Histone Modifications in NAFLD: Mechanisms and Potential Therapy
Abstract
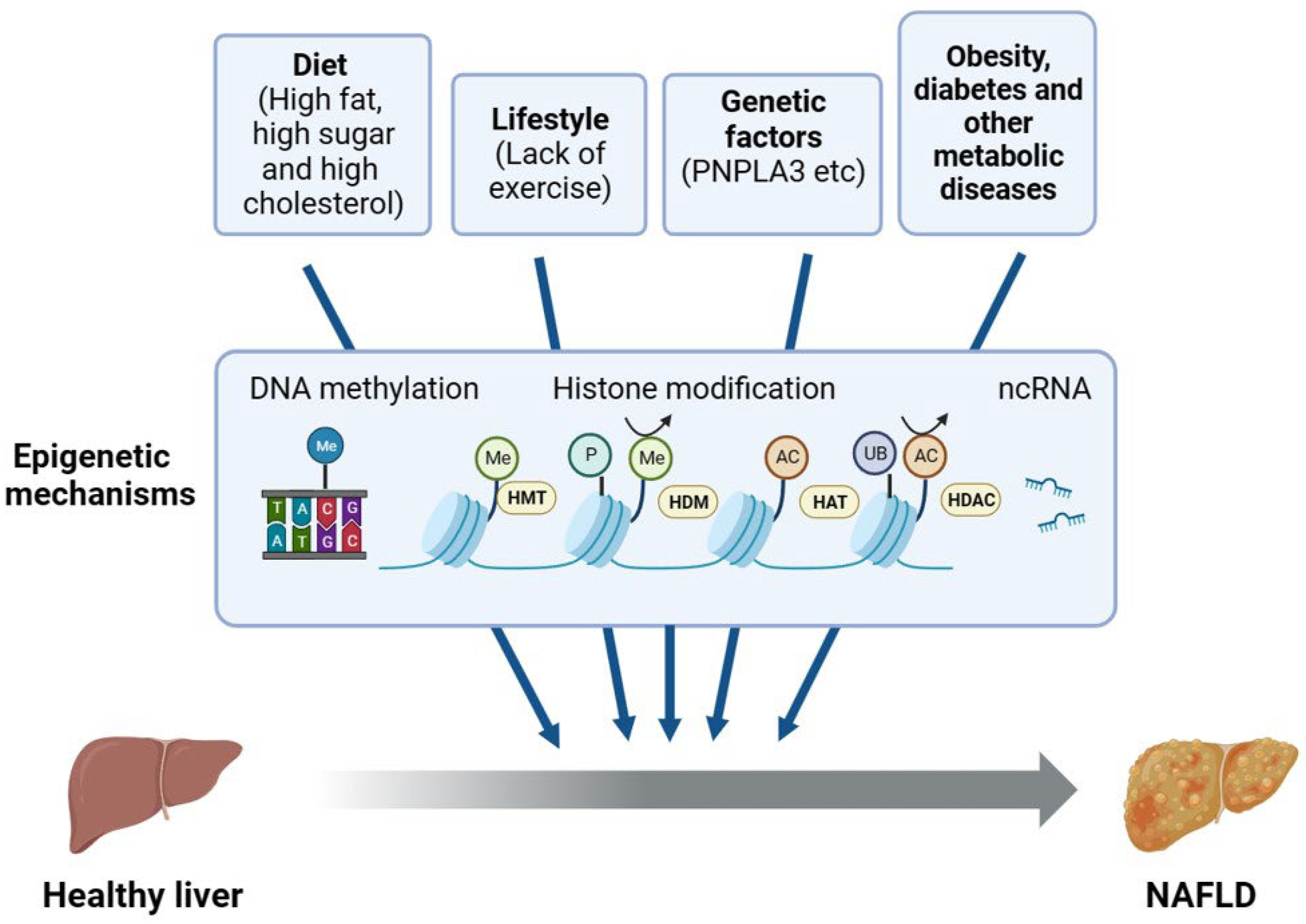
:1. Introduction
1.1. Pathogenesis of Nonalcoholic Fatty Liver Disease
1.2. Factors Influencing NAFLD
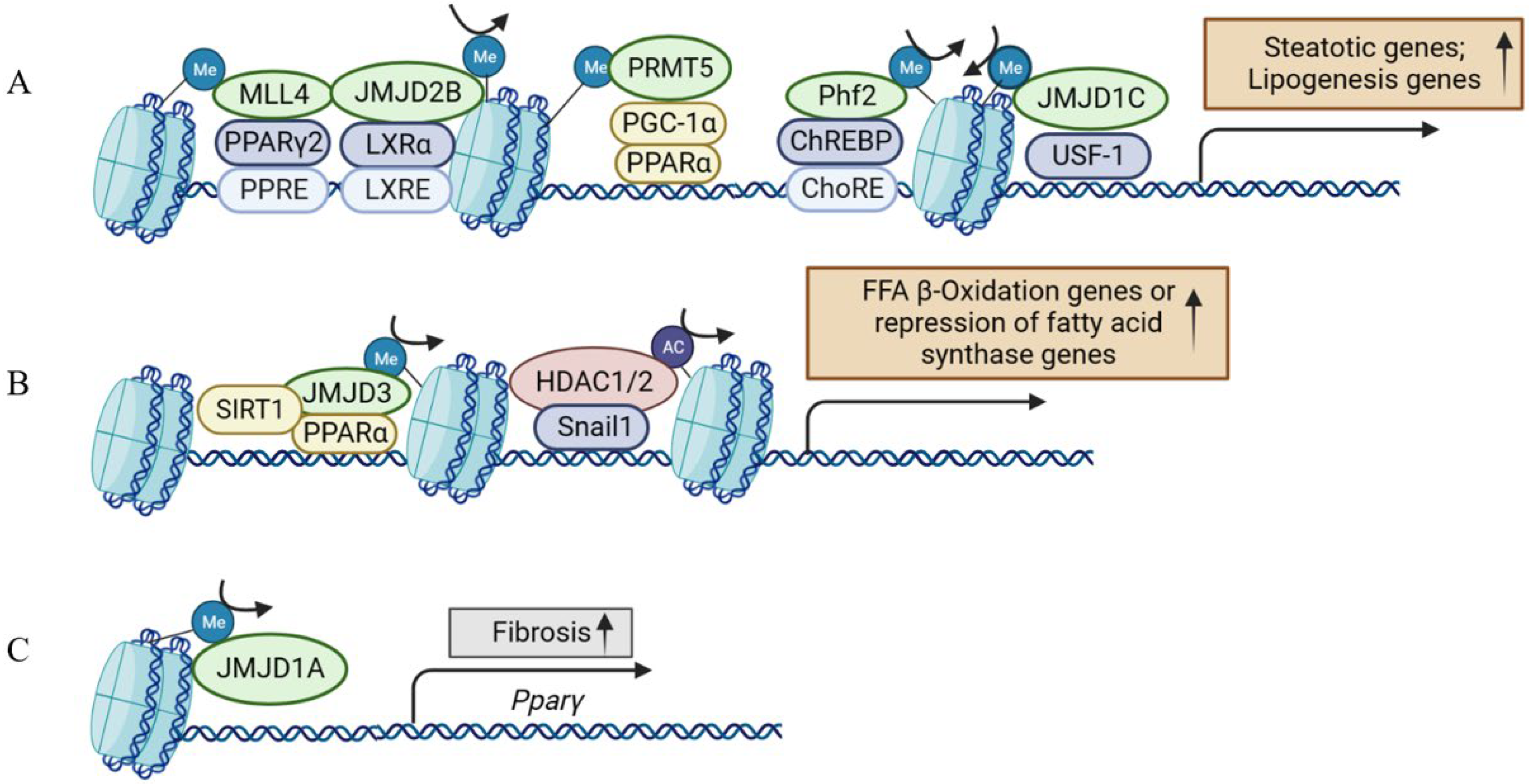
2. Regulation of NAFLD by Histone Modifications
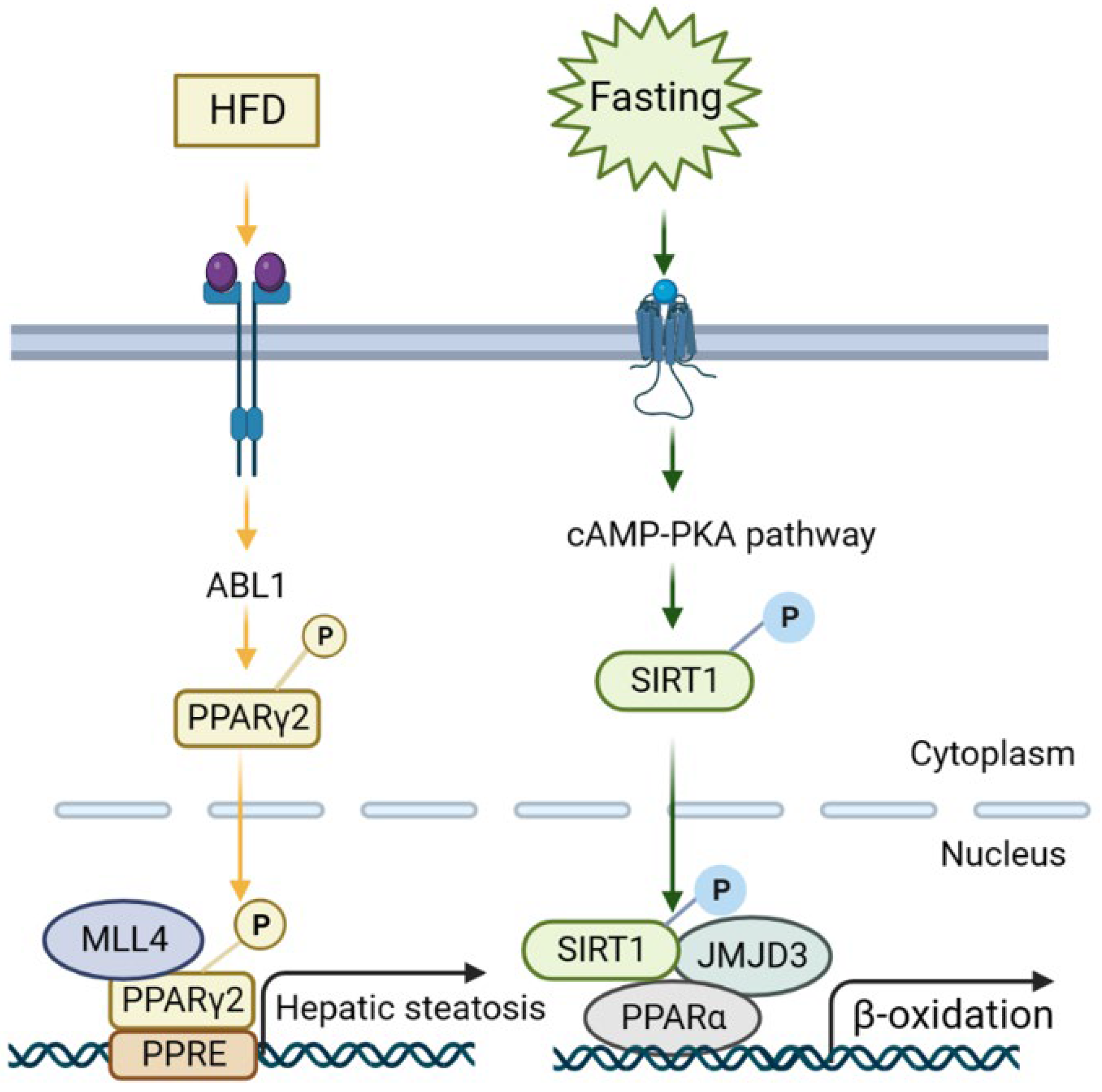
2.1. Histone Methylation in NAFLD and its Therapeutic Implications
Potential Targets and Compounds Modulating Histone Methyltransferases in NAFLD
2.2. Histone Demethylation in NAFLD and its Therapeutic Implications
Potential Targets and Compounds Modulating Histone Demethylase in NAFLD
2.3. Histone Acetylation in NAFLD and Its Therapeutic Implications
Potential Targets and Compounds Modulating Histone Acetylase in NAFLD
2.4. Histone Deacetylation in NAFLD and Its Therapeutic Implications
Potential Targets and Compounds Modulating Histone Deacetylase in NAFLD
2.5. Histone Ubiquitination and Phosphorylation in NAFLD
3. Conclusions and Future Perspectives
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Hazlehurst, J.M.; Woods, C.; Marjot, T.; Cobbold, J.F.; Tomlinson, J.W. Non-alcoholic fatty liver disease and diabetes. Metabolism 2016, 65, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Pais, R.; Giral, P.; Khan, J.F.; Rosenbaum, D.; Housset, C.; Poynard, T.; Ratziu, V. Fatty liver is an independent predictor of early carotid atherosclerosis. J. Hepatol. 2016, 65, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef]
- Ascha, M.S.; Hanouneh, I.A.; Lopez, R.; Tamimi, T.A.; Feldstein, A.F.; Zein, N.N. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology 2010, 51, 1972–1978. [Google Scholar] [CrossRef]
- Brunt, E.M. Pathology of nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 195–203. [Google Scholar] [CrossRef]
- Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Human fatty liver disease: Old questions and new insights. Science 2011, 332, 1519–1523. [Google Scholar] [CrossRef]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef]
- Uyeda, K.; Repa, J.J. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006, 4, 107–110. [Google Scholar] [CrossRef]
- Mucinski, J.M.; Manrique-Acevedo, C.; Kasumov, T.; Garrett, T.J.; Gaballah, A.; Parks, E.J. Relationships between Very Low-Density Lipoproteins-Ceramides, -Diacylglycerols, and -Triacylglycerols in Insulin-Resistant Men. Lipids 2020, 55, 387–393. [Google Scholar] [CrossRef]
- Huby, T.; Gautier, E.L. Immune cell-mediated features of non-alcoholic steatohepatitis. Nat. Rev. Immunol. 2022, 22, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Umemura, A.; Taniguchi, K.; Font-Burgada, J.; Dhar, D.; Ogata, H.; Zhong, Z.; Valasek, M.A.; Seki, E.; Hidalgo, J.; et al. ER stress cooperates with hypernutrition to trigger TNF-dependent spontaneous HCC development. Cancer Cell 2014, 26, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.S.; Harrison, S.A.; Abdelmalek, M.F.; Anstee, Q.M.; Bedossa, P.; Castera, L.; Dimick-Santos, L.; Friedman, S.L.; Greene, K.; Kleiner, D.E.; et al. Case definitions for inclusion and analysis of endpoints in clinical trials for nonalcoholic steatohepatitis through the lens of regulatory science. Hepatology 2018, 67, 2001–2012. [Google Scholar] [CrossRef]
- Neuschwander-Tetri, B.A. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: The central role of nontriglyceride fatty acid metabolites. Hepatology 2010, 52, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Yilma, M.; Saxena, V.; Mehta, N. Models to Predict Development or Recurence of Hepatocellular Carcinoma (HCC) in Patients with Advanced Hepatic Fibrosis. Curr. Gastroenterol. Rep. 2022, 24, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hannah, W.N., Jr.; Harrison, S.A. Lifestyle and Dietary Interventions in the Management of Nonalcoholic Fatty Liver Disease. Dig. Dis. Sci. 2016, 61, 1365–1374. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, J.; Sanyal, A.J. Insulin resistance and the pathogenesis of nonalcoholic fatty liver disease. Clin. Liver Dis. 2004, 8, 575–594. [Google Scholar] [CrossRef]
- Volynets, V.; Küper, M.A.; Strahl, S.; Maier, I.B.; Spruss, A.; Wagnerberger, S.; Königsrainer, A.; Bischoff, S.C.; Bergheim, I. Nutrition, intestinal permeability, and blood ethanol levels are altered in patients with nonalcoholic fatty liver disease (NAFLD). Dig. Dis. Sci. 2012, 57, 1932–1941. [Google Scholar] [CrossRef]
- Membrez, M.; Blancher, F.; Jaquet, M.; Bibiloni, R.; Cani, P.D.; Burcelin, R.G.; Corthesy, I.; Macé, K.; Chou, C.J. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008, 22, 2416–2426. [Google Scholar] [CrossRef]
- Mazagova, M.; Wang, L.; Anfora, A.T.; Wissmueller, M.; Lesley, S.A.; Miyamoto, Y.; Eckmann, L.; Dhungana, S.; Pathmasiri, W.; Sumner, S.; et al. Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. FASEB J. 2015, 29, 1043–1055. [Google Scholar] [CrossRef]
- Henao-Mejia, J.; Elinav, E.; Jin, C.; Hao, L.; Mehal, W.Z.; Strowig, T.; Thaiss, C.A.; Kau, A.L.; Eisenbarth, S.C.; Jurczak, M.J.; et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012, 482, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Valenti, L.; Romeo, S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J. Hepatol. 2018, 68, 268–279. [Google Scholar] [CrossRef] [PubMed]
- He, S.; McPhaul, C.; Li, J.Z.; Garuti, R.; Kinch, L.; Grishin, N.V.; Cohen, J.C.; Hobbs, H.H. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J. Biol. Chem. 2010, 285, 6706–6715. [Google Scholar] [CrossRef] [PubMed]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Huang, Y.; Karaman, R.; Ivanova, P.T.; Brown, H.A.; Roddy, T.; Castro-Perez, J.; Cohen, J.C.; Hobbs, H.H. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J. Clin. Investig. 2012, 122, 4130–4144. [Google Scholar] [CrossRef]
- Romeo, S.; Sentinelli, F.; Dash, S.; Yeo, G.S.; Savage, D.B.; Leonetti, F.; Capoccia, D.; Incani, M.; Maglio, C.; Iacovino, M.; et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int. J. Obes. 2010, 34, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Trépo, E.; Valenti, L. Update on NAFLD genetics: From new variants to the clinic. J. Hepatol. 2020, 72, 1196–1209. [Google Scholar] [CrossRef]
- Balcar, L.; Scheiner, B.; Urheu, M.; Weinberger, P.; Paternostro, R.; Simbrunner, B.; Semmler, G.; Willheim, C.; Pinter, M.; Ferenci, P.; et al. The impact of transmembrane 6 superfamily 2 (TM6SF2) rs58542926 on liver-related events in patients with advanced chronic liver disease. Dig. Liver Dis. 2023, 55, 1072–1080. [Google Scholar] [CrossRef]
- Franzago, M.; Pilenzi, L.; Di Rado, S.; Vitacolonna, E.; Stuppia, L. The epigenetic aging, obesity, and lifestyle. Front. Cell Dev. Biol. 2022, 10, 985274. [Google Scholar] [CrossRef]
- Lee, J.H.; Friso, S.; Choi, S.W. Epigenetic mechanisms underlying the link between non-alcoholic fatty liver diseases and nutrition. Nutrients 2014, 6, 3303–3325. [Google Scholar] [CrossRef]
- Pirola, C.J.; Gianotti, T.F.; Burgueño, A.L.; Rey-Funes, M.; Loidl, C.F.; Mallardi, P.; Martino, J.S.; Castaño, G.O.; Sookoian, S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 2013, 62, 1356–1363. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Jang, H.J.; Kim, S.; Choi, S.S.; Khim, K.W.; Eom, H.J.; Hyun, J.; Shin, K.J.; Chae, Y.C.; Kim, H.; et al. Hepatic MIR20B promotes nonalcoholic fatty liver disease by suppressing PPARA. Elife 2021, 10, e70472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Lepack, A.E.; Werner, C.T.; Stewart, A.F.; Fulton, S.L.; Zhong, P.; Farrelly, L.A.; Smith, A.C.W.; Ramakrishnan, A.; Lyu, Y.; Bastle, R.M.; et al. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 2020, 368, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Goldsworthy, M.; Absalom, N.L.; Schröter, D.; Matthews, H.C.; Bogani, D.; Moir, L.; Long, A.; Church, C.; Hugill, A.; Anstee, Q.M.; et al. Mutations in Mll2, an H3K4 methyltransferase, result in insulin resistance and impaired glucose tolerance in mice. PLoS ONE 2013, 8, e61870. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, J.; Kwon, J.S.; Sandhu, J.; Tontonoz, P.; Lee, S.K.; Lee, S.; Lee, J.W. Critical Roles of the Histone Methyltransferase MLL4/KMT2D in Murine Hepatic Steatosis Directed by ABL1 and PPARγ2. Cell Rep. 2016, 17, 1671–1682. [Google Scholar] [CrossRef]
- Kim, J.; Lee, B.; Kim, D.H.; Yeon, J.G.; Lee, J.; Park, Y.; Lee, Y.; Lee, S.K.; Lee, S.; Lee, J.W. UBE3A Suppresses Overnutrition-Induced Expression of the Steatosis Target Genes of MLL4 by Degrading MLL4. Hepatology 2019, 69, 1122–1134. [Google Scholar] [CrossRef]
- Perugorria, M.J.; Wilson, C.L.; Zeybel, M.; Walsh, M.; Amin, S.; Robinson, S.; White, S.A.; Burt, A.D.; Oakley, F.; Tsukamoto, H.; et al. Histone methyltransferase ASH1 orchestrates fibrogenic gene transcription during myofibroblast transdifferentiation. Hepatology 2012, 56, 1129–1139. [Google Scholar] [CrossRef]
- Atta, H.; El-Rehany, M.; Hammam, O.; Abdel-Ghany, H.; Ramzy, M.; Roderfeld, M.; Roeb, E.; Al-Hendy, A.; Raheim, S.A.; Allam, H.; et al. Mutant MMP-9 and HGF gene transfer enhance resolution of CCl4-induced liver fibrosis in rats: Role of ASH1 and EZH2 methyltransferases repression. PLoS ONE 2014, 9, e112384. [Google Scholar] [CrossRef]
- Huang, L.; Liu, J.; Zhang, X.O.; Sibley, K.; Najjar, S.M.; Lee, M.M.; Wu, Q. Inhibition of protein arginine methyltransferase 5 enhances hepatic mitochondrial biogenesis. J. Biol. Chem. 2018, 293, 10884–10894. [Google Scholar] [CrossRef]
- Motojima, K.; Passilly, P.; Peters, J.M.; Gonzalez, F.J.; Latruffe, N. Expression of putative fatty acid transporter genes are regulated by peroxisome proliferator-activated receptor alpha and gamma activators in a tissue- and inducer-specific manner. J. Biol. Chem. 1998, 273, 16710–16714. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Peters, J.M.; Iritani, N.; Nakajima, T.; Furihata, K.; Hashimoto, T.; Gonzalez, F.J. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J. Biol. Chem. 1998, 273, 5678–5684. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Verwilligen, R.A.F.; Van Eck, M.; Hoekstra, M. PRMT5 inhibition induces pro-inflammatory macrophage polarization and increased hepatic triglyceride levels without affecting atherosclerosis in mice. J. Cell. Mol. Med. 2023, 27, 1056–1068. [Google Scholar] [CrossRef] [PubMed]
- Vella, S.; Gnani, D.; Crudele, A.; Ceccarelli, S.; De Stefanis, C.; Gaspari, S.; Nobili, V.; Locatelli, F.; Marquez, V.E.; Rota, R.; et al. EZH2 down-regulation exacerbates lipid accumulation and inflammation in in vitro and in vivo NAFLD. Int. J. Mol. Sci. 2013, 14, 24154–24168. [Google Scholar] [CrossRef]
- Au, S.L.; Wong, C.C.; Lee, J.M.; Fan, D.N.; Tsang, F.H.; Ng, I.O.; Wong, C.M. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology 2012, 56, 622–631. [Google Scholar] [CrossRef]
- Xu, L.; Lin, J.; Deng, W.; Luo, W.; Huang, Y.; Liu, C.Q.; Zhang, F.P.; Qin, Y.F.; Wong, P.P.; Liu, C. EZH2 facilitates BMI1-dependent hepatocarcinogenesis through epigenetically silencing microRNA-200c. Oncogenesis 2020, 9, 101. [Google Scholar] [CrossRef]
- Brower-Toland, B.; Riddle, N.C.; Jiang, H.; Huisinga, K.L.; Elgin, S.C. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 2009, 181, 1303–1319. [Google Scholar] [CrossRef]
- Shao, J.; Li, L.; Xu, H.; Yang, L.; Bian, Y.; Fang, M.; Xu, Y. Suv39h2 deficiency ameliorates diet-induced steatosis in mice. Biochem. Biophys. Res. Commun. 2017, 485, 658–664. [Google Scholar] [CrossRef]
- Fan, Z.; Li, L.; Li, M.; Zhang, X.; Hao, C.; Yu, L.; Zeng, S.; Xu, H.; Fang, M.; Shen, A.; et al. The histone methyltransferase Suv39h2 contributes to nonalcoholic steatohepatitis in mice. Hepatology 2017, 65, 1904–1919. [Google Scholar] [CrossRef]
- Lim, H.J.; Kim, M. EZH2 as a Potential Target for NAFLD Therapy. Int. J. Mol. Sci. 2020, 21, 8617. [Google Scholar] [CrossRef]
- Mann, J.; Chu, D.C.; Maxwell, A.; Oakley, F.; Zhu, N.L.; Tsukamoto, H.; Mann, D.A. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology 2010, 138, 705–714.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, Z.; Tang, F.; Zhao, Y.; Feng, D.; Li, Y.; Hu, Y.; Wang, C.; Zhou, J.; Tian, X.; et al. Carnosol-mediated Sirtuin 1 activation inhibits Enhancer of Zeste Homolog 2 to attenuate liver fibrosis. Pharmacol. Res. 2018, 128, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Martin-Mateos, R.; De Assuncao, T.M.; Arab, J.P.; Jalan-Sakrikar, N.; Yaqoob, U.; Greuter, T.; Verma, V.K.; Mathison, A.J.; Cao, S.; Lomberk, G.; et al. Enhancer of Zeste Homologue 2 Inhibition Attenuates TGF-β Dependent Hepatic Stellate Cell Activation and Liver Fibrosis. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 197–209. [Google Scholar] [CrossRef]
- Zeybel, M.; Luli, S.; Sabater, L.; Hardy, T.; Oakley, F.; Leslie, J.; Page, A.; Moran Salvador, E.; Sharkey, V.; Tsukamoto, H.; et al. A Proof-of-Concept for Epigenetic Therapy of Tissue Fibrosis: Inhibition of Liver Fibrosis Progression by 3-Deazaneplanocin A. Mol. Ther. 2017, 25, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.D.; Chiang, Y.M.; Higashiyama, R.; Asahina, K.; Mann, D.A.; Mann, J.; Wang, C.C.; Tsukamoto, H. Rosmarinic acid and baicalin epigenetically derepress peroxisomal proliferator-activated receptor γ in hepatic stellate cells for their antifibrotic effect. Hepatology 2012, 55, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Barcena-Varela, M.; Paish, H.; Alvarez, L.; Uriarte, I.; Latasa, M.U.; Santamaria, E.; Recalde, M.; Garate, M.; Claveria, A.; Colyn, L.; et al. Epigenetic mechanisms and metabolic reprogramming in fibrogenesis: Dual targeting of G9a and DNMT1 for the inhibition of liver fibrosis. Gut 2021, 70, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Bricambert, J.; Alves-Guerra, M.C.; Esteves, P.; Prip-Buus, C.; Bertrand-Michel, J.; Guillou, H.; Chang, C.J.; Vander Wal, M.N.; Canonne-Hergaux, F.; Mathurin, P.; et al. The histone demethylase Phf2 acts as a molecular checkpoint to prevent NAFLD progression during obesity. Nat. Commun. 2018, 9, 2092. [Google Scholar] [CrossRef]
- Huang, C.; Xiang, Y.; Wang, Y.; Li, X.; Xu, L.; Zhu, Z.; Zhang, T.; Zhu, Q.; Zhang, K.; Jing, N.; et al. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 2010, 20, 154–165. [Google Scholar] [CrossRef]
- Kim, J.H.; Nagappan, A.; Jung, D.Y.; Suh, N.; Jung, M.H. Histone Demethylase KDM7A Contributes to the Development of Hepatic Steatosis by Targeting Diacylglycerol Acyltransferase 2. Int. J. Mol. Sci. 2021, 22, 11085. [Google Scholar] [CrossRef]
- Dong, F.; Jiang, S.; Li, J.; Wang, Y.; Zhu, L.; Huang, Y.; Jiang, X.; Hu, X.; Zhou, Q.; Zhang, Z.; et al. The histone demethylase KDM4D promotes hepatic fibrogenesis by modulating Toll-like receptor 4 signaling pathway. EBioMedicine 2019, 39, 472–483. [Google Scholar] [CrossRef]
- Chen, Z.; Zang, J.; Whetstine, J.; Hong, X.; Davrazou, F.; Kutateladze, T.G.; Simpson, M.; Mao, Q.; Pan, C.H.; Dai, S.; et al. Structural insights into histone demethylation by JMJD2 family members. Cell 2006, 125, 691–702. [Google Scholar] [CrossRef]
- Li, M.; Deng, Y.; Zhuo, M.; Zhou, H.; Kong, X.; Xia, X.; Su, Z.; Chen, Q.; Guo, P.; Mo, P.; et al. Demethylase-independent function of JMJD2D as a novel antagonist of p53 to promote Liver Cancer initiation and progression. Theranostics 2020, 10, 8863–8879. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Li, M.; Zhuo, M.; Guo, P.; Chen, Q.; Mo, P.; Li, W.; Yu, C. Histone demethylase JMJD2D promotes the self-renewal of liver cancer stem-like cells by enhancing EpCAM and Sox9 expression. J. Biol. Chem. 2021, 296, 100121. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Wu, J.; Fan, Z.; Chen, B.; Wu, T.; Xu, Y. The histone demethylase Kdm4 suppresses activation of hepatic stellate cell by inducing MiR-29 transcription. Biochem. Biophys. Res. Commun. 2019, 514, 16–23. [Google Scholar] [CrossRef]
- Kim, J.H.; Jung, D.Y.; Nagappan, A.; Jung, M.H. Histone H3K9 demethylase JMJD2B induces hepatic steatosis through upregulation of PPARγ2. Sci. Rep. 2018, 8, 13734. [Google Scholar] [CrossRef]
- Kim, J.H.; Jung, D.Y.; Kim, H.R.; Jung, M.H. Histone H3K9 Demethylase JMJD2B Plays a Role in LXRα-Dependent Lipogenesis. Int. J. Mol. Sci. 2020, 21, 8313. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, S.; Zhao, Y.; Lin, C.; Zhong, F.; Jin, L.; He, F.; Wang, H. Histone H3K9 demethylase JMJD1A modulates hepatic stellate cells activation and liver fibrosis by epigenetically regulating peroxisome proliferator-activated receptor γ. FASEB J. 2015, 29, 1830–1841. [Google Scholar] [CrossRef]
- Viscarra, J.A.; Wang, Y.; Nguyen, H.P.; Choi, Y.G.; Sul, H.S. Histone demethylase JMJD1C is phosphorylated by mTOR to activate de novo lipogenesis. Nat. Commun. 2020, 11, 796. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Xiang, C.; Zhong, F.; Zhang, Y.; Wang, L.; Zhao, Y.; Wang, J.; Ding, C.; Jin, L.; He, F.; et al. Histone H3K27 methyltransferase EZH2 and demethylase JMJD3 regulate hepatic stellate cells activation and liver fibrosis. Theranostics 2021, 11, 361–378. [Google Scholar] [CrossRef] [PubMed]
- Seok, S.; Kim, Y.C.; Byun, S.; Choi, S.; Xiao, Z.; Iwamori, N.; Zhang, Y.; Wang, C.; Ma, J.; Ge, K.; et al. Fasting-induced JMJD3 histone demethylase epigenetically activates mitochondrial fatty acid β-oxidation. J. Clin. Investig. 2018, 128, 3144–3159. [Google Scholar] [CrossRef]
- Byun, S.; Seok, S.; Kim, Y.C.; Zhang, Y.; Yau, P.; Iwamori, N.; Xu, H.E.; Ma, J.; Kemper, B.; Kemper, J.K. Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase. Nat. Commun. 2020, 11, 807. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin, H.; Jiang, L.; Shang, Q.; Yin, L.; Lin, J.D.; Wu, W.S.; Rui, L. Hepatic Slug epigenetically promotes liver lipogenesis, fatty liver disease, and type 2 diabetes. J. Clin. Investig. 2020, 130, 2992–3004. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.R.; Kim, J.H.; Kim, J.H.; Jung, M.H. Protective effects of gomisin N against hepatic steatosis through AMPK activation. Biochem. Biophys. Res. Commun. 2017, 482, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.K.; Yun, Y.R.; Kim, S.H.; Kim, J.H.; Jung, M.H. Protective Effect of Gomisin N against Endoplasmic Reticulum Stress-Induced Hepatic Steatosis. Biol. Pharm. Bull. 2016, 39, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Cai, C.; Yao, J.; Zhou, Y.; Yu, H.; Shen, W. Histone modifications in FASN modulated by sterol regulatory element-binding protein 1c and carbohydrate responsive-element binding protein under insulin stimulation are related to NAFLD. Biochem. Biophys. Res. Commun. 2017, 483, 409–417. [Google Scholar] [CrossRef]
- Zhou, B.; Jia, L.; Zhang, Z.; Xiang, L.; Yuan, Y.; Zheng, P.; Liu, B.; Ren, X.; Bian, H.; Xie, L.; et al. The Nuclear Orphan Receptor NR2F6 Promotes Hepatic Steatosis through Upregulation of Fatty Acid Transporter CD36. Adv. Sci. 2020, 7, 2002273. [Google Scholar] [CrossRef]
- Becares, N.; Gage, M.C.; Voisin, M.; Shrestha, E.; Martin-Gutierrez, L.; Liang, N.; Louie, R.; Pourcet, B.; Pello, O.M.; Luong, T.V.; et al. Impaired LXRα Phosphorylation Attenuates Progression of Fatty Liver Disease. Cell Rep. 2019, 26, 984–995.e6. [Google Scholar] [CrossRef]
- Rohrbach, T.D.; Asgharpour, A.; Maczis, M.A.; Montefusco, D.; Cowart, L.A.; Bedossa, P.; Sanyal, A.J.; Spiegel, S. FTY720/fingolimod decreases hepatic steatosis and expression of fatty acid synthase in diet-induced nonalcoholic fatty liver disease in mice. J. Lipid Res. 2019, 60, 1311–1322. [Google Scholar] [CrossRef]
- Chung, M.Y.; Song, J.H.; Lee, J.; Shin, E.J.; Park, J.H.; Lee, S.H.; Hwang, J.T.; Choi, H.K. Tannic acid, a novel histone acetyltransferase inhibitor, prevents non-alcoholic fatty liver disease both in vivo and in vitro model. Mol. Metab. 2019, 19, 34–48. [Google Scholar] [CrossRef]
- Sharma, M.; Nazareth, I.; Petersen, I. Trends in incidence, prevalence and prescribing in type 2 diabetes mellitus between 2000 and 2013 in primary care: A retrospective cohort study. BMJ Open 2016, 6, e010210. [Google Scholar] [CrossRef]
- Hundal, R.S.; Krssak, M.; Dufour, S.; Laurent, D.; Lebon, V.; Chandramouli, V.; Inzucchi, S.E.; Schumann, W.C.; Petersen, K.F.; Landau, B.R.; et al. Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes 2000, 49, 2063–2069. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Meng, S.; Chang, E.; Beckwith-Fickas, K.; Xiong, L.; Cole, R.N.; Radovick, S.; Wondisford, F.E.; He, L. Low concentrations of metformin suppress glucose production in hepatocytes through AMP-activated protein kinase (AMPK). J. Biol. Chem. 2014, 289, 20435–20446. [Google Scholar] [CrossRef] [PubMed]
- Marin, T.L.; Gongol, B.; Zhang, F.; Martin, M.; Johnson, D.A.; Xiao, H.; Wang, Y.; Subramaniam, S.; Chien, S.; Shyy, J.Y. AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Sci. Signal. 2017, 10, eaaf7478. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Sabet, A.; Djedjos, S.; Miller, R.; Sun, X.; Hussain, M.A.; Radovick, S.; Wondisford, F.E. Metformin and insulin suppress hepatic gluconeogenesis through phosphorylation of CREB binding protein. Cell 2009, 137, 635–646. [Google Scholar] [CrossRef]
- Caton, P.W.; Nayuni, N.K.; Kieswich, J.; Khan, N.Q.; Yaqoob, M.M.; Corder, R. Metformin suppresses hepatic gluconeogenesis through induction of SIRT1 and GCN5. J. Endocrinol. 2010, 205, 97–106. [Google Scholar] [CrossRef]
- Li, X.; Wu, X.Q.; Xu, T.; Li, X.F.; Yang, Y.; Li, W.X.; Huang, C.; Meng, X.M.; Li, J. Role of histone deacetylases(HDACs) in progression and reversal of liver fibrosis. Toxicol. Appl. Pharmacol. 2016, 306, 58–68. [Google Scholar] [CrossRef]
- Tian, Y.; Wong, V.W.; Wong, G.L.; Yang, W.; Sun, H.; Shen, J.; Tong, J.H.; Go, M.Y.; Cheung, Y.S.; Lai, P.B.; et al. Histone Deacetylase HDAC8 Promotes Insulin Resistance and β-Catenin Activation in NAFLD-Associated Hepatocellular Carcinoma. Cancer Res. 2015, 75, 4803–4816. [Google Scholar] [CrossRef]
- Wang, L.T.; Chiou, S.S.; Chai, C.Y.; Hsi, E.; Wang, S.N.; Huang, S.K.; Hsu, S.H. Aryl hydrocarbon receptor regulates histone deacetylase 8 expression to repress tumor suppressive activity in hepatocellular carcinoma. Oncotarget 2017, 8, 7489–7501. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, L.; Sun, C.; Ireland, N.; Shah, Y.M.; Liu, Y.; Rui, L. Insulin/Snail1 axis ameliorates fatty liver disease by epigenetically suppressing lipogenesis. Nat. Commun. 2018, 9, 2751. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Li, C.; Zhu, T.; Gao, J.; Zhou, H.; Zheng, Y.; Chang, Q.; Wang, M.; Wu, J.; et al. S100A11 Promotes Liver Steatosis via FOXO1-Mediated Autophagy and Lipogenesis. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 697–724. [Google Scholar] [CrossRef]
- Hou, X.; Xu, S.; Maitland-Toolan, K.A.; Sato, K.; Jiang, B.; Ido, Y.; Lan, F.; Walsh, K.; Wierzbicki, M.; Verbeuren, T.J.; et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J. Biol. Chem. 2008, 283, 20015–20026. [Google Scholar] [CrossRef]
- Ponugoti, B.; Kim, D.H.; Xiao, Z.; Smith, Z.; Miao, J.; Zang, M.; Wu, S.Y.; Chiang, C.M.; Veenstra, T.D.; Kemper, J.K. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 2010, 285, 33959–33970. [Google Scholar] [CrossRef]
- Purushotham, A.; Schug, T.T.; Xu, Q.; Surapureddi, S.; Guo, X.; Li, X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009, 9, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.K.; Yang, F.; Jiang, K.; Ji, J.Y.; Watts, J.L.; Purushotham, A.; Boss, O.; Hirsch, M.L.; Ribich, S.; Smith, J.J.; et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010, 24, 1403–1417. [Google Scholar] [CrossRef] [PubMed]
- Schug, T.T.; Xu, Q.; Gao, H.; Peres-da-Silva, A.; Draper, D.W.; Fessler, M.B.; Purushotham, A.; Li, X. Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Mol. Cell. Biol. 2010, 30, 4712–4721. [Google Scholar] [CrossRef] [PubMed]
- Escande, C.; Chini, C.C.; Nin, V.; Dykhouse, K.M.; Novak, C.M.; Levine, J.; van Deursen, J.; Gores, G.J.; Chen, J.; Lou, Z.; et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J. Clin. Investig. 2010, 120, 545–558. [Google Scholar] [CrossRef]
- Ka, S.O.; Bang, I.H.; Bae, E.J.; Park, B.H. Hepatocyte-specific sirtuin 6 deletion predisposes to nonalcoholic steatohepatitis by up-regulation of Bach1, an Nrf2 repressor. FASEB J. 2017, 31, 3999–4010. [Google Scholar] [CrossRef]
- Kim, H.S.; Xiao, C.; Wang, R.H.; Lahusen, T.; Xu, X.; Vassilopoulos, A.; Vazquez-Ortiz, G.; Jeong, W.I.; Park, O.; Ki, S.H.; et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010, 12, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Naiman, S.; Huynh, F.K.; Gil, R.; Glick, Y.; Shahar, Y.; Touitou, N.; Nahum, L.; Avivi, M.Y.; Roichman, A.; Kanfi, Y.; et al. SIRT6 Promotes Hepatic Beta-Oxidation via Activation of PPARα. Cell Rep. 2019, 29, 4127–4143.e8. [Google Scholar] [CrossRef]
- Hou, T.; Tian, Y.; Cao, Z.; Zhang, J.; Feng, T.; Tao, W.; Sun, H.; Wen, H.; Lu, X.; Zhu, Q.; et al. Cytoplasmic SIRT6-mediated ACSL5 deacetylation impedes nonalcoholic fatty liver disease by facilitating hepatic fatty acid oxidation. Mol. Cell 2022, 82, 4099–4115.e9. [Google Scholar] [CrossRef]
- Dominy, J.E., Jr.; Lee, Y.; Jedrychowski, M.P.; Chim, H.; Jurczak, M.J.; Camporez, J.P.; Ruan, H.B.; Feldman, J.; Pierce, K.; Mostoslavsky, R.; et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol. Cell 2012, 48, 900–913. [Google Scholar] [CrossRef]
- Mannaerts, I.; Nuytten, N.R.; Rogiers, V.; Vanderkerken, K.; van Grunsven, L.A.; Geerts, A. Chronic administration of valproic acid inhibits activation of mouse hepatic stellate cells in vitro and in vivo. Hepatology 2010, 51, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, L.; Jiao, F.Z.; Zhang, W.B.; Chen, Q.; Gong, Z.J. Histone deacetylase inhibitor suberoylanilide hydroxamic acid alleviates liver fibrosis by suppressing the transforming growth factor-β1 signal pathway. Hepatobiliary Pancreat. Dis. Int. 2018, 17, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Jia, Y.; Hong, J.; Sun, Q.; Gao, S.; Hu, Y.; Zhao, N.; Zhao, R. Sodium Butyrate Ameliorates High-Fat-Diet-Induced Non-alcoholic Fatty Liver Disease through Peroxisome Proliferator-Activated Receptor α-Mediated Activation of β Oxidation and Suppression of Inflammation. J. Agric. Food. Chem. 2018, 66, 7633–7642. [Google Scholar] [CrossRef] [PubMed]
- Berman, A.Y.; Motechin, R.A.; Wiesenfeld, M.Y.; Holz, M.K. The therapeutic potential of resveratrol: A review of clinical trials. NPJ Precis. Oncol. 2017, 1, 35. [Google Scholar] [CrossRef]
- Zhou, R.; Yi, L.; Ye, X.; Zeng, X.; Liu, K.; Qin, Y.; Zhang, Q.; Mi, M. Resveratrol Ameliorates Lipid Droplet Accumulation in Liver Through a SIRT1/ ATF6-Dependent Mechanism. Cell. Physiol. Biochem. 2018, 51, 2397–2420. [Google Scholar] [CrossRef] [PubMed]
- Faghihzadeh, F.; Adibi, P.; Rafiei, R.; Hekmatdoost, A. Resveratrol supplementation improves inflammatory biomarkers in patients with nonalcoholic fatty liver disease. Nutr. Res. 2014, 34, 837–843. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, X.; Ran, L.; Wan, J.; Wang, X.; Qin, Y.; Shu, F.; Gao, Y.; Yuan, L.; Zhang, Q.; et al. Resveratrol improves insulin resistance, glucose and lipid metabolism in patients with non-alcoholic fatty liver disease: A randomized controlled trial. Dig. Liver Dis. 2015, 47, 226–232. [Google Scholar] [CrossRef]
- Chachay, V.S.; Macdonald, G.A.; Martin, J.H.; Whitehead, J.P.; O’Moore-Sullivan, T.M.; Lee, P.; Franklin, M.; Klein, K.; Taylor, P.J.; Ferguson, M.; et al. Resveratrol does not benefit patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2014, 12, 2092–2103.e6. [Google Scholar] [CrossRef]
- Feige, J.N.; Lagouge, M.; Canto, C.; Strehle, A.; Houten, S.M.; Milne, J.C.; Lambert, P.D.; Mataki, C.; Elliott, P.J.; Auwerx, J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008, 8, 347–358. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Usui, I.; Kanatani, Y.; Matsuya, Y.; Tsuneyama, K.; Fujisaka, S.; Bukhari, A.; Suzuki, H.; Senda, S.; Imanishi, S.; et al. Treatment with SRT1720, a SIRT1 activator, ameliorates fatty liver with reduced expression of lipogenic enzymes in MSG mice. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E1179–E1186. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Horváth, B.; Rajesh, M.; Varga, Z.V.; Gariani, K.; Ryu, D.; Cao, Z.; Holovac, E.; Park, O.; Zhou, Z.; et al. PARP inhibition protects against alcoholic and non-alcoholic steatohepatitis. J. Hepatol. 2017, 66, 589–600. [Google Scholar] [CrossRef]
- Baser, K.H. Biological and pharmacological activities of carvacrol and carvacrol bearing essential oils. Curr. Pharm. Des. 2008, 14, 3106–3119. [Google Scholar] [CrossRef] [PubMed]
- Ezhumalai, M.; Radhiga, T.; Pugalendi, K.V. Antihyperglycemic effect of carvacrol in combination with rosiglitazone in high-fat diet-induced type 2 diabetic C57BL/6J mice. Mol. Cell. Biochem. 2014, 385, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Nagasawa, K.; Münch, C.; Xu, Y.; Satterstrom, K.; Jeong, S.; Hayes, S.D.; Jedrychowski, M.P.; Vyas, F.S.; Zaganjor, E.; et al. Mitochondrial Sirtuin Network Reveals Dynamic SIRT3-Dependent Deacetylation in Response to Membrane Depolarization. Cell 2016, 167, 985–1000.e21. [Google Scholar] [CrossRef]
- Sroka, Z.; Cisowski, W. Hydrogen peroxide scavenging, antioxidant and anti-radical activity of some phenolic acids. Food. Chem. Toxicol. 2003, 41, 753–758. [Google Scholar] [CrossRef]
- Min, S.W.; Ryu, S.N.; Kim, D.H. Anti-inflammatory effects of black rice, cyanidin-3-O-beta-D-glycoside, and its metabolites, cyanidin and protocatechuic acid. Int. Immunopharmacol. 2010, 10, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Kang, X.; Zhao, Y.; Wang, Z.; Wang, R.; Fu, R.; Li, Y.; Hu, Y.; Wang, Z.; Shan, W.; et al. Sirtuin 3-mediated deacetylation of acyl-CoA synthetase family member 3 by protocatechuic acid attenuates non-alcoholic fatty liver disease. Br. J. Pharmacol. 2020, 177, 4166–4180. [Google Scholar] [CrossRef]
- Chen, S.; Dai, X.; Li, H.; Gong, Y.; Zhao, Y.; Huang, H. Overexpression of ring finger protein 20 inhibits the progression of liver fibrosis via mediation of histone H2B lysine 120 ubiquitination. Hum. Cell 2021, 34, 759–770. [Google Scholar] [CrossRef]
- Cai, C.; Yu, H.; Huang, G.; Du, X.; Yu, X.; Zhou, Y.; Shen, W. Histone modifications in fatty acid synthase modulated by carbohydrate responsive element binding protein are associated with non-alcoholic fatty liver disease. Int. J. Mol. Med. 2018, 42, 1215–1228. [Google Scholar] [CrossRef]
- Zarei, M.; Pizarro-Delgado, J.; Barroso, E.; Palomer, X.; Vázquez-Carrera, M. Targeting FGF21 for the Treatment of Nonalcoholic Steatohepatitis. Trends Pharmacol. Sci. 2020, 41, 199–208. [Google Scholar] [CrossRef] [PubMed]

| Epigenetic Regulators | Histone Modification | Effect | Reference |
|---|---|---|---|
| MLL4 | H3K4me1 | Resulting in the induction of the steatosis target genes of PPARγ2; UBE3A degrades MLL4 to suppress steatosis | [36,37] |
| ASH1 | H3K4me3 | ASH1 inhibition results in the downregulation of fibrogenic gene expression | [38,39] |
| PRMT5 | Symmetric dimethylation of H4R3, H3R8, and H2AR3 | PRMT5 inhibition increases hepatic triglyceride levels | [40,43] |
| SUV39H2 | H3K9me3 | Facilitating hepatic steatosis by repressing SIRT1 transcription | [48,49] |
| PHF2 | H3K9me2 demethylation | Preventing lipotoxity via upregulating SCD1 | [57] |
| KDM7A | H3K9me2 and H3K27me2 demethylation | Inducing hepatic steatosis by increasing the expression of DGAT2 | [59] |
| KDM4D | H3K9me demethylation | Triggering hepatic fibrogenesis by activating NF-κB signaling pathways; Promoting HCC progression | [60,62,63] |
| KDM4A, KDM4B, KDM4C | H3K9me demethylation | SREBP2-KDM4 pathway leads to the maintenance of HSC quiescence by promoting miR-29 transcription | [64] |
| KDM4B | H3K9 demethylation | Upregulating PPARγ and target genes related to fatty acid uptake | [65,66] |
| KDM3A | H3K9 demethylation | Downregulating the KDM3A increases liver fibrosis through modulating the expression of PPARγ | [67] |
| KDM3C | H3K9 demethylation | Elevating TG levels in the liver and plasma by upregulating lipogenesis genes | [68] |
| KDM6B | H3K27me3 demethylation | Liver-specific downregulation of KDM6B reduces fatty acid oxidation and leads to NAFLD; KDM6B inhibition enhances HSCs activation | [69,70] |
| HDAC8 | H4 deacetylation | Suppressing Wnt antagonists together with EZH2; AHR-HDAC8 axis promotes HCC | [87,88] |
| HDAC1/2 | H3K9 and H3K27 deacetylation | Recruited to fatty acid synthase promoter by snail1 and decrease insulin-stimulated lipogenesis | [89] |
| Chemical Compounds | Target | Effect | Reference |
|---|---|---|---|
| GSK-503 | EZH2 | Preventing the progression of liver fibrosis in mice | [53] |
| DZNep | [54] | ||
| Yang-Gan-Wan (YGW) | EZH2 | Inhibiting liver fibrosis by de-repressing Pparγ in mice | [55] |
| CM272 | G9a and DNMT1 | Inhibiting liver fibrogenesis in mice and human | [56] |
| GSK2879552 | LSD1 | Downregulating LSD1-mediated Slug stimulation of FAS expression in mice | [72] |
| Gomisin N | JMJD2B | Alleviating HFD-induced hepatic steatosis and reducing hepatic ER stress in mice | [65,74] |
| Tannic acid | HAT | Inhibiting lipid accumulation in mice | [79] |
| Metformin | HATs (p300/CBP/ GCN5) | Reducing the blood glucose level and inhibiting hepatic gluconeogenesis in mice | [83,84,85] |
| Valproic acid | HDAC | Inhibiting HSC activation in mice | [102] |
| SAHA | HDAC | Reducing liver fibrosis in rats | [103] |
| NaB | HDAC | Enhancing fatty acid oxidation and inhibiting NF-κB inflammatory pathway in rats | [104] |
| Resveratrol | SIRT1 | Alleviating hepatic steatosis in mice; reducing ALT/AST and improving insulin resistance in human with NAFLD | [106,107,108] |
| SRT1720 | SIRT1 | Decreasing the expressions of marker genes for oxidative stress and inflammatory cytokines and ameliorating fatty liver in mice | [110,111] |
| Olaparib | SIRT1 | Decreasing hepatic triglyceride accumulation, inflammation and fibrosis in NASH mice | [112] |
| Carvacrol | SIRT1 | Decreasing hepatic marker enzymes (ALT/AST) activities in combination with rosiglitazone in mice | [114] |
| Protocatechuic acid | SIRT3 | Preventing liver damage and steatosis in mice and rats | [118] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Qi, W. Histone Modifications in NAFLD: Mechanisms and Potential Therapy. Int. J. Mol. Sci. 2023, 24, 14653. https://doi.org/10.3390/ijms241914653
Shi Y, Qi W. Histone Modifications in NAFLD: Mechanisms and Potential Therapy. International Journal of Molecular Sciences. 2023; 24(19):14653. https://doi.org/10.3390/ijms241914653
Chicago/Turabian StyleShi, Yulei, and Wei Qi. 2023. "Histone Modifications in NAFLD: Mechanisms and Potential Therapy" International Journal of Molecular Sciences 24, no. 19: 14653. https://doi.org/10.3390/ijms241914653





