Identifying Serum Metabolomic Markers Associated with Skin Disease Activity in Patients with Psoriatic Arthritis
Abstract
:1. Introduction
2. Results
2.1. Principal Component Analysis
2.2. Machine Learning Models
2.3. Tentative Metabolite Identification
2.4. Overlapping Features
3. Discussion
3.1. Models
3.2. Metabolite Identification and Biological Significance
3.2.1. Overlapping Features
3.2.2. Positive Mode
3.2.3. Negative Mode
3.2.4. Limitations
4. Materials and Methods
4.1. Patients
4.2. Materials
4.3. Quality Assurance and Quality Control
4.4. Preparation of Solid-Phase Microextraction (SPME) Devices
4.5. Solid-Phase Microextraction (SPME) for Sample Preparation
4.6. Instrumental Analysis
4.7. Data Pre-Processing and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lebwohl, M.G.; Bachelez, H.; Barker, J.; Girolomoni, G.; Kavanaugh, A.; Langley, R.G.; Paul, C.F.; Puig, L.; Reich, K.; Van De Kerkhof, P.C.M. Patient Perspectives in the Management of Psoriasis: Results from the Population-Based Multinational Assessment of Psoriasis and Psoriatic Arthritis Survey. J. Am. Acad. Dermatol. 2014, 70, 871–881.e30. [Google Scholar] [CrossRef] [PubMed]
- Mrowietz, U.; Reich, K. Psoriasis-New Insights into Pathogenesis and Treatment. Dtsch. Arztebl. 2009, 106, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Alinaghi, F.; Calov, M.; Kristensen, L.E.; Gladman, D.D.; Coates, L.C.; Jullien, D.; Gottlieb, A.B.; Gisondi, P.; Wu, J.J.; Thyssen, J.P.; et al. Prevalence of Psoriatic Arthritis in Patients with Psoriasis: A Systematic Review and Meta-Analysis of Observational and Clinical Studies. J. Am. Acad. Dermatol. 2019, 80, 251–265.e19. [Google Scholar] [CrossRef] [PubMed]
- Ritchlin, C.T.; Colbert, R.A.; Gladman, D.D. Psoriatic Arthritis. N. Engl. J. Med. 2017, 376, 957–970. [Google Scholar] [CrossRef]
- Schieir, O.; Tosevski, C.; Glazier, R.H.; Hogg-Johnson, S.; Badley, E.M. Incident Myocardial Infarction Associated with Major Types of Arthritis in the General Population: A Systematic Review and Meta-Analysis. Ann. Rheum. Dis. 2017, 76, 1396–1404. [Google Scholar] [CrossRef]
- Polachek, A.; Touma, Z.; Anderson, M.; Eder, L. Risk of Cardiovascular Morbidity in Patients with Psoriatic Arthritis: A Meta-Analysis of Observational Studies. Arthritis Care Res. 2017, 69, 67–74. [Google Scholar] [CrossRef]
- Binus, A.M.; Han, J.; Qamar, A.A.; Mody, E.A.; Holt, E.W.; Qureshi, A.A. Associated Comorbidities in Psoriasis and Inflammatory Bowel Disease. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 644–650. [Google Scholar] [CrossRef]
- Dal, G.; Paolo, B.; Luca, G.; Giampiero Girolomoni, I. Psoriatic Arthritis and Diabetes Mellitus: A Narrative Review. Rheumatol. Ther. 2020, 7, 271–285. [Google Scholar] [CrossRef]
- Sterry, W.; Strober, B.E.; Menter, A. Obesity in Psoriasis: The Metabolic, Clinical and Therapeutic Implications. Report of an Interdisciplinary Conference and Review. Br. J. Dermatol. 2007, 157, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Herron, M.D.; Hinckley, M.; Hoffman, M.S.; Papenfuss, J.; Hansen, C.B.; Callis, K.P.; Krueger, G.G. Impact of Obesity and Smoking on Psoriasis Presentation and Management. Arch. Dermatol. 2005, 141, 1527–1534. [Google Scholar] [CrossRef]
- Jones, S.M.; Harris, P.D.; Lloyd, J.; Stirling, C.A.; Reckless, J.P.D.; Mchugh, N.J. Lipoproteins and Their Subfractions in Psoriatic Arthritis: Identification of an Atherogenic Profile with Active Joint Disease. Ann. Rheum. Dis. 2000, 59, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, A.; Chodorowska, G.; Szepietowski, J.; Zalewska-Janowska, A.; Krasowska, D.; Hercogová, J. Psoriasis and Serum Lipid Abnormalities. Dermatol. Ther. 2010, 23, 160–173. [Google Scholar] [PubMed]
- Azuaga, A.B.; Ramírez, J.; Cañete, J.D. Psoriatic Arthritis: Pathogenesis and Targeted Therapies. Int. J. Mol. Sci. 2023, 24, 4901. [Google Scholar] [CrossRef]
- Tucker Laura C Coates Philip S Helliwell, L.J. Assessing Disease Activity in Psoriatic Arthritis: A Literature Review. Rheumatol. Ther. 2019, 6, 23–32. [Google Scholar] [CrossRef]
- Clinical Review Report: Guselkumab (Tremfya): (Janssen Inc.): Indication: For the Treatment of Adult Patients with Moderate-to-Severe Plaque Psoriasis Who Are Candidates for Systemic Therapy or Phototherapy. Appendix 5: Validity of Outcome Measures; Canadian Agency for Drugs and Technologies in Health Common Drug Reviews: Ottawa, ON, Canada, 2018.
- Helliwell, P.S.; FitzGerald, O.; Fransen, J.; Gladman, D.D.; Kreuger, G.G.; Callis-Duffin, K.; McHugh, N.; Mease, P.J.; Strand, V.; Waxman, R.; et al. The Development of Candidate Composite Disease Activity and Responder Indices for Psoriatic Arthritis (GRACE Project). Ann. Rheum. Dis. 2013, 72, 986–991. [Google Scholar] [CrossRef]
- Kell, D.B.; Oliver, S.G. The Metabolome 18 Years on: A Concept Comes of Age. Metabolomics 2016, 12, 148. [Google Scholar] [CrossRef]
- Jutley, G.S.; Young, S.P. Metabolomics to Identify Biomarkers and as a Predictive Tool in Inflammatory Diseases. Best. Pract. Res. Clin. Rheumatol. 2015, 29, 770–782. [Google Scholar] [CrossRef]
- Koussiouris, J.; Looby, N.; Kulasingam, V.; Chandran, V. A Solid-Phase Microextraction—Liquid Chromatography-Mass Spectrometry Method for Analyzing Serum Lipids in Psoriatic Disease. Metabolites 2023, 13, 963. [Google Scholar] [CrossRef]
- Jollife, I.T.; Cadima, J. Principal Component Analysis: A Review and Recent Developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Galal, A.; Talal, M.; Moustafa, A. Applications of Machine Learning in Metabolomics: Disease Modeling and Classification. Front. Genet. 2022, 13, 1017340. [Google Scholar] [CrossRef]
- Kelesidis, T.; Papakonstantinou, V.; Detopoulou, P.; Fragopoulou, E.; Chini, M.; Lazanas, M.C.; Antonopoulou, S. The Role of Platelet-Activating Factor in Chronic Inflammation, Immune Activation, and Comorbidities Associated with HIV Infection. AIDS Rev. 2015, 17, 191–201. [Google Scholar] [PubMed]
- Bochkov, V.N.; Oskolkova, O.V.; Birukov, K.G.; Levonen, A.-L.; Binder, C.J.; Stöckl, J. Comprehensive Invited Review Generation and Biological Activities of Oxidized Phospholipids. Antioxid. Redox Signal. 2010, 12, 1009–1059. [Google Scholar] [CrossRef]
- Sorokin, A.V.; Remaley, A.T.; Mehta, N.N. Oxidized Lipids and Lipoprotein Dysfunction in Psoriasis. J. Psoriasis Psoriatic Arthritis 2020, 5, 139–146. [Google Scholar] [CrossRef]
- Canpolat, F.; Akpınar, H.; Eskioğlu, F. Mean Platelet Volume in Psoriasis and Psoriatic Arthritis. Clin. Rheumatol. 2010, 29, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Dobrzyńska, I.; Szachowicz-Petelska, B.; Wroński, A.; Jarocka-Karpowicz, I.; Skrzydlewska, E. Changes in the Physicochemical Properties of Blood and Skin Cell Membranes as a Result of Psoriasis Vulgaris and Psoriatic Arthritis Development. Int. J. Mol. Sci. 2020, 21, 9129. [Google Scholar] [CrossRef]
- Chen, C.; Hou, G.; Zeng, C.; Ren, Y.; Chen, X.; Peng, C. Metabolomic Profiling Reveals Amino Acid and Carnitine Alterations as Metabolic Signatures in Psoriasis. Theranostics 2020, 11, 754–767. [Google Scholar] [CrossRef] [PubMed]
- Pohla, L.; Ottas, A.; Kaldvee, B.; Abram, K.; Soomets, U.; Zilmer, M.; Reemann, P.; Jaks, V.; Kingo, K. Hyperproliferation Is the Main Driver of Metabolomic Changes in Psoriasis Lesional Skin. Sci. Rep. 2020, 10, 3081. [Google Scholar] [CrossRef]
- Coras, R.; Kavanaugh, A.; Kluzniak, A.; Holt, D.; Weilgosz, A.; Aaron, A.; Quehenberger, O.; Ritchlin, C.; Guma, M. Differences in Oxylipin Profile in Psoriasis versus Psoriatic Arthritis. Arthritis Res. Ther. 2021, 23, 200. [Google Scholar] [CrossRef]
- Coras, R.; Kavanaugh, A.; Boyd, T.; Huynh, Q.; Pedersen, B.; Armando, A.M.; Dahlberg-Wright, S.; Marsal, S.; Jain, M.; Paravar, T.; et al. Pro- and Anti-Inflammatory Eicosanoids in Psoriatic Arthritis. Metabolomics 2019, 15, 65. [Google Scholar] [CrossRef]
- Chen, M.; Lam, B.K.; Kanaoka, Y.; Nigrovic, P.A.; Audoly, L.P.; Austen, K.F.; Lee, D.M. Neutrophil-Derived Leukotriene B4 Is Required for Inflammatory Arthritis. J. Exp. Med. 2006, 203, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Nadler, J.L.; Mirmira, R.G.; Casimiro, I. Regulation of Tissue Inflammation by 12-Lipoxygenases. Biomolecules 2021, 11, 717. [Google Scholar] [CrossRef]
- Kamleh, M.A.; Snowden, S.G.; Grapov, D.; Blackburn, G.J.; Watson, D.G.; Xu, N.; Ståhle, M.; Wheelock, C.E. LC-MS Metabolomics of Psoriasis Patients Reveals Disease Severity-Dependent Increases in Circulating Amino Acids That Are Ameliorated by Anti-TNFα Treatment. J. Proteome Res. 2015, 14, 557–566. [Google Scholar] [CrossRef]
- Schett, G.; Loza, M.J.; Palanichamy, A.; FitzGerald, O.; Ritchlin, C.; Bay-Jensen, A.C.; Nielsen, S.H.; Gao, S.; Hsia, E.C.; Kollmeier, A.P.; et al. Collagen Turnover Biomarkers Associate with Active Psoriatic Arthritis and Decrease with Guselkumab Treatment in a Phase 3 Clinical Trial (DISCOVER-2). Rheumatol. Ther. 2022, 9, 1017–1030. [Google Scholar] [CrossRef]
- Singh, B.K.; Kambayashi, T. The Immunomodulatory Functions of Diacylglycerol Kinase ζ. Front. Cell Dev. Biol. 2016, 4, 96. [Google Scholar] [CrossRef]
- Poursharifi, P.; Madiraju, S.R.M.; Prentki, M. Monoacylglycerol Signalling and ABHD6 in Health and Disease. Diabetes Obes. Metab. 2017, 19, 76–89. [Google Scholar] [CrossRef]
- Mysliwiec, H.; Harasim-Symbor, E.; Baran, A.; Szterling-Jaworowska, M.; Milewska, A.J.; Chabowski, A.; Flisiak, I. Abnormal Serum Fatty Acid Profile in Psoriatic Arthritis. Arch. Med. Sci. 2019, 15, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, S.; Svahn, S.L.; Johansson, M.E. Effects of Omega-3 Fatty Acids on Immune Cells. Int. J. Mol. Sci. 2019, 20, 5028. [Google Scholar] [CrossRef] [PubMed]
- Paine, A.; Brookes, P.S.; Bhattacharya, S.; Li, D.; De La Luz Garcia-Hernandez, M.; Tausk, F.; Ritchlin, C. Dysregulation of Bile Acids, Lipids, and Nucleotides in Psoriatic Arthritis Revealed by Unbiased Profiling of Serum Metabolites. Arthritis Rheumatol. 2023, 75, 53–63. [Google Scholar] [CrossRef]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile Acids Activated Receptors Regulate Innate Immunity. Front. Immunol. 2018, 9, 1853. [Google Scholar] [CrossRef]
- Enoki, Y.; Watanabe, H.; Arake, R.; Sugimoto, R.; Imafuku, T.; Tominaga, Y.; Ishima, Y.; Kotani, S.; Nakajima, M.; Tanaka, M.; et al. Indoxyl Sulfate Potentiates Skeletal Muscle Atrophy by Inducing the Oxidative Stress-Mediated Expression of Myostatin and Atrogin-1. Sci. Rep. 2016, 6, 32084. [Google Scholar] [CrossRef] [PubMed]
- Dias, G.F.; Bonan, N.B.; Steiner, T.M.; Tozoni, S.S.; Rodrigues, S.; Nakao, L.S.; Kuntsevich, V.; Filho, R.P.; Kotanko, P.; Moreno-Amaral, A.N. Indoxyl Sulfate, a Uremic Toxin, Stimulates Reactive Oxygen Species Production and Erythrocyte Cell Death Supposedly by an Organic Anion Transporter 2 (OAT2) and NADPH Oxidase Activity-Dependent Pathways. Toxins 2018, 10, 280. [Google Scholar] [CrossRef]
- Watanabe, K.; Tominari, T.; Hirata, M.; Matsumoto, C.; Hirata, J.; Murphy, G.; Nagase, H.; Miyaura, C.; Inada, M. Indoxyl Sulfate, a Uremic Toxin in Chronic Kidney Disease, Suppresses Both Bone Formation and Bone Resorption. FEBS Open Bio. 2017, 7, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Toussirot, E.; Gallais-Sérézal, I.; Aubin, F. The Cardiometabolic Conditions of Psoriatic Disease. Front. Immunol. 2022, 13, 970371. [Google Scholar] [CrossRef]
- Ghosh, S. Metabolomic Studies for Metabolic Alterations Induced by Non-Steroidal Anti-Inflammatory Drugs: Mini Review. Biomolecules 2021, 11, 1456. [Google Scholar] [CrossRef]
- Liu, J.W.; Murtada, K.; Reyes-Garcés, N.; Pawliszyn, J. Systematic Evaluation of Different Coating Chemistries Used in Thin-Film Microextraction. Molecules 2020, 25, 3448. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.; Gladman, D.; Helliwell, P.; Marchesoni, A.; Mease, P.; Mielants, H. Classification Criteria for Psoriatic Arthritis: Development of New Criteria from a Large International Study. Arthritis Rheum. 2006, 54, 2665–2673. [Google Scholar] [CrossRef] [PubMed]
- Mirnaghi, F.S.; Pawliszyn, J. Development of Coatings for Automated 96-Blade Solid Phase Microextraction-Liquid Chromatography-Tandem Mass Spectrometry System, Capable of Extracting a Wide Polarity Range of Analytes from Biological Fluids. J. Chromatogr. A 2012, 1261, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Thermo Fisher Compound DiscovererTM Software. Available online: https://www.thermofisher.com/order/catalog/product/OPTON-31061 (accessed on 29 May 2023).
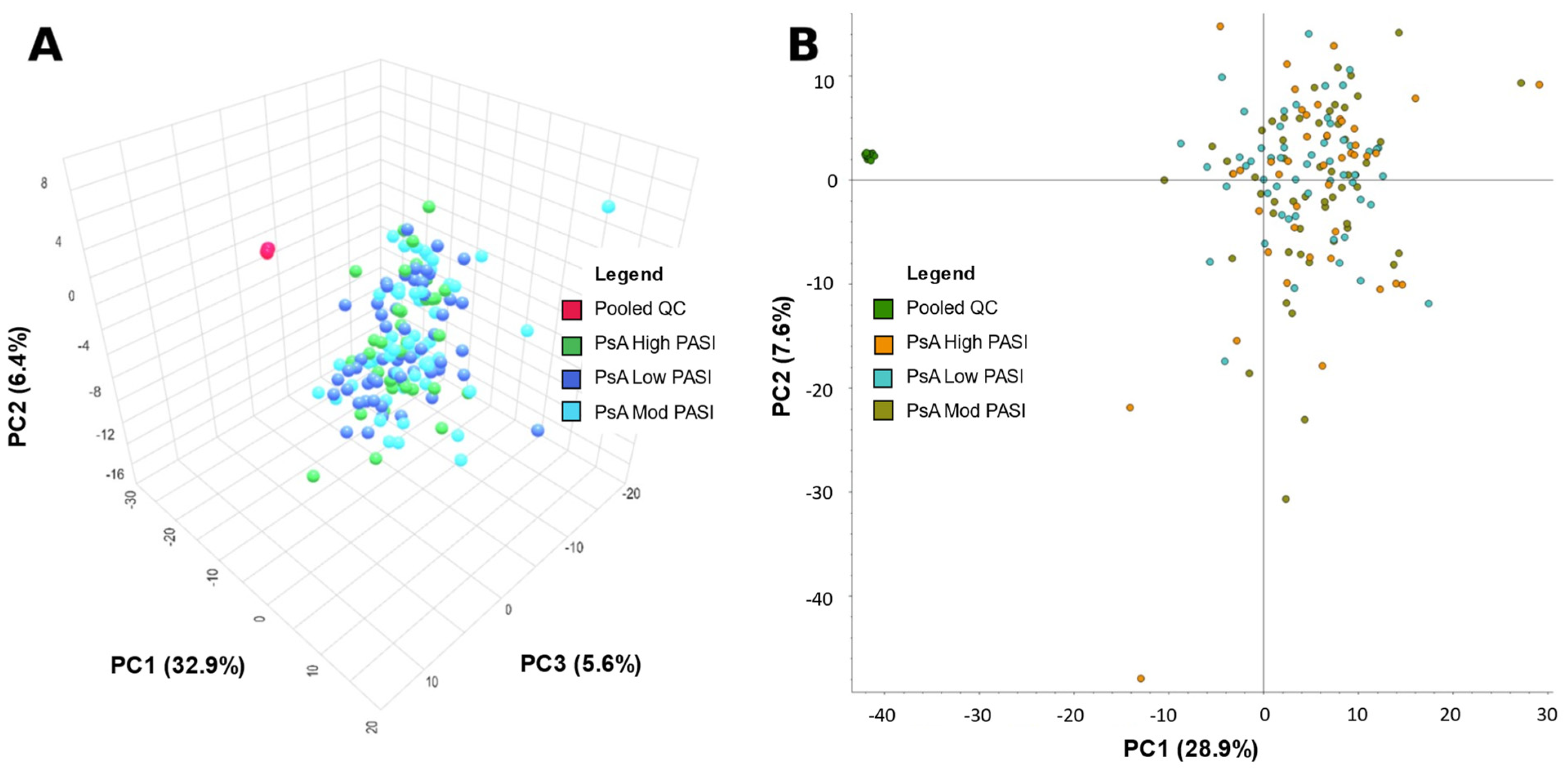
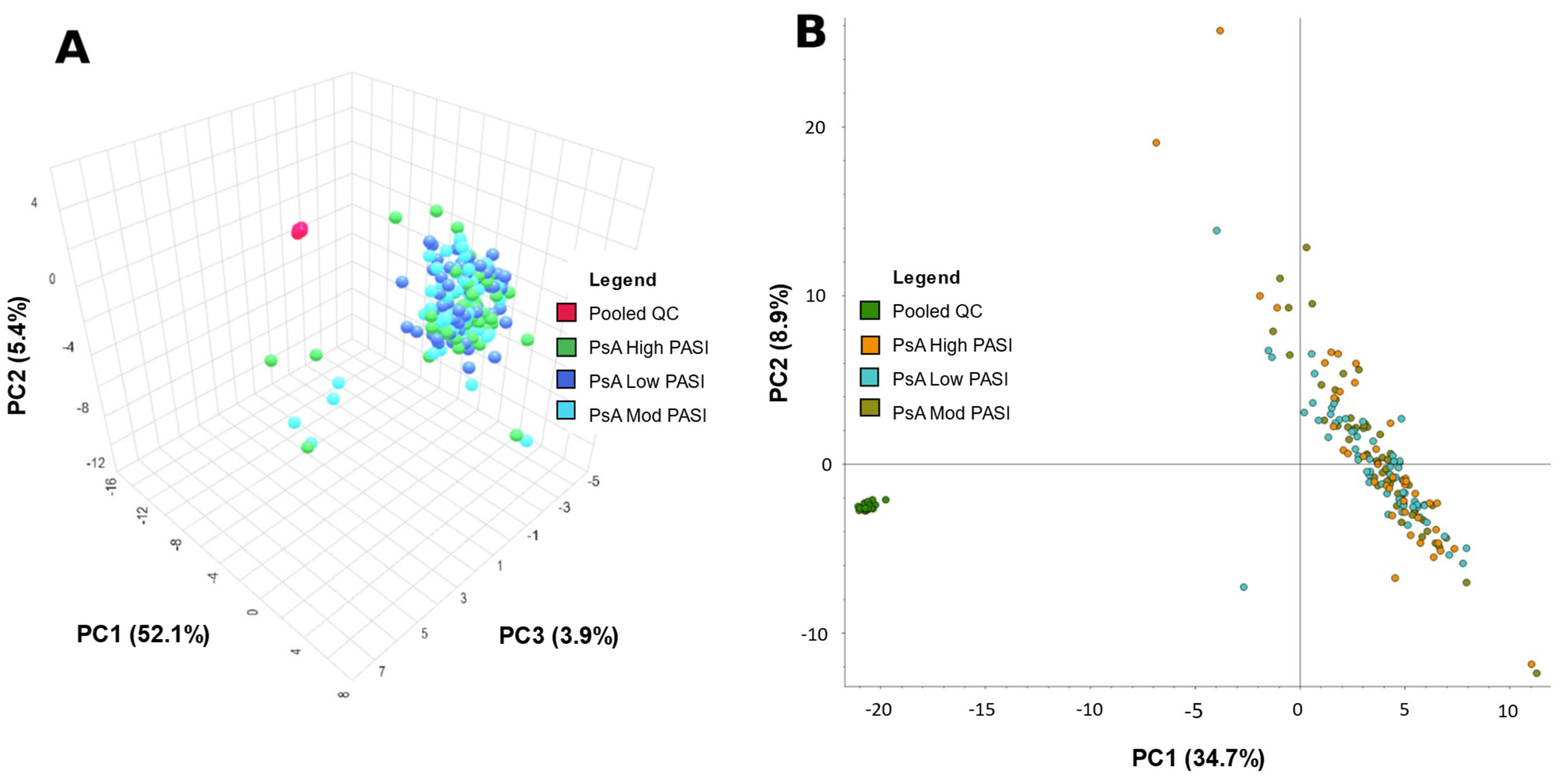
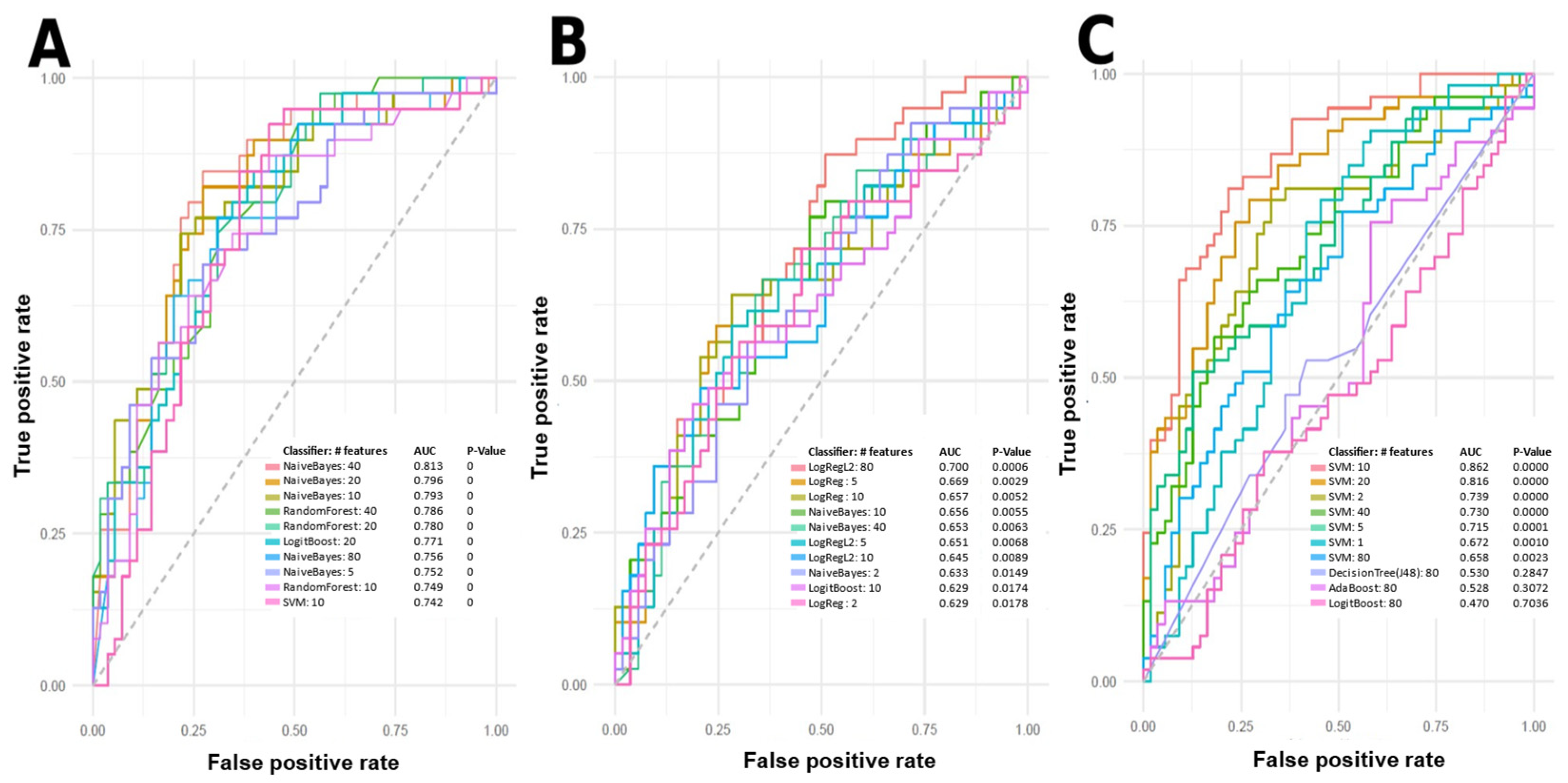
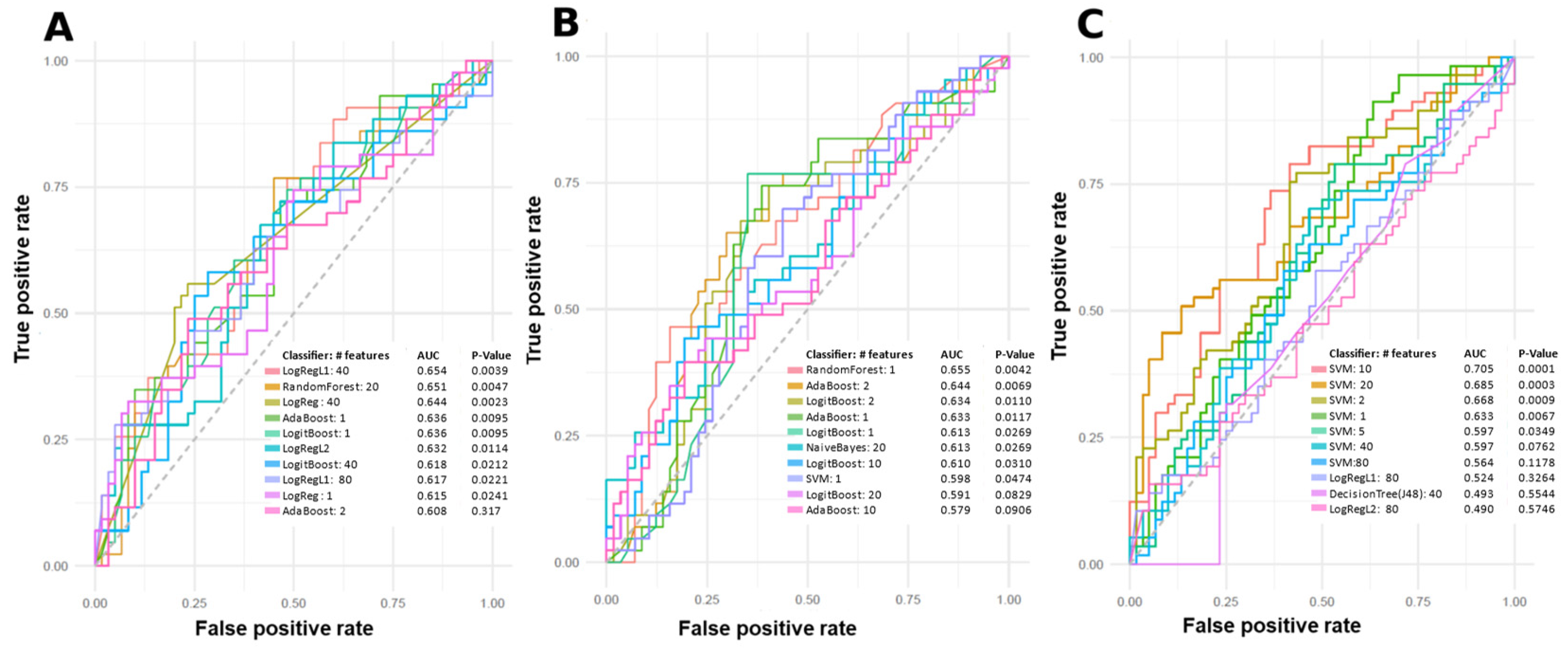

| Group | Sex (No. of Patients) | Mean Age Standard Deviation (Age Range) | Mean BMI Standard Deviation (BMI Range) | PASI Score Mean Standard Deviation (Range) | Dur. Psoriasis Mean Standard Deviation (Range) (Years) | Dur. PsA Mean Standard Deviation (Range) (Years) | Treatment (No. Patients) | Comorbidities (No. Patients in Group) |
|---|---|---|---|---|---|---|---|---|
| PsA | M (28) | 44.4 | 27.6 | 2.0 | 17.6 | 9.3 | NSAIDs (17) | Hypertension (5) |
| (Low) | 15.4 | 5.4 | 1.3 | 11.5 | 9.1 | DMARDs (9) | Depression (1) | |
| (18.1–74.6) | (16.2–42.6) | (0.3–4.8) | (0.5–53.6) | (0.0–25.9) | Steroids (1) | Hyperlipidemia (6) | ||
| Diabetes (3) | ||||||||
| F (28) | 47.9 | 28.5 | 1.8 | 22.1 | 9.5 | NSAIDs (19) | Hypertension (2) | |
| 10.0 | 7.4 | 1.4 | 13.6 | 8.5 | DMARDs (10) | Depression (3) | ||
| (30.6–67.6) | (19.8–49.3) | (0.1–4.8) | (0.5–46.8) | (0.5–28.8) | Steroids (1) | Hyperlipidemia (6) | ||
| PsA | M (27) | 51.0 | 28.5 | 6.4 | 22.5 | 13.6 | NSAIDs (19) | Hypertension (1) |
| (Moderate) | 10.1 | 6.9 | 1.4 | 12.7 | 10.5 | DMARDs (11) | Depression (2) | |
| (28.9–73.7) | (19.8–52.1) | (5.0–9.8) | (3.1–50.1) | (0.6–34.7) | Hyperlipidemia (3) | |||
| Diabetes (1) | ||||||||
| F (27) | 47.1 | 29.1 | 7.2 | 23.7 | 10.2 | NSAIDs (15) | Hypertension (1) | |
| 12.1 | 6.6 | 1.4 | 11.0 | 10.0 | DMARDs (14) | Depression (3) | ||
| (19.3–71.6) | (18.2–41.7) | (5.1–9.4) | (3.3–44.6) | (0.2–37.1) | Steroids (1) | Hyperlipidemia (3) | ||
| PsA | M (28) | 46.5 | 27.7 | 21.1 | 18.8 | 7.9 | NSAIDs (15) | Hypertension (3) |
| (High) | 13.5 | 4.6 | 9.6 | 11.0 | 9.7 | DMARDs (11) | Depression (2) | |
| (22.5–81.2) | (18.8–34.3) | (10.1–40.7) | (5.5–45.2) | (0.0–37.3) | Steroids (1) | Diabetes (3) | ||
| Hyperlipidemia (5) | ||||||||
| F (12) | 48.0 | 31.0 | 22.5 | 19.4 | 14.0 | NSAIDs (3) | Hyperlipidemia (2) | |
| 12.7 | 6.6 | 13.3 | 11.4 | 12.6 | DMARDs (7) | Diabetes (3) | ||
| (27.2–71.7) | (19.1–40.1) | (10.2–54.6) | (0.4–36.5) | (0.2–32.2) | Steroids (2) |
| m/z | Retention Time (min) | Adduct | Monoisotopic Mass | AUC | Tentative Identification |
|---|---|---|---|---|---|
| 546.3528 | 14.59 | [M + Na]+ | 523.3638 | 0.59 | Platelet-activating factor; 6 other hits |
| 544.3373 | 13.35 | [M + Na]+ | 521.3481 | 0.58 | LysoPC(0:0/18:1(9Z)); 5 other hits |
| 116.0708 | 0.60 | [M + H]+ | 115.0633 | 0.58 | Proline |
| 427.1938 | 5.74 | [M + NH4]+ | 409.1584 | 0.58 | dermatan L-iduronate; 1 other hit |
| 496.3397 | 12.95 | [M + Na]+ | 473.3505 | 0.57 | Clupanodonyl carnitine; 5 other hits |
| 373.2735 | 9.79 | [M + H]+ | 372.2664 | 0.57 | Cervonoyl ethanolamide |
| 520.3398 | 12.33 | [M + H]+ | 519.3325 | 0.57 | LysoPC(0:0/18:2(9Z,12Z)); 1 other hit |
| 370.2950 | 8.20 | [M + NH4]+ | 352.2614 | 0.57 | MG(18:3(6Z,9Z,12Z)/0:0/0:0); 8 other hits |
| 289.1409 | 5.12 | [M + Na]+ | 266.1518 | 0.57 | pentadeca-5,7,9-trienedioic acid; 8 other hits * |
| 356.2793 | 7.49 | [M + NH4]+ | 338.2457 | 0.57 | 11,12-DiHETrE; 8 other hits |
| m/z | Retention Time (min) | Adduct | Monoisotopic Mass | AUC | Tentative Identification |
|---|---|---|---|---|---|
| 211.1441 | 3.90 | [M + NH4]+ | 193.1103 | 0.60 | (R)-N-Methylsalsolinol; 3 other hits |
| 215.1279 | 4.78 | [M + H]+ | 214.1205 | 0.58 | undec-3-enedioic acid; 3 other hits * |
| 769.4224 | 21.08 | [M + Na]+ | 746.4370 | 0.57 | PG(20:4(8Z,11Z,14Z,17Z)-2OH(5S,6R)/i-12:0); 5 other hits |
| 760.5845 | 20.22 | [M + H]+ | 759.5778 | 0.57 | Pe-NMe2(20:1(11Z)/15:0); 26 other hits |
| 802.5350 | 19.32 | [M + Na]+ | 779.5465 | 0.57 | PC(20:5(6E,8Z,11Z,14Z,17Z)-OH(5)/P-16:0); 89 other hits |
| 496.3397 | 12.95 | [M + Na]+ | 473.3505 | 0.57 | Clupanodonyl carnitine; 5 other hits |
| 544.3373 | 13.35 | [M + Na]+ | 521.3481 | 0.56 | LysoPC(18:1(9Z)/0:0); 5 other hits |
| 518.3215 | 12.95 | [M + Na]+ | 495.3325 | 0.56 | LysoPC(16:0/0:0); 2 other hits |
| 351.2504 | 12.67 | [M + Na]+ | 328.2614 | 0.56 | MG(0:0/16:1(9Z)/0:0); 1 other hit |
| 828.5507 | 19.67 | [M + Na]+ | 805.5621555 | 0.56 | PC(20:5(6E,8Z,11Z,14Z,17Z)-OH(5)/P-18:1(9Z)); 107 other hits |
| m/z | Retention Time (min) | Adduct | Monoisotopic Mass | AUC | Tentative Identification |
|---|---|---|---|---|---|
| 311.1464 | 3.40 | [M + NH4]+ | 293.1111 | 0.75 | 4-Hydroxyproline galactoside; 3 other hits |
| 524.3710 | 14.21 | [M + H]+ | 523.3638 | 0.72 | Platelet-activating factor; 4 other hits |
| 263.0887 | 6.70 | [M + Na]+ | 240.0998 | 0.72 | 3-Carboxy-4-methyl-5-propyl-2-furanpropionic acid; 4 other hits |
| 641.5110 | 20.65 | [M + NH4]+ | 623.4761 | 0.71 | Cer(d16:1/6 keto-PGF1alpha); 58 other hits |
| 830.5664 | 19.88 | [M + Na]+ | 807.5778 | 0.71 | PC(20:4(5Z,7E,11Z,14Z)-OH(9)/P-18:1(9Z)); 151 other hits |
| 769.4224 | 21.08 | [M + Na]+ | 746.4370 | 0.70 | PG(i-12:0/20:4(6Z,8E,10E,14Z)-2OH(5S,12R)); 5 other hits |
| 188.0707 | 2.35 | [M + H]+ | 187.0633 | 0.70 | Indoleacrylic acid |
| 813.6839 | 21.40 | [M + H]+ | 812.6771 | 0.68 | SM(d18:2(4E,14Z)/24:0); 1 other hit |
| 377.2659 | 13.29 | [M + Na]+ | 354.2770 | 0.68 | Glyceryl monolinoleate |
| 343.2241 | 11.17 | [M + Na]+ | 320.2351 | 0.67 | 12-HETE; 39 other hits |
| m/z | Retention Time (min) | Adduct | Monoisotopic Mass | AUC | Tentative ID |
|---|---|---|---|---|---|
| 212.0026 | 3.97 | [M − H]− | 213.0096 | 0.72 | Indoxyl sulfate; 3 other hits |
| 729.1788 | 11.02 | [M + Cl]− | 694.2109 | 0.69 | Neocuscutoside C; 3 other hits |
| 464.3020 | 7.39 | [M − H]− | 465.3090 | 0.67 | Glyco-beta-muricholic acid; 5 other hits |
| 802.5614 | 13.14 | [M − H]− | 803.5676 | 0.67 | PE(PGF1alpha/P-18:0); 38 other hits |
| 343.1707 | 8.15 | [M − H]− | 344.1776 | 0.64 | Tamoxifen-ol |
| 667.1419 | 11.98 | [M − H]− | 668.1506 | 0.63 | Etoposide Phosphate; 1 other hit |
| 448.3073 | 8.18 | [M − H]− | 449.3141 | 0.63 | Glycohyodeoxycholic acid; 7 other hits |
| 388.1559 | 8.00 | [M + Cl]− | 353.1852 | 0.63 | Epiroprim |
| 203.08299 | 3.58 | [M − H]− | 204.0899 | 0.63 | L-Tryptophan; 13 other hits |
| Moderate vs. High (m/z) | Low vs. Moderate (m/z) | Low vs. High (m/z) | Tentative Identification |
|---|---|---|---|
| 736.2599 | 736.2599 | N/A | |
| 215.1279 | 215.1279 | Medium chain fatty acids | |
| 680.3549 | 680.3549 | N/A | |
| 496.3397 | 496.3397 | Phospholipid, Acylcarnitine | |
| 159.1168 | 159.1168 | Exposome—Benzene and derivatives | |
| 544.3373 | 544.3373 | Lysophosphatidylcholine | |
| 688.3047 | 688.3047 | N/A | |
| 116.0708 | 116.0708 | Proline; Exposome-related Metabolites | |
| 415.2536 | 415.2536 | Exposome—Peptides | |
| 769.4224 | 769.4224 | Oxidized phospholipid | |
| 646.2575 | 646.2575 | Exposome—Drug for increasing blood glucose concentration | |
| 668.5443 | 668.5443 | DAGs | |
| 549.1857 | 549.1857 | 549.1857 | Exposome—Neoacrimarine I/F |
| Time (min) | % Mobile Phase B (Methanol + 0.1% Formic Acid) |
| 0 | 5 |
| 1 | 5 |
| 20 | 100 (curve 3) |
| 22.5 | 100 |
| 25 | 5 |
| 30 | 5 |
| Time (min) | % Mobile Phase B (Methanol) |
| 0 | 0 |
| 2 | 0 |
| 12 | 100 (curve 3) |
| 15 | 100 |
| 18 | 0 |
| 20 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choksi, H.; Li, S.; Looby, N.; Kotlyar, M.; Jurisica, I.; Kulasingam, V.; Chandran, V. Identifying Serum Metabolomic Markers Associated with Skin Disease Activity in Patients with Psoriatic Arthritis. Int. J. Mol. Sci. 2023, 24, 15299. https://doi.org/10.3390/ijms242015299
Choksi H, Li S, Looby N, Kotlyar M, Jurisica I, Kulasingam V, Chandran V. Identifying Serum Metabolomic Markers Associated with Skin Disease Activity in Patients with Psoriatic Arthritis. International Journal of Molecular Sciences. 2023; 24(20):15299. https://doi.org/10.3390/ijms242015299
Chicago/Turabian StyleChoksi, Hani, Shenghan Li, Nikita Looby, Max Kotlyar, Igor Jurisica, Vathany Kulasingam, and Vinod Chandran. 2023. "Identifying Serum Metabolomic Markers Associated with Skin Disease Activity in Patients with Psoriatic Arthritis" International Journal of Molecular Sciences 24, no. 20: 15299. https://doi.org/10.3390/ijms242015299






