In Silico Gene Prioritization Highlights the Significance of Bone Morphogenetic Protein 4 (BMP4) Promoter Methylation across All Methylation Clusters in Colorectal Cancer
Abstract
1. Introduction
2. Results
2.1. Classification of CIMP Status
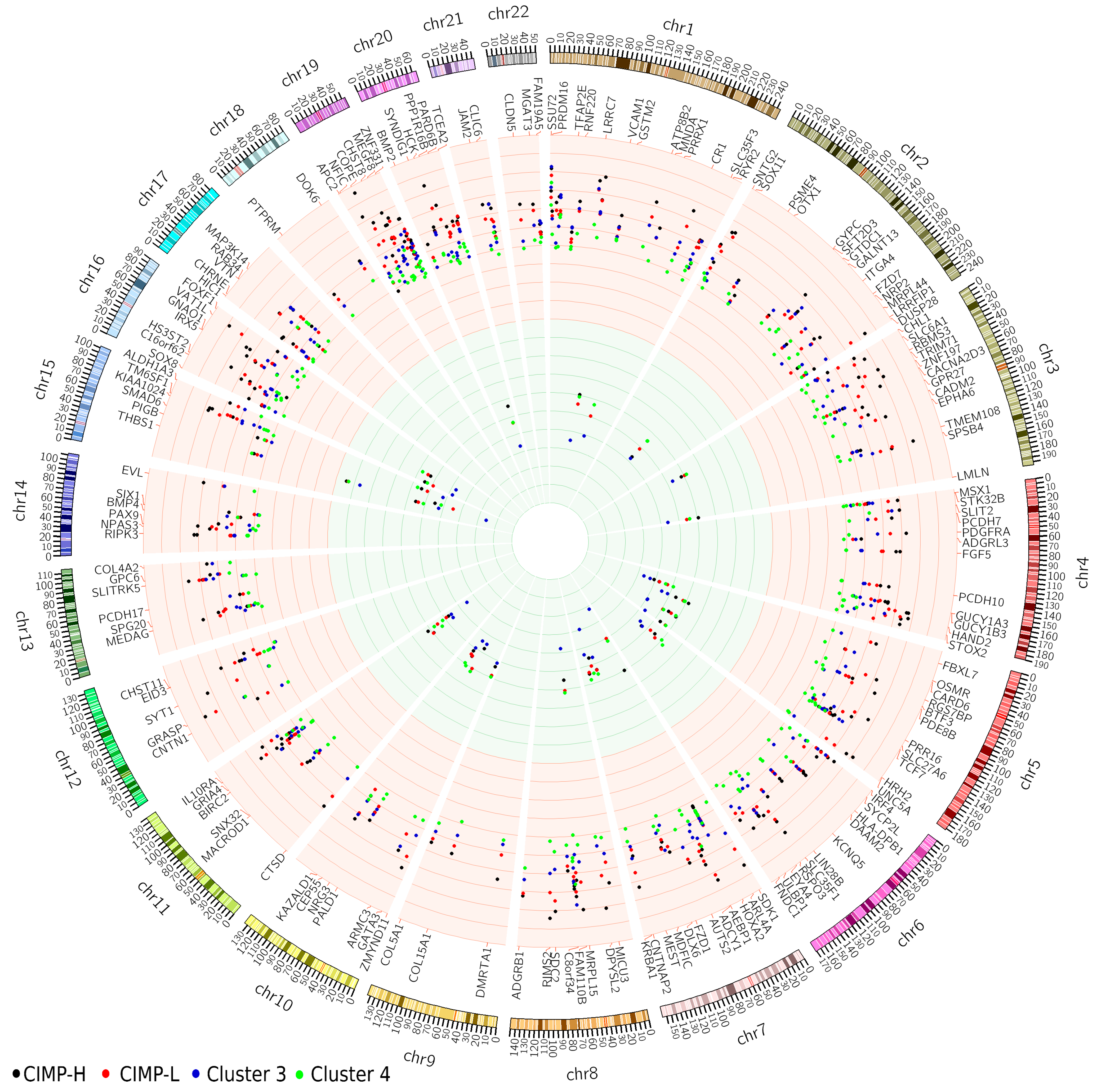
2.2. Promoters Methylated in All Clusters
2.3. Functional Analysis
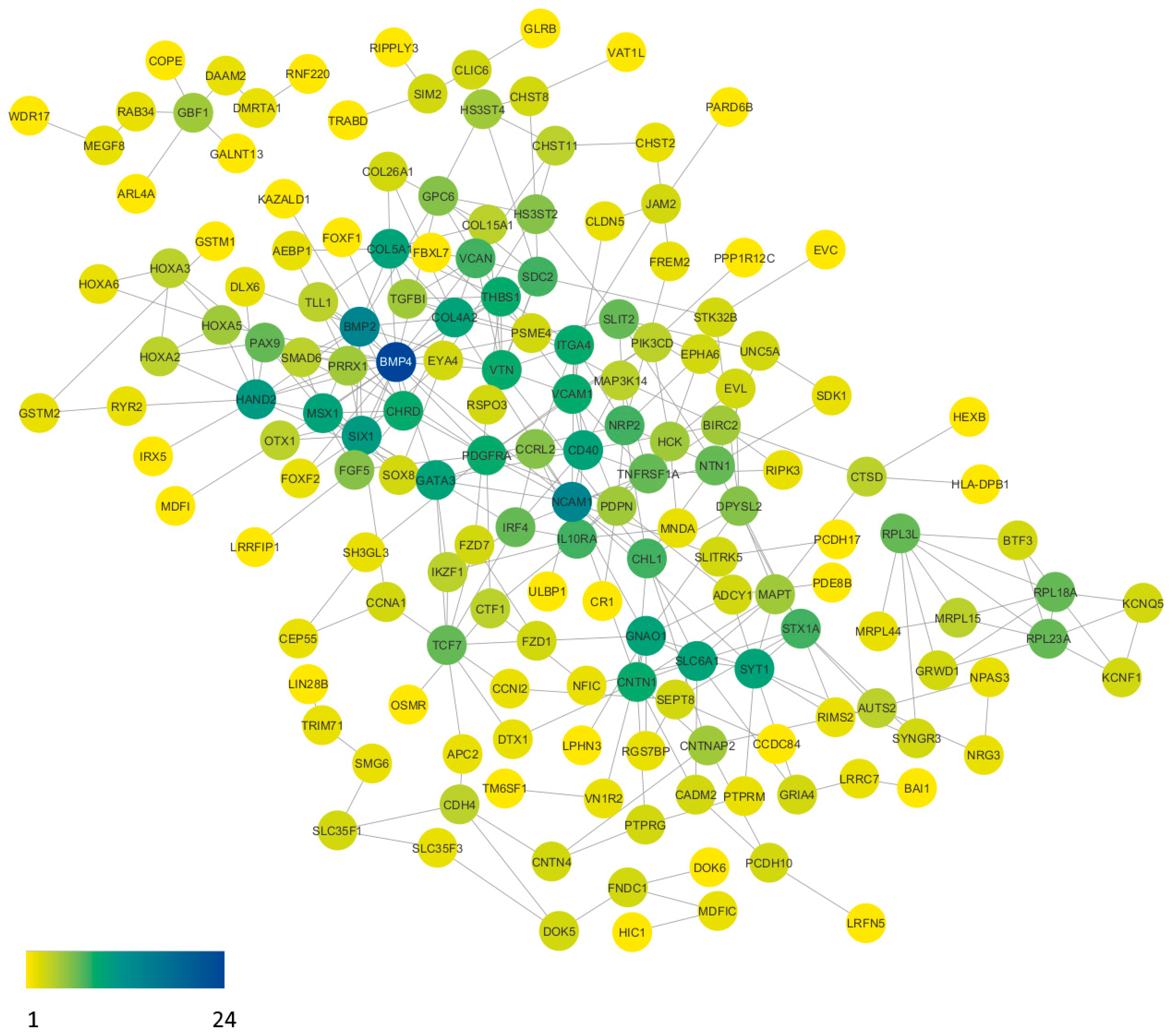
2.4. Protein–Protein Interactions
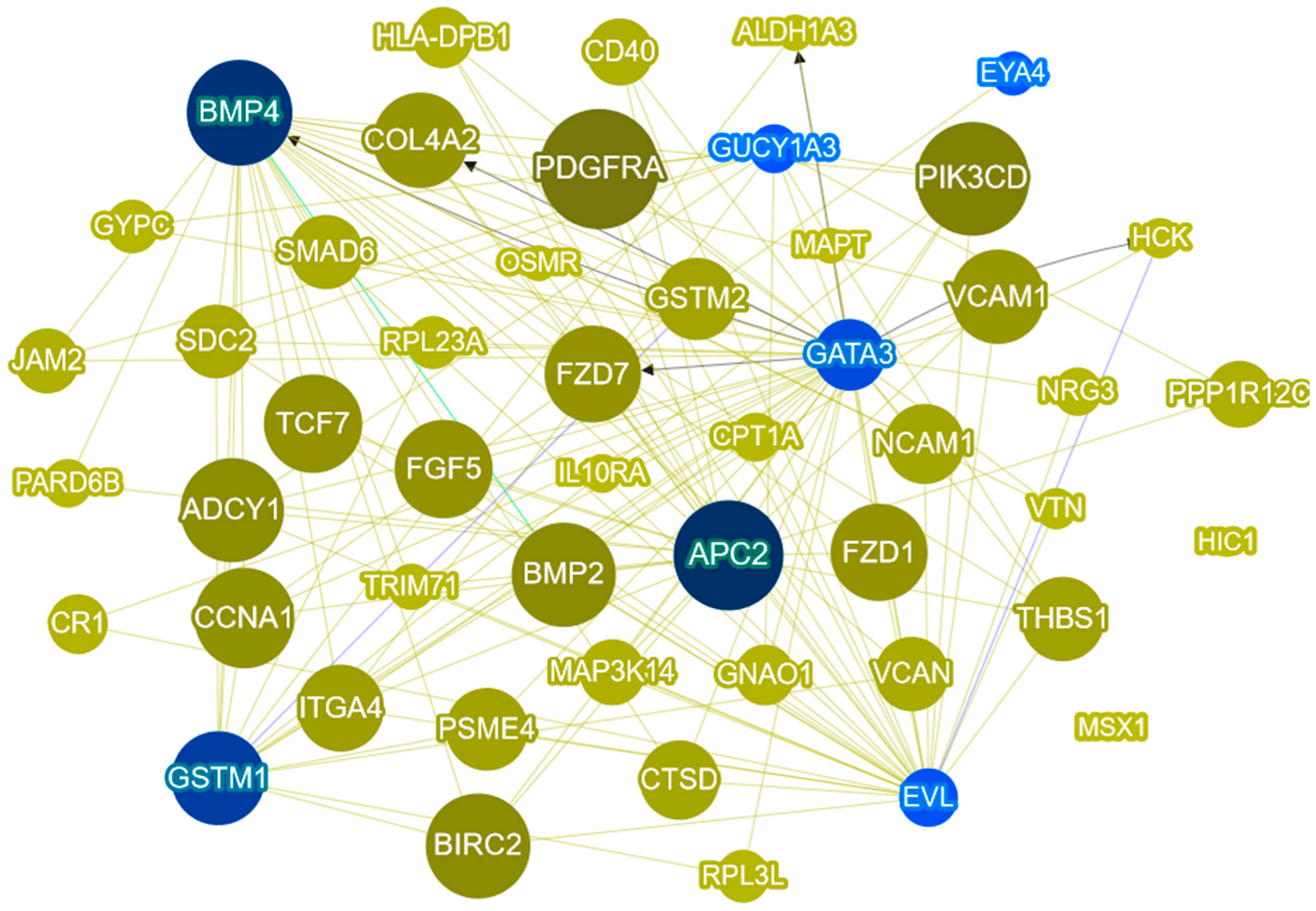
2.5. Gene Prioretization Using Phenolyzer
3. Materials and Methods
3.1. Patients and Data
3.2. Probes and Genes
3.3. Unsupervised Clustering
3.4. Statistical Analysis and Data Visualization
3.5. Functional Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Laird, P.W. Interplay between the cancer genome and epigenome. Cell 2013, 153, 38–55. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef]
- Laird, P.W. Cancer epigenetics. Hum. Mol. Genet. 2005, 14, R65–R76. [Google Scholar] [CrossRef]
- Issa, J.P. CpG island methylator phenotype in cancer. Nat. Rev. Cancer 2004, 4, 988–993. [Google Scholar] [CrossRef]
- Rashid, A.; Issa, J.P. CpG island methylation in gastroenterologic neoplasia: A maturing field. Gastroenterology 2004, 127, 1578–1588. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, W.; Li, J. Identifying molecular subtypes in human colon cancer using gene expression and DNA methylation microarray data. Int. J. Oncol. 2016, 48, 690–702. [Google Scholar] [CrossRef]
- Toyota, M.; Ahuja, N.; Ohe-Toyota, M.; Herman, J.G.; Baylin, S.B.; Issa, J.P. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA 1999, 96, 8681–8686. [Google Scholar] [CrossRef]
- Yagi, K.; Akagi, K.; Hayashi, H.; Nagae, G.; Tsuji, S.; Isagawa, T.; Midorikawa, Y.; Nishimura, Y.; Sakamoto, H.; Seto, Y.; et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin. Cancer Res. 2010, 16, 21–33. [Google Scholar] [CrossRef]
- Weisenberger, D.J.; Siegmund, K.D.; Campan, M.; Young, J.; Long, T.I.; Faasse, M.A.; Kang, G.H.; Widschwendter, M.; Weener, D.; Buchanan, D.; et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006, 38, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Kawasaki, T.; Kirkner, G.J.; Loda, M.; Fuchs, C.S. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: Possible associations with male sex and KRAS mutations. J. Mol. Diagn. 2006, 8, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Toyota, M.; Kondo, Y.; Lin, E.; Zhang, L.; Guo, Y.; Hernandez, N.S.; Chen, X.; Ahmed, S.; Konishi, K.; et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 18654–18659. [Google Scholar] [CrossRef] [PubMed]
- Hinoue, T.; Weisenberger, D.J.; Lange, C.P.; Shen, H.; Byun, H.M.; Van Den Berg, D.; Malik, S.; Pan, F.; Noushmehr, H.; van Dijk, C.M.; et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012, 22, 271–282. [Google Scholar] [CrossRef]
- Guo, M.; Peng, Y.; Gao, A.; Du, C.; Herman, J.G. Epigenetic heterogeneity in cancer. Biomark. Res. 2019, 7, 23. [Google Scholar] [CrossRef]
- Michailidi, C.; Theocharis, S.; Tsourouflis, G.; Pletsa, V.; Kouraklis, G.; Patsouris, E.; Papavassiliou, A.G.; Troungos, C. Expression and promoter methylation status of hMLH1, MGMT, APC, and CDH1 genes in patients with colon adenocarcinoma. Exp. Biol. Med. 2015, 240, 1599–1605. [Google Scholar] [CrossRef]
- Imperiale, T.F.; Ransohoff, D.F.; Itzkowitz, S.H.; Levin, T.R.; Lavin, P.; Lidgard, G.P.; Ahlquist, D.A.; Berger, B.M. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef]
- Park, S.-K.; Baek, H.L.; Yu, J.; Kim, J.Y.; Yang, H.-J.; Jung, Y.S.; Choi, K.Y.; Kim, H.; Kim, H.O.; Jeong, K.U.; et al. Is methylation analysis of SFRP2, TFPI2, NDRG4, and BMP3 promoters suitable for colorectal cancer screening in the Korean population? Intest. Res. 2017, 15, 495–501. [Google Scholar] [CrossRef]
- Xiao, W.; Zhao, H.; Dong, W.; Li, Q.; Zhu, J.; Li, G.; Zhang, S.; Ye, M. Quantitative detection of methylated NDRG4 gene as a candidate biomarker for diagnosis of colorectal cancer. Oncol. Lett. 2015, 9, 1383–1387. [Google Scholar] [CrossRef]
- Rokni, P.; Shariatpanahi, A.M.; Sakhinia, E.; Kerachian, M.A. BMP3 promoter hypermethylation in plasma-derived cell-free DNA in colorectal cancer patients. Genes Genom. 2018, 40, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Joosten, S.C.; Feng, Z.; de Ruijter, T.C.; Draht, M.X.; Melotte, V.; Smits, K.M.; Veeck, J.; Herman, J.G.; Van Neste, L.; et al. Analysis of DNA methylation in cancer: Location revisited. Nat. Rev. Clin. Oncol. 2018, 15, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Weisenberger, D.J.; Levine, A.J.; Long, T.I.; Buchanan, D.D.; Walters, R.; Clendenning, M.; Rosty, C.; Joshi, A.D.; Stern, M.C.; LeMarchand, L.; et al. Association of the colorectal CpG island methylator phenotype with molecular features, risk factors, and family history. Cancer Epidemiol. Biomark. Prev. 2015, 24, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Oster, B.; Thorsen, K.; Lamy, P.; Wojdacz, T.K.; Hansen, L.L.; Birkenkamp-Demtroder, K.; Sorensen, K.D.; Laurberg, S.; Orntoft, T.F.; Andersen, C.L. Identification and validation of highly frequent CpG island hypermethylation in colorectal adenomas and carcinomas. Int. J. Cancer 2011, 129, 2855–2866. [Google Scholar] [CrossRef]
- Avraham, A.; Sandbank, J.; Yarom, N.; Shalom, A.; Karni, T.; Pappo, I.; Sella, A.; Fich, A.; Walfisch, S.; Gheber, L.; et al. A similar cell-specific pattern of HOXA methylation in normal and in cancer tissues. Epigenetics 2010, 5, 41–46. [Google Scholar] [CrossRef][Green Version]
- Zhang, C.; Zhao, H.; Li, J.; Liu, H.; Wang, F.; Wei, Y.; Su, J.; Zhang, D.; Liu, T.; Zhang, Y. The identification of specific methylation patterns across different cancers. PLoS ONE 2015, 10, e0120361. [Google Scholar] [CrossRef]
- Yang, H.; Robinson, P.N.; Wang, K. Phenolyzer: Phenotype-based prioritization of candidate genes for human diseases. Nat. Methods 2015, 12, 841–843. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Wilder, S.P.; Johnson, N.; Juettemann, T.; Flicek, P.R. The ensembl regulatory build. Genome Biol 2015, 16, 56. [Google Scholar] [CrossRef]
- Zhou, W.; Laird, P.W.; Shen, H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017, 45, e22. [Google Scholar] [CrossRef]
- Houseman, E.A.; Christensen, B.C.; Yeh, R.F.; Marsit, C.J.; Karagas, M.R.; Wrensch, M.; Nelson, H.H.; Wiemels, J.; Zheng, S.; Wiencke, J.K.; et al. Model-based clustering of DNA methylation array data: A recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinform. 2008, 9, 365. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Von Bubnoff, A.; Cho, K.W. Intracellular BMP signaling regulation in vertebrates: Pathway or network? Dev. Biol. 2001, 239, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K.; Kamiya, Y.; Morikawa, M. Bone morphogenetic protein receptors and signal transduction. J. Biochem. 2010, 147, 35–51. [Google Scholar] [CrossRef]
- Ji, T.; Takabayashi, H.; Mao, M.; Han, X.; Xue, X.; Brazil, J.C.; Eaton, K.A.; Shah, Y.M.; Todisco, A. Regulation and function of bone morphogenetic protein signaling in colonic injury and inflammation. Am. J. Physiol. Gastrointest. Liver. Physiol. 2017, 312, G24–G33. [Google Scholar] [CrossRef]
- Hu, L.; Xu, J.; Wang, X.; Feng, L.; Zhang, C.; Wang, J.; Wang, S. Bone Morphogenetic Protein 4 Alleviates DSS-Induced Ulcerative Colitis Through Activating Intestinal Stem Cell by Target ID3. Front. Cell Dev. Biol. 2021, 9, 700864. [Google Scholar] [CrossRef]
- Koppens, M.A.J.; Davis, H.; Valbuena, G.N.; Mulholland, E.J.; Nasreddin, N.; Colombe, M.; Antanaviciute, A.; Biswas, S.; Friedrich, M.; Lee, L.; et al. Bone Morphogenetic Protein Pathway Antagonism by Grem1 Regulates Epithelial Cell Fate in Intestinal Regeneration. Gastroenterology 2021, 161, 239–254.e9. [Google Scholar] [CrossRef]
- Yu, X.; Li, S.; Xu, Y.; Zhang, Y.; Ma, W.; Liang, C.; Lu, H.; Ji, Y.; Liu, C.; Chen, D.; et al. Androgen Maintains Intestinal Homeostasis by Inhibiting BMP Signaling via Intestinal Stromal Cells. Stem Cell Rep. 2020, 15, 912–925. [Google Scholar] [CrossRef]
- Xie, Z.; Zhou, G.; Zhang, M.; Han, J.; Wang, Y.; Li, X.; Wu, Q.; Li, M.; Zhang, S. Recent developments on BMPs and their antagonists in inflammatory bowel diseases. Cell Death Discov. 2023, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.; Raja, E.; Miyazono, K.; Tsubakihara, Y.; Moustakas, A. Mechanisms of action of bone morphogenetic proteins in cancer. Cytokine Growth Factor Rev. 2016, 27, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Wakefield, L.M.; Hill, C.S. Beyond TGFβ: Roles of other TGFβ superfamily members in cancer. Nat. Rev. Cancer 2013, 13, 328–341. [Google Scholar] [CrossRef]
- Ehata, S.; Yokoyama, Y.; Takahashi, K.; Miyazono, K. Bi-directional roles of bone morphogenetic proteins in cancer: Another molecular Jekyll and Hyde? Pathol. Int. 2013, 63, 287–296. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.; Peng, W.; Li, Y.; Yao, J. Identification of hub genes and pathways in colitis-associated colon cancer by integrated bioinformatic analysis. BMC Genom. Data 2022, 23, 48. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Watanabe, T.; Tamura, Y.; Hashizume, Y.; Miyazono, K.; Ehata, S. Autocrine BMP-4 Signaling Is a Therapeutic Target in Colorectal Cancer. Cancer Res. 2017, 77, 4026–4038. [Google Scholar] [CrossRef]
- Deng, H.; Ravikumar, T.S.; Yang, W.-L. Bone morphogenetic protein-4 inhibits heat-induced apoptosis by modulating MAPK pathways in human colon cancer HCT116 cells. Cancer Lett. 2007, 256, 207–217. [Google Scholar] [CrossRef]
- Uhan, S.; Zidar, N.; Tomažič, A.; Hauptman, N. Hypermethylated promoters of genes UNC5D and KCNA1 as potential novel diagnostic biomarkers in colorectal cancer. Epigenomics 2020, 12, 1677–1688. [Google Scholar] [CrossRef]
- Kang, M.H.; Kang, H.N.; Kim, J.L.; Kim, J.S.; Oh, S.C.; Yoo, Y.A. Inhibition of PI3 kinase/Akt pathway is required for BMP2-induced EMT and invasion. Oncol. Rep. 2009, 22, 525–534. [Google Scholar] [CrossRef][Green Version]
- Kim, B.R.; Oh, S.C.; Lee, D.H.; Kim, J.L.; Lee, S.Y.; Kang, M.H.; Lee, S.I.; Kang, S.; Joung, S.Y.; Min, B.W. BMP-2 induces motility and invasiveness by promoting colon cancer stemness through STAT3 activation. Tumour Biol. 2015, 36, 9475–9486. [Google Scholar] [CrossRef]
- Lorente-Trigos, A.; Varnat, F.; Melotti, A.; Ruiz i Altaba, A. BMP signaling promotes the growth of primary human colon carcinomas in vivo. J. Mol. Cell Biol. 2010, 2, 318–332. [Google Scholar] [CrossRef]
- Nørgaard, K.; Müller, C.; Christensen, N.; Chiloeches, M.L.; Madsen, C.L.; Nielsen, S.S.; Thingholm, T.E.; Belcheva, A. Loss of mismatch repair signaling impairs the WNT-bone morphogenetic protein crosstalk and the colonic homeostasis. J. Mol. Cell Biol. 2020, 12, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, C.; Yuan, Y.; Liu, L.; Xiong, G.; Wu, J. Bone morphogenetic protein-4 polymorphism and colorectal cancer risk: A meta analysis. Mol. Biol. Rep. 2012, 39, 5239–5251. [Google Scholar] [CrossRef]
- Fernandez-Rozadilla, C.; Palles, C.; Carvajal-Carmona, L.; Peterlongo, P.; Nici, C.; Veneroni, S.; Pinheiro, M.; Teixeira, M.R.; Moreno, V.; Lamas, M.J.; et al. BMP2/BMP4 colorectal cancer susceptibility loci in northern and southern European populations. Carcinogenesis 2013, 34, 314–318. [Google Scholar] [CrossRef][Green Version]
- Motoyama, K.; Tanaka, F.; Kosaka, Y.; Mimori, K.; Uetake, H.; Inoue, H.; Sugihara, K.; Mori, M. Clinical significance of BMP7 in human colorectal cancer. Ann. Surg. Oncol. 2008, 15, 1530–1537. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-L.; Makizumi, R.; Dong, H.; Ravikumar, T.S. Associated changes of bone morphogenetic proteins levels in human colorectal cancer during tumor progression. J. Am. Coll. Surg. 2005, 201, S86. [Google Scholar] [CrossRef]
- Fan, Y.; Guo, L.; Zheng, H.; Ji, C.; Wang, W.; Sun, H. BMP-9 is a novel marker for colorectal tumorigenesis undergoing the normal mucosa-adenoma-adenocarcinoma sequence and is associated with colorectal cancer prognosis. Oncol. Lett. 2020, 19, 271–282. [Google Scholar] [CrossRef]
- Wen, J.; Liu, X.; Qi, Y.; Niu, F.; Niu, Z.; Geng, W.; Zou, Z.; Huang, R.; Wang, J.; Zou, H. BMP3 suppresses colon tumorigenesis via ActRIIB/SMAD2-dependent and TAK1/JNK signaling pathways. J. Exp. Clin. Cancer Res. 2019, 38, 428. [Google Scholar] [CrossRef]
- Sobanski, T.; Arantes, L.; Dos Santos, W.; Matsushita, M.; de Oliveira, M.A.; Costa, M.; de Carvalho, A.C.; Berardinelli, G.N.; Syrjänen, K.; Reis, R.M.; et al. Methylation profile of colon cancer genes in colorectal precursor lesions and tumor tissue: Perspectives for screening. Scand. J. Gastroenterol. 2021, 56, 920–928. [Google Scholar] [CrossRef]
- Jevšinek Skok, D.; Hauptman, N.; Boštjančič, E.; Zidar, N. The integrative knowledge base for miRNA-mRNA expression in colorectal cancer. Sci. Rep. 2019, 9, 18065. [Google Scholar] [CrossRef]
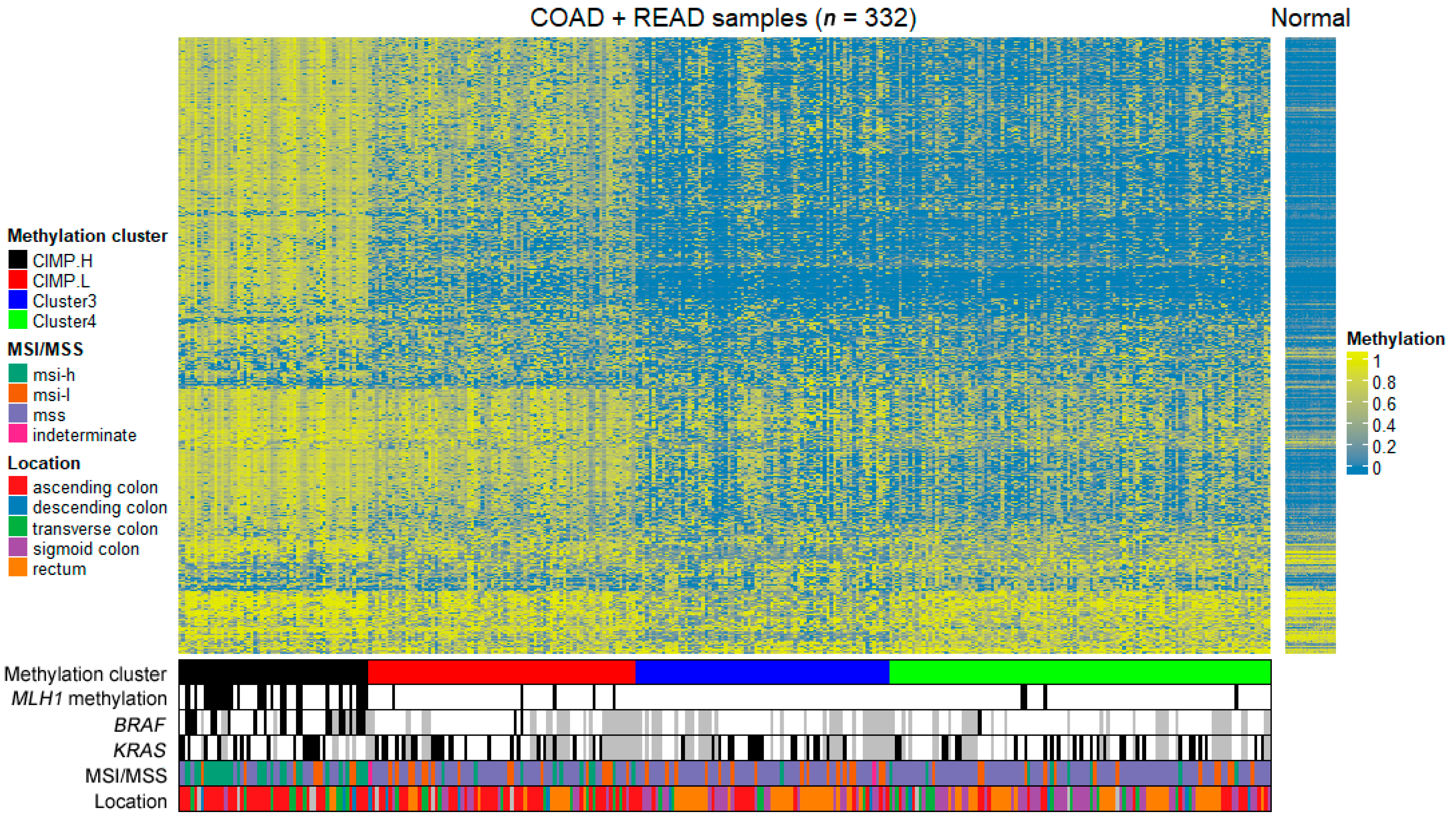
| Overall n = 332 | CIMP-H n = 58 | CIMP-L n = 81 | Cluster 3 n = 77 | Cluster 4 n = 116 | ||
|---|---|---|---|---|---|---|
| Gender | Female | 146 (44%) | 25 (43%) | 35 (43%) | 36 (47%) | 50 (43%) |
| Male | 186 (56%) | 33 (57%) | 46 (57%) | 41 (53%) | 66 (57%) | |
| Age | Median | 66 | 69 | 68 | 63 | 63 |
| Range | (31–90) | (33–88) | (37–90) | (37–90) | (31–90) | |
| Subsite | Ascending | 111 (34%) | 38 (70%) | 40 (51%) | 15 (19%) | 18 (16%) |
| Transverse | 41 (13%) | 10 (19%) | 11 (14%) | 5 (6%) | 15 (13%) | |
| Descending | 13 (4%) | 3 (6%) | 4 (5%) | 2 (3%) | 4 (4%) | |
| Sigmoid | 77 (24%) | 1 (2%) | 11 (14%) | 23 (30%) | 42 (36%) | |
| Rectum | 80 (25%) | 2 (4%) | 12 (15%) | 32 (42%) | 34 (29%) | |
| No data | 10 | 4 | 3 | 0 | 3 | |
| MSI status | MSI-H | 48 (14%) | 29 (50%) | 10 (12%) | 1 (1%) | 8 (7%) |
| MSI-L | 54 (16%) | 8 (14%) | 16 (20%) | 16 (21%) | 14 (12%) | |
| Intermediate | 2 (1%) | 0 | 1 (1%) | 1 (1%) | 0 | |
| MSS | 228 (69%) | 21 (36%) | 54 (67%) | 59 (77%) | 94 (81%) | |
| BRAF | Mutant | 24 (10%) | 21 (43%) | 2 (4%) | 0 (0%) | 1 (1%) |
| Wild-type | 208 (90%) | 28 (57%) | 52 (96%) | 46 (100%) | 82 (99%) | |
| No data | 100 | 9 | 27 | 31 | 33 | |
| KRAS | Mutant | 83 (36%) | 16 (33%) | 23 (43%) | 17 (37%) | 27 (33%) |
| Wild-type | 149 (64%) | 33 (67%) | 31 (57%) | 29 (63%) | 56 (67%) | |
| No data | 100 | 9 | 27 | 31 | 33 | |
| MLH1 | Methylated | 36 (11%) | 28 (48%) | 4 (5%) | 0 (0%) | 4 (3%) |
| Unmethylated | 296 (89%) | 30 (52%) | 77 (95%) | 77 (100%) | 112 (97%) |
| Gene Name | Mean Normal | Mean CIMP-H | Mean CIMP-L | Mean Cluster 3 | Mean Cluster 4 | Status | Probe Location | Number of Probes in Region |
|---|---|---|---|---|---|---|---|---|
| EYA4 | 0.09 | 0.62 | 0.59 | 0.59 | 0.48 | Hypermethylated | promoter bound region | 29 |
| EYA4 | 0.09 | 0.62 | 0.599 | 0.60 | 0.49 | Hypermethylated | predicted promoter region | 25 |
| HOXA3 | 0.33 | 0.73 | 0.72 | 0.74 | 0.69 | Hypermethylated | promoter bound region | 19 |
| PCDHGC5 | 0.20 | 0.65 | 0.63 | 0.70 | 0.60 | Hypermethylated | promoter bound region | 16 |
| PCDHGC4 | 0.20 | 0.65 | 0.63 | 0.70 | 0.60 | Hypermethylated | promoter bound region | 16 |
| HOXA2 | 0.38 | 0.80 | 0.79 | 0.81 | 0.76 | Hypermethylated | predicted promoter region | 14 |
| HOXA3 | 0.38 | 0.80 | 0.79 | 0.81 | 0.76 | Hypermethylated | predicted promoter region | 14 |
| HOXA6 | 0.31 | 0.65 | 0.71 | 0.71 | 0.69 | Hypermethylated | promoter bound region | 12 |
| RIPK3 | 0.10 | 0.67 | 0.52 | 0.51 | 0.45 | Hypermethylated | promoter bound region | 10 |
| PCDHGC5 | 0.18 | 0.66 | 0.64 | 0.71 | 0.62 | Hypermethylated | predicted promoter region | 10 |
| PCDHGC4 | 0.18 | 0.66 | 0.64 | 0.71 | 0.62 | Hypermethylated | predicted promoter region | 10 |
| RIPK3 | 0.10 | 0.67 | 0.52 | 0.51 | 0.45 | Hypermethylated | promoter bound region | 10 |
| MAP3K14 | 0.08 | 0.61 | 0.55 | 0.61 | 0.52 | Hypermethylated | promoter bound region | 9 |
| ADHFE1 | 0.11 | 0.69 | 0.67 | 0.73 | 0.64 | Hypermethylated | predicted promoter region | 9 |
| ADHFE1 | 0.11 | 0.69 | 0.67 | 0.73 | 0.64 | Hypermethylated | promoter bound region | 9 |
| Pathway ID | Pathway Description | Observed Gene Count | Background Gene Count | Strength | False Discovery Rate |
|---|---|---|---|---|---|
| Gene ontology: Biological process | |||||
| GO:0072197 | Ureter morphogenesis | 3 | 7 | 1.51 | 0.02 |
| GO:0072189 | Ureter development | 5 | 17 | 1.35 | 0.0013 |
| GO:0010463 | Mesenchymal cell proliferation | 4 | 16 | 1.28 | 0.0114 |
| GO:0048557 | Embryonic digestive tract morphogenesis | 4 | 18 | 1.23 | 0.0152 |
| GO:0042474 | Middle ear morphogenesis | 4 | 20 | 1.18 | 0.02 |
| GO:0003177 | Pulmonary valve development | 4 | 21 | 1.16 | 0.022 |
| GO:0048485 | Sympathetic nervous system development | 4 | 21 | 1.16 | 0.022 |
| GO:0002053 | Positive regulation of mesenchymal cell proliferation | 4 | 25 | 1.08 | 0.0333 |
| GO:0045992 | Negative regulation of embryonic development | 4 | 25 | 1.08 | 0.0333 |
| GO:0061217 | Regulation of mesonephros development | 4 | 25 | 1.08 | 0.0333 |
| GO:0003176 | Aortic valve development | 5 | 32 | 1.07 | 0.0113 |
| GO:0060037 | Pharyngeal system development | 4 | 26 | 1.07 | 0.0364 |
| GO:0010464 | Regulation of mesenchymal cell proliferation | 5 | 33 | 1.06 | 0.0124 |
| GO:0048566 | Embryonic digestive tract development | 5 | 34 | 1.05 | 0.0137 |
| GO:0042481 | Regulation of odontogenesis | 4 | 27 | 1.05 | 0.0385 |
| GO:0048701 | Embryonic cranial skeleton morphogenesis | 7 | 48 | 1.04 | 0.0011 |
| GO:0003180 | Aortic valve morphogenesis | 4 | 28 | 1.03 | 0.043 |
| GO:0031128 | Developmental induction | 4 | 28 | 1.03 | 0.043 |
| GO:0048704 | Embryonic skeletal system morphogenesis | 13 | 97 | 1.01 | 1.01 × 10−6 |
| GO:0048483 | Autonomic nervous system development | 6 | 44 | 1.01 | 0.0057 |
| Gene ontology: Molecular function | |||||
| GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | 25 | 672 | 0.45 | 0.0041 |
| GO:0000987 | Cis-regulatory region sequence-specific DNA binding | 26 | 701 | 0.45 | 0.0041 |
| GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | 30 | 878 | 0.41 | 0.0041 |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 32 | 1022 | 0.38 | 0.0041 |
| GO:0000976 | Transcription regulatory region sequence-specific DNA binding | 32 | 1028 | 0.37 | 0.0041 |
| GO:0003690 | Double-stranded DNA binding | 36 | 1156 | 0.37 | 0.0041 |
| GO:1990837 | Sequence-specific double-stranded DNA binding | 33 | 1068 | 0.37 | 0.0041 |
| GO:0003700 | DNA-binding transcription factor activity | 37 | 1238 | 0.36 | 0.0041 |
| GO:0043565 | Sequence-specific DNA binding | 40 | 1331 | 0.36 | 0.0041 |
| GO:0140110 | Transcription regulator activity | 43 | 1657 | 0.29 | 0.0054 |
| GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | 25 | 672 | 0.45 | 0.0041 |
| GO:0000987 | Cis-regulatory region sequence-specific DNA binding | 26 | 701 | 0.45 | 0.0041 |
| Gene ontology: Cellular component | |||||
| GO:0062023 | Collagen-containing extracellular matrix | 16 | 396 | 0.49 | 0.0453 |
| GO:0098797 | Plasma membrane protein complex | 21 | 547 | 0.46 | 0.0196 |
| GO:0031226 | Intrinsic component of plasma membrane | 44 | 1703 | 0.29 | 0.0196 |
| GO:0030054 | Cell junction | 51 | 2075 | 0.27 | 0.0196 |
| KEGG pathways | |||||
| hsa05144 | Malaria | 6 | 46 | 1 | 0.0095 |
| hsa05217 | Basal cell carcinoma | 6 | 62 | 0.87 | 0.0286 |
| hsa04514 | Cell adhesion molecules | 13 | 137 | 0.86 | 2.87 × 10−5 |
| hsa04390 | Hippo signaling pathway | 9 | 153 | 0.65 | 0.0286 |
| Node 1 | Node 2 | Homology | Coexpression | Experimentally Determined Interaction | Database Annotated | Automated Textmining | Combined Score |
|---|---|---|---|---|---|---|---|
| BMP4 | CHRD | 0 | 0.062 | 0.723 | 0.9 | 0.988 | 0.999 |
| BMP4 | BMP2 | 0.961 | 0.076 | 0 | 0.9 | 0.901 | 0.907 |
| BMP4 | MSX1 | 0 | 0.062 | 0 | 0 | 0.854 | 0.857 |
| BMP4 | SMAD6 | 0 | 0.097 | 0.148 | 0 | 0.738 | 0.781 |
| BMP4 | FGF5 | 0 | 0 | 0 | 0 | 0.7 | 0.7 |
| BMP4 | PAX9 | 0 | 0 | 0 | 0 | 0.672 | 0.672 |
| BMP4 | HAND2 | 0 | 0.062 | 0.056 | 0 | 0.636 | 0.649 |
| BMP4 | SIX1 | 0 | 0.07 | 0.139 | 0 | 0.568 | 0.624 |
| BMP4 | GATA3 | 0 | 0.099 | 0.105 | 0 | 0.539 | 0.596 |
| BMP4 | VCAN | 0 | 0 | 0 | 0 | 0.577 | 0.577 |
| BMP4 | FOXF1 | 0 | 0.07 | 0.056 | 0 | 0.551 | 0.571 |
| BMP4 | TLL1 | 0 | 0.064 | 0.243 | 0 | 0.406 | 0.542 |
| BMP4 | GPC6 | 0 | 0.096 | 0.153 | 0 | 0.441 | 0.535 |
| BMP4 | THBS1 | 0 | 0 | 0 | 0 | 0.532 | 0.532 |
| BMP4 | PDGFRA | 0 | 0.062 | 0.07 | 0 | 0.48 | 0.507 |
| BMP4 | NCAM1 | 0 | 0.062 | 0.057 | 0 | 0.483 | 0.503 |
| BMP4 | COL4A2 | 0 | 0.088 | 0.246 | 0 | 0.304 | 0.479 |
| BMP4 | OTX1 | 0 | 0 | 0 | 0 | 0.455 | 0.455 |
| BMP4 | COL5A1 | 0 | 0.089 | 0.213 | 0 | 0.289 | 0.446 |
| BMP4 | FOXF2 | 0 | 0.075 | 0.056 | 0 | 0.407 | 0.437 |
| BMP4 | VTN | 0 | 0.062 | 0 | 0 | 0.41 | 0.423 |
| BMP4 | SDC2 | 0 | 0 | 0 | 0 | 0.422 | 0.422 |
| BMP4 | HOXA5 | 0 | 0.053 | 0.057 | 0 | 0.403 | 0.42 |
| BMP4 | SLIT2 | 0 | 0.099 | 0.056 | 0 | 0.355 | 0.404 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jevšinek Skok, D.; Hauptman, N. In Silico Gene Prioritization Highlights the Significance of Bone Morphogenetic Protein 4 (BMP4) Promoter Methylation across All Methylation Clusters in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 12692. https://doi.org/10.3390/ijms241612692
Jevšinek Skok D, Hauptman N. In Silico Gene Prioritization Highlights the Significance of Bone Morphogenetic Protein 4 (BMP4) Promoter Methylation across All Methylation Clusters in Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(16):12692. https://doi.org/10.3390/ijms241612692
Chicago/Turabian StyleJevšinek Skok, Daša, and Nina Hauptman. 2023. "In Silico Gene Prioritization Highlights the Significance of Bone Morphogenetic Protein 4 (BMP4) Promoter Methylation across All Methylation Clusters in Colorectal Cancer" International Journal of Molecular Sciences 24, no. 16: 12692. https://doi.org/10.3390/ijms241612692
APA StyleJevšinek Skok, D., & Hauptman, N. (2023). In Silico Gene Prioritization Highlights the Significance of Bone Morphogenetic Protein 4 (BMP4) Promoter Methylation across All Methylation Clusters in Colorectal Cancer. International Journal of Molecular Sciences, 24(16), 12692. https://doi.org/10.3390/ijms241612692






