Integration of Metabolomic and Transcriptomic Provides Insights into Anti-Inflammatory Response to trans-10-Hydroxy-2-decenoic Acid on LPS-Stimulated RAW 264.7 Cells
Abstract
:1. Introduction
2. Results
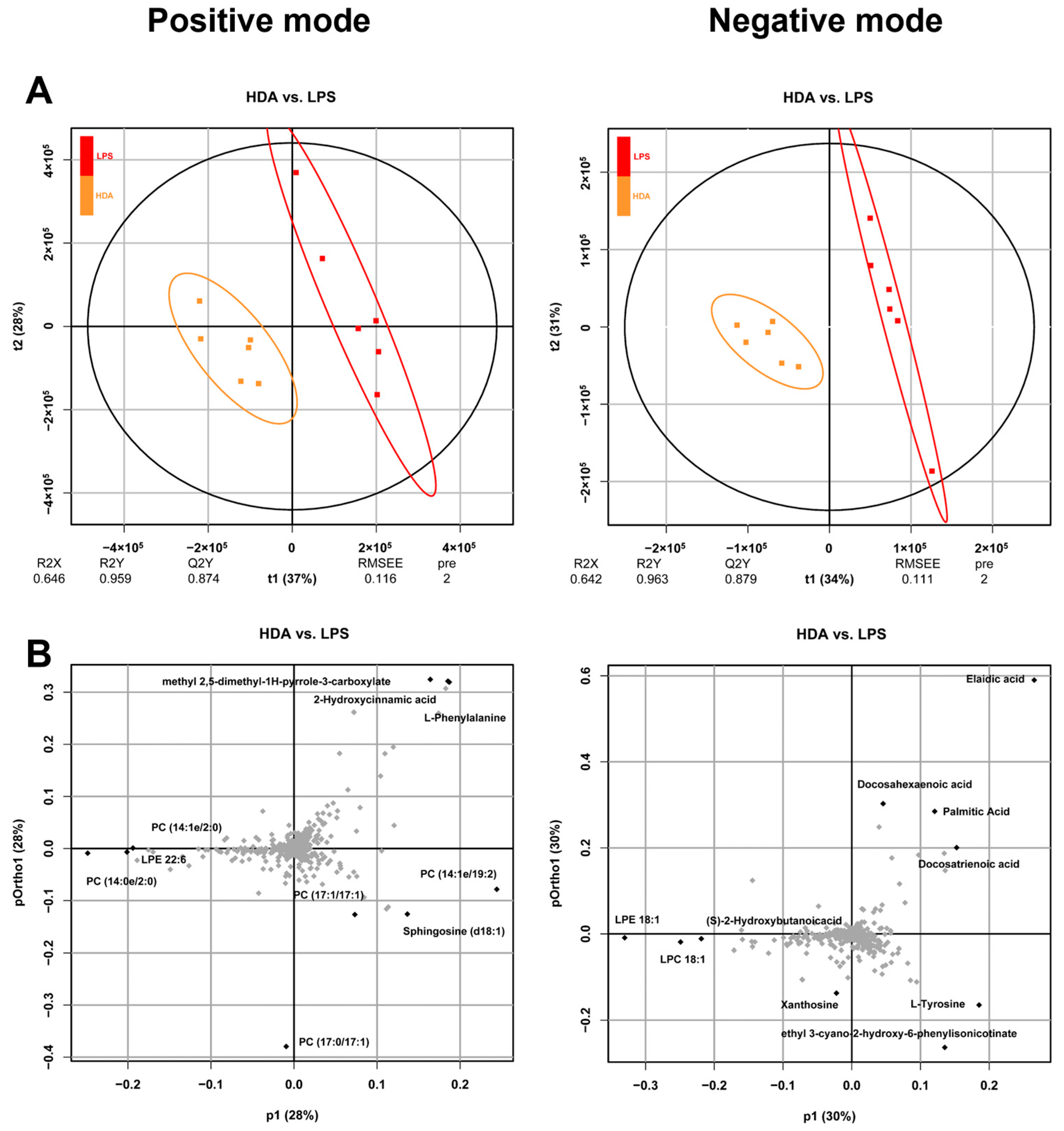
2.1. Intracellular Metabolomic Changes Associated with 10-HDA Treated
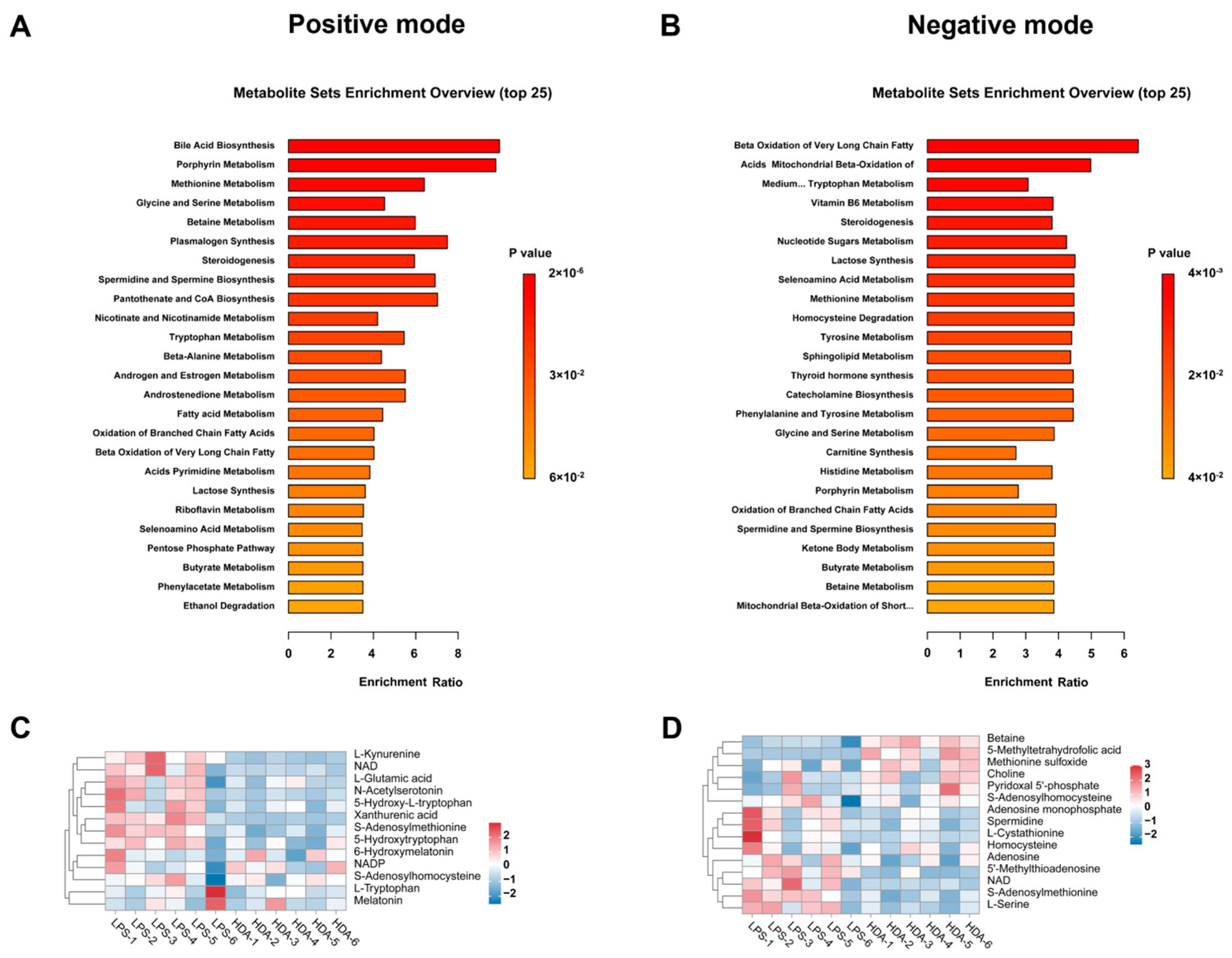
2.2. Impacted Metabolic Pathways
2.3. Genes Alteration and Function Analysis Associated with 10-HDA Treated
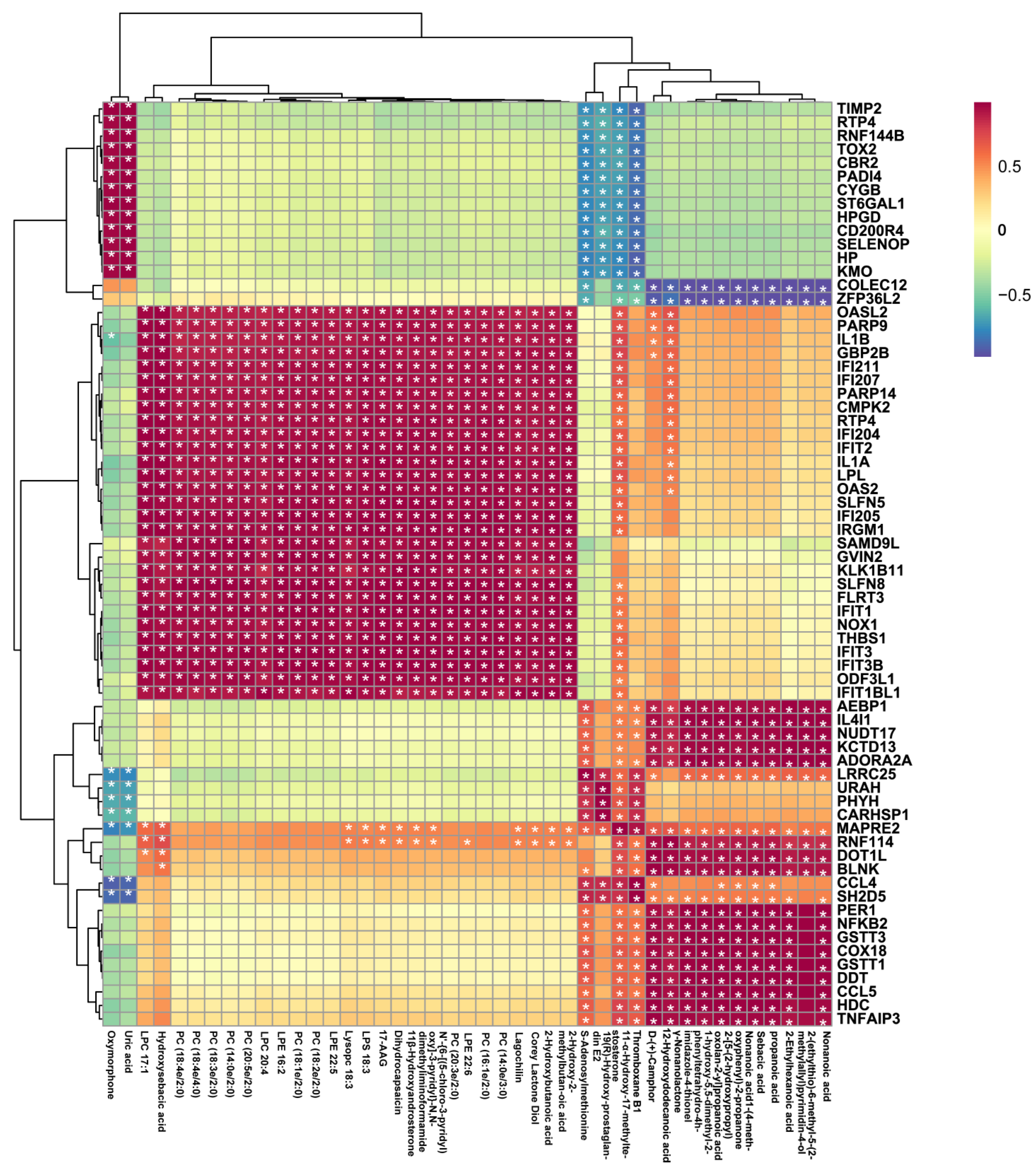
2.4. Combined Analysis of the Mechanism of LPS-Stimulated RAW 264.7 Cells Treated with 10-HDA
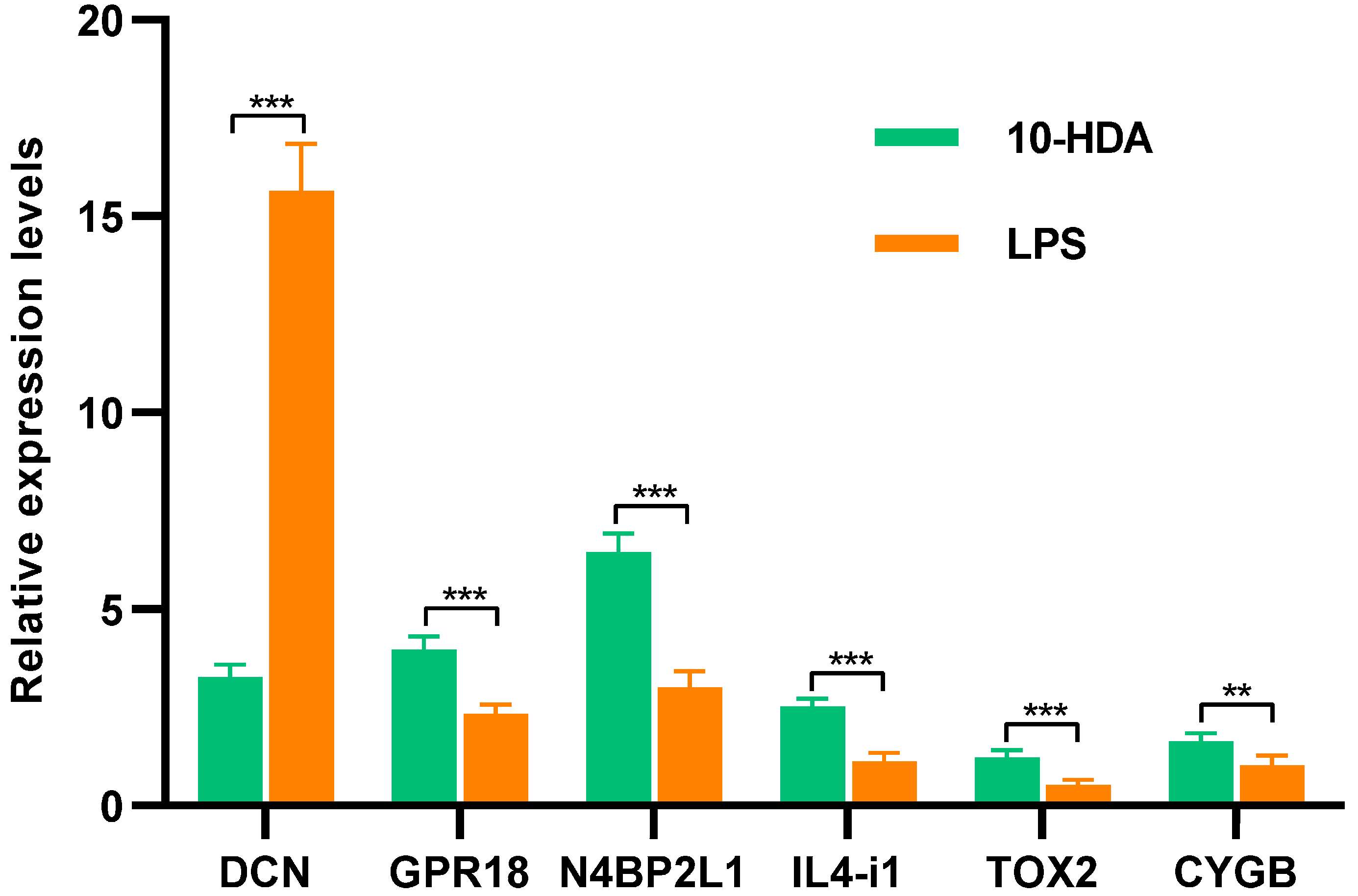
2.5. Quantitative Real-PCR Validation
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture and Treatment
4.3. Metabolite Extraction
4.4. UHPLC-MS/MS and Metabolites Analyses
4.5. UHPLC-MS/MS and Metabolites Analyses
4.6. Gene Differential Expression and Enrichment Analyses
4.7. Gene Differential Expression and Enrichment Analyses
4.8. Real-Time Quantitative Polymerase Chain Reaction Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hotamisligil, G.S. Inflammation, metaflammation and immunometabolic disorders. Nature 2017, 542, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, Inflammation, and Cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccolo, V.; Curina, A.; Genua, M.; Ghisletti, S.; Simonatto, M.; Sabò, A.; Amati, B.; Ostuni, R.; Natoli, G. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat. Immunol. 2017, 18, 530–540. [Google Scholar] [CrossRef] [Green Version]
- Chung, S.J.; Lee, C.H.; Lee, H.S.; Kim, S.T.; Sohn, U.D.; Park, E.S.; Bang, J.S.; Lee, J.H.; Chung, Y.H.; Jeong, J.H. The role of phosphatidylcholine and deoxycholic acid in inflammation. Life Sci. 2014, 108, 88–93. [Google Scholar] [CrossRef]
- Jung, J.; Zeng, H.; Horng, T. Metabolism as a guiding force for immunity. Nat. Cell Biol. 2019, 21, 85–93. [Google Scholar] [CrossRef]
- Ma, B.; Yang, Y.; Li, Z.; Zhao, D.; Zhang, W.; Jiang, Y.; Xue, D. Modular bioinformatics analysis demonstrates that a Toll-like receptor signaling pathway is involved in the regulation of macrophage polarization. Mol. Med. Rep. 2018, 18, 4313–4320. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Cao, X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 2019, 19, 417–432. [Google Scholar] [CrossRef]
- Medzhitov, R.; Horng, T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 2009, 9, 692–703. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Pearce, E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016, 213, 15–23. [Google Scholar] [CrossRef]
- O’Neill, L.A.J.; Hardie, D.G. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 2013, 493, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Wu, L.; Wang, K. Chemical Composition of Royal Jelly. In Bee Products–Chemical and Biological Properties; Alvarez-Suarez, J.M., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 181–190. [Google Scholar]
- Isidorov, V.A.; Czyżewska, U.; Jankowska, E.; Bakier, S. Determination of royal jelly acids in honey. Food Chem. 2011, 124, 387–391. [Google Scholar] [CrossRef]
- Eslami-Kaliji, F.; Mirahmadi-Zare, S.Z.; Nazem, S.; Shafie, N.; Ghaedi, R.; Asadian-esfahani, M.h. A label-free SPR biosensor for specific detection of TLR4 expression; introducing of 10-HDA as an antagonist. Int. J. Biol. Macromol. 2022, 217, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Takahashi, K.; Tokoro, S.; Gotou, T.; Neri, P.; Mori, H. Inhibitory effect of 10-hydroxy-trans-2-decenoic acid on LPS-induced IL-6 production via reducing IκB-ζ expression. Innate Immun. 2012, 18, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Xiao, M.; Dong, J.; Huang, Y.; Sun, H.; Wang, D. Synergistic anti-inflammatory effects of graphene oxide quantum dots and trans-10-hydroxy-2-decenoic acid on LPS-stimulated RAW 264.7 macrophage cells. Biomater. Adv. 2022, 136, 212774. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Mantovani, A. Orchestration of Metabolism by Macrophages. Cell Metab. 2012, 15, 432–437. [Google Scholar] [CrossRef] [Green Version]
- Geiß, C.; Salas, E.; Guevara-Coto, J.; Régnier-Vigouroux, A.; Mora-Rodríguez, R.A. Multistability in Macrophage Activation Pathways and Metabolic Implications. Cells 2022, 11, 404. [Google Scholar] [CrossRef] [PubMed]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of glutathione in cancer progression and chemoresistance. Oxid. Med. Cell Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef] [Green Version]
- Farzaneh, H.; Ebrahimi Nik, M.; Mashreghi, M.; Saberi, Z.; Jaafari, M.R.; Teymouri, M. A study on the role of cholesterol and phosphatidylcholine in various features of liposomal doxorubicin: From liposomal preparation to therapy. Inter. J. Pharmaceut. 2018, 551, 300–308. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Chakraborty, K.; Shukla, A. Cellular copper homeostasis: Current concepts on its interplay with glutathione homeostasis and its implication in physiology and human diseases. Metallomics 2017, 9, 1376–1388. [Google Scholar] [CrossRef]
- Lewerenz, J.; Hewett, S.J.; Huang, Y.; Lambros, M.; Gout, P.W.; Kalivas, P.W.; Massie, A.; Smolders, I.; Methner, A.; Pergande, M.; et al. The cystine/glutamate antiporter system x(c)(-) in health and disease: From molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Sign 2013, 18, 522–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanderson, S.M.; Gao, X.; Dai, Z.; Locasale, J.W. Methionine metabolism in health and cancer: A nexus of diet and precision medicine. Nat. Rev. Cancer 2019, 19, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bazer, F.W.; Burghardt, R.C.; Johnson, G.A.; Kim, S.W.; Knabe, D.A.; Li, P.; Li, X.; McKnight, J.R.; Satterfield, M.C.; et al. Proline and hydroxyproline metabolism: Implications for animal and human nutrition. Amino Acids 2011, 40, 1053–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Xu, R.; Gu, H.; Zhang, E.; Qu, J.; Cao, W.; Huang, X.; Yan, H.; He, J.; Cai, Z. Metabolic reprogramming in macrophage responses. Biomark. Res. 2021, 9, 1. [Google Scholar] [CrossRef]
- dos Santos, L.M.; da Silva, T.M.; Azambuja, J.H.; Ramos, P.T.; Oliveira, P.S.; da Silveira, E.F.; Pedra, N.S.; Galdino, K.; do Couto, C.A.T.; Soares, M.S.P.; et al. Methionine and methionine sulfoxide treatment induces M1/classical macrophage polarization and modulates oxidative stress and purinergic signaling parameters. Mol. Cell Biochem. 2017, 424, 69–78. [Google Scholar] [CrossRef]
- Ouyang, Y.; Wu, Q.; Li, J.; Sun, S.; Sun, S. S-adenosylmethionine: A metabolite critical to the regulation of autophagy. Cell Proliferat 2020, 53, e12891. [Google Scholar] [CrossRef]
- Fall, F.; Lamy, E.; Brollo, M.; Naline, E.; Lenuzza, N.; Thévenot, E.; Devillier, P.; Grassin-Delyle, S. Metabolic reprograming of LPS-stimulated human lung macrophages involves tryptophan metabolism and the aspartate-arginosuccinate shunt. PLoS ONE 2020, 15, e0230813. [Google Scholar] [CrossRef] [Green Version]
- Sorgdrager, F.J.H.; Naudé, P.J.W.; Kema, I.P.; Nollen, E.A.; Deyn, P.P. Tryptophan Metabolism in Inflammaging: From Biomarker to Therapeutic Target. Front. Immunol. 2019, 10, 2565. [Google Scholar] [CrossRef]
- Mándi, Y.; Vécsei, L. The kynurenine system and immunoregulation. J. Neural Transm. 2012, 119, 197–209. [Google Scholar] [CrossRef]
- Rock, K.L.; Farfán-Arribas, D.J.; Shen, L. Proteases in MHC Class I Presentation and Cross-Presentation. J. Immunol. 2009, 184, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Bogdan, C. Nitric oxide synthase in innate and adaptive immunity: An update. Trends Immunol. 2015, 36, 161–178. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.H.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.; Liberal, J.; Martins, J.D.; Silva, A.; Neves, B.M.; Cruz, M.T. Inflammasome in Dendritic Cells Immunobiology: Implications to Diseases and Therapeutic Strategies. Curr. Drug Targets 2017, 18, 1003–1018. [Google Scholar] [CrossRef] [PubMed]
- Roudaire, T.; Héloir, M.-C.; Wendehenne, D.; Zadoroznyj, A.; Dubrez, L.; Poinssot, B. Cross Kingdom Immunity: The Role of Immune Receptors and Downstream Signaling in Animal and Plant Cell Death. Front. Immunol. 2021, 11, 612452. [Google Scholar] [CrossRef]
- Treede, I.; Braun, A.; Sparla, R.; Kühnel, M.; Giese, T.; Turner, J.R.; Anes, E.; Kulaksiz, H.; Füllekrug, J.; Stremmel, W.; et al. Anti-inflammatory Effects of Phosphatidylcholine*. J. Biol. Chem. 2007, 282, 27155–27164. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.; Houweling, M. Phosphatidylcholine and cell death. BBA-Mol. Cell Biol. Lipids 2002, 1585, 87–96. [Google Scholar] [CrossRef]
- Pfalzer, A.C.; Choi, S.-W.; Tammen, S.A.; Park, L.K.; Bottiglieri, T.; Parnell, L.D.; Lamon-Fava, S. S-adenosylmethionine mediates inhibition of inflammatory response and changes in DNA methylation in human macrophages. Physiol. Genom. 2014, 46, 617–623. [Google Scholar] [CrossRef]
- Sağraç, D.; Şenkal, S.; Hayal, T.B.; Demirci, S.; Şişli, H.B.; Asutay, A.B.; Doğan, A. Protective role of Cytoglobin and Neuroglobin against the Lipopolysaccharide (LPS)-induced inflammation in Leydig cells ex vivo. Reprod. Biol. 2022, 22, 100595. [Google Scholar] [CrossRef]
- Stylianou, E.; Saklatvala, J. Interleukin-1. Int. J. Biochem. Cell Biol. 1998, 30, 1075–1079. [Google Scholar] [CrossRef]
- Lin, X.M.; Liu, S.B.; Luo, Y.H.; Xu, W.T.; Zhang, Y.; Zhang, T.; Xue, H.; Zuo, W.B.; Li, Y.N.; Lu, B.X.; et al. 10-HDA Induces ROS-Mediated Apoptosis in A549 Human Lung Cancer Cells by Regulating the MAPK, STAT3, NF-κB, and TGF-β1 Signaling Pathways. Biomed. Res. Int. 2020, 2020, 3042636. [Google Scholar] [CrossRef]
- Chen, Y.-F.; You, M.-M.; Liu, Y.-C.; Shi, Y.-Z.; Wang, K.; Lu, Y.-Y.; Hu, F.-L. Potential protective effect of Trans-10-hydroxy-2-decenoic acid on the inflammation induced by Lipoteichoic acid. J. Funct. Foods 2018, 45, 491–498. [Google Scholar] [CrossRef]
- Albalawi, A.E.; Althobaiti, N.A.; Alrdahe, S.S.; Alhasani, R.H.; Alaryani, F.S.; BinMowyna, M.N. Anti-Tumor Effects of Queen Bee Acid (10-Hydroxy-2-Decenoic Acid) Alone and in Combination with Cyclophosphamide and Its Cellular Mechanisms against Ehrlich Solid Tumor in Mice. Molecules 2021, 26, 7021. [Google Scholar] [CrossRef] [PubMed]
- Weiser, M.J.; Grimshaw, V.; Wynalda, K.M.; Mohajeri, M.H.; Butt, C.M. Long-Term Administration of Queen Bee Acid (QBA) to Rodents Reduces Anxiety-Like Behavior, Promotes Neuronal Health and Improves Body Composition. Nutrients 2018, 10, 13. [Google Scholar] [CrossRef] [Green Version]
- Bylesjo, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemometr. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, 71–77. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Bartel, J.; Krumsiek, J.; Schramm, K.; Adamski, J.; Gieger, C.; Herder, C.; Carstensen, M.; Peters, A.; Rathmann, W.; Roden, M.; et al. The Human Blood Metabolome-Transcriptome Interface. PLoS Genet. 2015, 11, e1005274. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Gene | Forward (5′-3′) | Reserve (5′-3′) | bp |
|---|---|---|---|
| GAPDH | TGACCTCAACTACATGGTCTACA | CTTCCCATTCTCGGCCTTG | 85 |
| DCN | TCTTGGGCTGGACCATTTGAA | CATCGGTAGGGGCACATAGA | 119 |
| GPR18 | CACCCTGAGCAATCACAACCA | AGTGACATTAACAAACAGCCCA | 121 |
| N4BP2L1 | TATTCCGGGAACCCGACACT | GAAGGACACTGTGAAAGGTAACA | 138 |
| IL4-i1 | AACACTTGTTGGTGGAAACGA | TCCTTGCGATTAGGAGTGGTC | 241 |
| TOX2 | TATAACGCCTCCCAATCTCCC | TTGCTTGGTACGAGTAGGCAG | 125 |
| CYGB | GGCGACATGGAGATAGAGCG | CTCGCAGTTGGCATACAGC | 102 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.; Dong, J.; Tan, X.; Yang, S.; Xiao, M.; Wang, D. Integration of Metabolomic and Transcriptomic Provides Insights into Anti-Inflammatory Response to trans-10-Hydroxy-2-decenoic Acid on LPS-Stimulated RAW 264.7 Cells. Int. J. Mol. Sci. 2023, 24, 12666. https://doi.org/10.3390/ijms241612666
Huang M, Dong J, Tan X, Yang S, Xiao M, Wang D. Integration of Metabolomic and Transcriptomic Provides Insights into Anti-Inflammatory Response to trans-10-Hydroxy-2-decenoic Acid on LPS-Stimulated RAW 264.7 Cells. International Journal of Molecular Sciences. 2023; 24(16):12666. https://doi.org/10.3390/ijms241612666
Chicago/Turabian StyleHuang, Minjie, Jie Dong, Xiaodong Tan, Shuyuan Yang, Minghui Xiao, and Deqian Wang. 2023. "Integration of Metabolomic and Transcriptomic Provides Insights into Anti-Inflammatory Response to trans-10-Hydroxy-2-decenoic Acid on LPS-Stimulated RAW 264.7 Cells" International Journal of Molecular Sciences 24, no. 16: 12666. https://doi.org/10.3390/ijms241612666






