barreiro-Alonso A, Rodríguez-Belmonte E, Quindós-Varela M, Cerdán ME. Identification of lncRNAs Deregulated in Epithelial Ovarian Cancer Based on a Gene Expression Profiling Meta-Analysis. International Journal of Molecular Sciences. 2023; 24(13):10798. https://doi.org/10.3390/ijms241310798
Chicago/Turabian StyleSalamini-Montemurri, Martín, Mónica Lamas-Maceiras, Lidia Lorenzo-Catoira, Ángel Vizoso-Vázquez, Aida Barreiro-Alonso, Esther Rodríguez-Belmonte, María Quindós-Varela, and M. Esperanza Cerdán. 2023. "Identification of lncRNAs Deregulated in Epithelial Ovarian Cancer Based on a Gene Expression Profiling Meta-Analysis" International Journal of Molecular Sciences 24, no. 13: 10798. https://doi.org/10.3390/ijms241310798
APA StyleSalamini-Montemurri, M., Lamas-Maceiras, M., Lorenzo-Catoira, L., Vizoso-Vázquez, Á., Barreiro-Alonso, A., Rodríguez-Belmonte, E., Quindós-Varela, M., & Cerdán, M. E. (2023). Identification of lncRNAs Deregulated in Epithelial Ovarian Cancer Based on a Gene Expression Profiling Meta-Analysis. International Journal of Molecular Sciences, 24(13), 10798. https://doi.org/10.3390/ijms241310798








