Abstract
Drought stress severely threatens the yield of cereal crops. Therefore, understanding the molecular mechanism of drought stress response of plants is crucial for developing drought-tolerant cultivars. NAC transcription factors (TFs) play important roles in abiotic stress of plants, but the functions of NAC TFs in sorghum are largely unknown. Here, we characterized a sorghum NAC gene, SbNAC9, and found that SbNAC9 can be highly induced by polyethylene glycol (PEG)-simulated dehydration treatments. We therefore investigated the function of SbNAC9 in drought stress response. Sorghum seedlings overexpressing SbNAC9 showed enhanced drought-stress tolerance with higher chlorophyll content and photochemical efficiency of PSII, stronger root systems, and higher reactive oxygen species (ROS) scavenging capability than wild-type. In contrast, sorghum seedlings with silenced SbNAC9 by virus-induced gene silencing (VIGS) showed weakened drought stress tolerance. Furthermore, SbNAC9 can directly activate a putative peroxidase gene SbC5YQ75 and a putative ABA biosynthesis gene SbNCED3. Silencing SbC5YQ75 and SbNCED3 led to compromised drought tolerance and reduced ABA content of sorghum seedlings, respectively. Therefore, our findings revealed the important role of SbNAC9 in response to drought stress in sorghum and may shed light on genetic improvement of other crop species under drought-stress conditions.
1. Introduction
Drought is one of the major environmental factors limiting the yield of cereal crops, and drought leads to a decrease in seed germination, retardation of vegetative development, and reduction of grain quantity and quality [1,2]. Sorghum (Sorghum bicolor (L.) Moench) is widely cultivated in arid and semiarid regions of developing countries and provides staple food for over 500 million people [3]. Although sorghum is generally tolerant to a drought environment, it is still susceptible to water scarcity at seedling stage, preflowering stage, and postflowering stage [3,4]. Water loss induced by drought stress may cause impaired cell division and elongation which may lead to severe inhibition of embryonic growth, accompanied with poor seedling establishment [2]. At postflowering stage, water deficiency directly influences grain size due to premature plant senescence [5]. Therefore, understanding the molecular mechanism of drought-stress tolerance of sorghum will be benefit to the yield of sorghum and even other cereal crops.
Drought stress can induce accumulation of reactive oxygen species (ROS) in plants, which include superoxide anions, hydroxyl radicals, hydrogen peroxide, and singlet oxygen [6]. ROS can harm molecular activities in cells and even may lead to programmed cell death [7,8]. Although low concentration of ROS is an important part of stress signaling [9], excess accumulation of ROS requires antioxidant enzymes such as superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) to eliminate ROS toxicity to cells [10,11]. ROS can also lead to an increase in lipid peroxidation in cell membranes, which can be indicated by malondialdehyde (MDA) content [12,13]. To cope with drought stress, plants adopt a series of physiological, morphological, and molecular strategies. There are abundant stress-responsive factors which can induce the protective enzyme activities to reduce ROS damage to cells.
The responses of plants to drought stress involve signaling cascades with a phytohormone ABA playing the central role [14]. For ABA biosynthesis, the small peptide CLAVATA3/EMBRYO-SURROUNDING REGIONRELATED 25 (CLE25) moves from the roots to the leaves to activate expression of the key enzyme gene NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3) in Arabidopsis [15]. Induction of NCED3 promotes rapid accumulation of ABA to trigger downstream signaling, which leads to stomatal closure to reduce water loss [16]. In addition to ABA signaling, transcription factors (TFs) also play vital roles under drought stress. In plants, NAC [no apical meristem (NAM), Arabidopsis transcription activation factor 1/2 (ATAF1/2), and cup-shaped cotyledon2 (CUC2)] family TFs [17,18], ethylene-responsive transcription factors, WRKY family TFs, heat stress transcription factors, and homeodomain leucine zipper family transcription factors all play important roles in drought-stress responses [3,19,20].
NAC TFs are one of the largest families of plant specific TFs. To date, 105 AtNAC TFs in Arabidopsis, 138 OsNAC TFs in rice, 131 SbNAC TFs in sorghum, and 147 ZmNAC TFs in maize have been identified [4,7,21,22]. Many NAC TFs are reported to be involved in abiotic stress responses in plants. NAC transcription factor JUNGBRUNNEN1 (JUB1) from Arabidopsis played a positive role in regulating Dehydration Responsive Element-Binding protein 1 (SlDREB1), Dehydration Responsive Element-Binding protein 2 (SlDREB2), and SlDELLA in tomato under drought stress [23]. Overexpression of OsNAC066 in rice upregulates the relative expression of stress-responsive genes including Early-Responsive to Dehydration 1 (OsERD1), Late Embryogenesis Abundant 3 (OsLEA3), and Dehydration Responsive Element-Binding protein 2A (OsDREB2A) [24]. Overexpression of SlNAC6 in tomato leads to efficient closure of stomata to reduce water loss under PEG-simulated drought treatment [20]. Overexpression of ZmNAC49 reduces not only stomatal conductance but also stomatal density to improve the plant stress tolerance in maize [22].
Although there are at least 131 NAC genes in sorghum, only a few SbNAC TFs have been functionally characterized [4]. SbSNAC1 is highly similar to OsSNAC1 of rice, which is responsive to multiple abiotic stresses and ABA treatment. Overexpression of SbSNAC1 in Arabidopsis can enhance plant drought- and salt-stress tolerance [25]. SbNAC2 can directly activate stress responsive gene SbAP37 and improve drought-stress tolerance of sorghum [26]. However, molecular functions of most sorghum SbNAC TFs remain to be elucidated.
In this study, we investigated the function of SbNAC9 and found that SbNAC9 can respond to various abiotic stresses and ABA treatment, especially highly to PEG-simulated dehydration stress. We overexpressed SbNAC9 in sorghum and found that the transgenic lines had enhanced plant drought-stress tolerance with increased photosynthesis, strengthened root architecture, and increased ROS scavenging ability. By contrast, in sorghum seedlings with virus-induced gene silencing (VIGS) of SbNAC9, drought-stress tolerance of the seedlings was obviously compromised. In addition, heterologous overexpression of SbNAC9 could also enhance drought tolerance of Arabidopsis. Moreover, SbNAC9 could directly activate the expression of putative peroxidase gene SbC5YQ75 [27] and putative enzyme gene SbNCED3 for ABA biosynthesis. VIGS-mediated silencing of SbC5YQ75 and SbNCED3 led to weakened drought tolerance and reduced ABA content in sorghum seedlings, respectively. Overall, our findings revealed the detailed molecular mechanism of SbNAC9 in response to drought stress in sorghum and shed light on genetic enhancement of sorghum and even other crop species.
2. Results
2.1. SbNAC9 Can Respond to PEG-Simulated Drought Stress
In a previous study, we constructed a phylogenetic tree based on 128 OsNAC TFs from rice and 119 SbNAC TFs from sorghum and noticed a subgroup containing 4 reported OsNAC TFs responsive to abiotic stress. Additionally, in this subgroup, we characterized a sorghum transcription factor, SbNAC2, which plays an important role under abiotic stresses [26]. However, the functions of other SbNAC TFs of this subgroup in abiotic stress need to be elucidated.
Here, we chose the SOBIC.005G064600.2 (SbNAC9) which showed the highest protein sequence similarity with OsNAC5 [28] for subsequent study. First, we found that the SbNAC9 promoter harbors many abiotic stress-related and hormone-responsive elements (Table S1), implying that SbNAC9 may respond to abiotic stress. The transcript level of SbNAC9 was upregulated under multiple abiotic stresses and phytohormone treatments. Among these treatments, SbNAC9 had the highest expression under PEG-simulated dehydration treatment (Figure 1A). Therefore, we presumed that SbNAC9 may play a potential role under drought stress in sorghum.
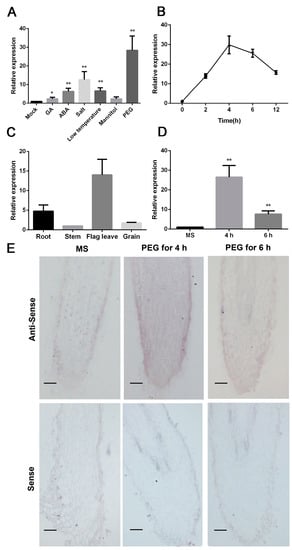
Figure 1.
SbNAC9 responds to PEG-simulated dehydration stress. (A) Relative expression of SbNAC9 in the fourth leaf of sorghum seedlings at four-leaf stage under hormone and abiotic stress treatments. Sorghum seedlings were used for water (Mock), 150 μM GA (gibberellin), 150 μM ABA, 200 mM NaCl, 200 mM mannitol, and 20% PEG6000 treatments. For low temperature treatment, sorghum seedlings were kept at 4 °C. SbEIF4A was used as the internal reference. Error bars indicate SD of three independent experiments. ** p < 0.01 and * p < 0.05 by Student’s t-test. (B) Time course transcript level of SbNAC9 in sorghum seedlings at four-leaf stage subjected to 20% PEG treatment. SbEIF4A was used as the internal reference. Error bars indicate SD of three independent experiments. (C) Relative transcript level of SbNAC9 in various tissues of sorghum at filling stage. SbEIF4A was used as the internal reference. Error bars indicate SD of three independent experiments. (D) Relative expression level of SbNAC9 in the three-day-old root tips of sorghum under MS media soaked into 20% PEG solution for 4 h and 6 h. SbEIF4A was used as the internal reference. Error bars indicate SD of three independent experiments. ** p < 0.01 by Student’s t-test. (E) In situ hybridization assay of SbNAC9 in root tips of the three-day-old sorghum seedlings subjected to 20 % PEG-simulated drought-stress treatment for 4 h and 6 h. Root tips grown on MS media served as negative control. Bars indicate 100 μm.
By PEG-simulated dehydration treatment, SbNAC9 transcript could be obviously induced and peaked at 4 h (Figure 1B). Meanwhile, SbNAC9 could also be induced and peaked at 1 h by ABA treatment (Figure S1). We also analyzed SbNAC9 expression in various sorghum tissues under normal growth condition. In roots and leaves, SbNAC9 transcript level was higher than in other tissues (Figure 1C). As roots may sense water deficiency in soil [29], we used sorghum root treated with 20% PEG6000 solution [25], which simulated water deficiency, for further analysis. After 4 h treatment, SbNAC9 activity was obviously induced in roots as shown by both qRT-PCR (quantitative real-time PCR) and in situ hybridization results (Figure 1D,E). These results showed that SbNAC9 can quickly respond to PEG-simulated drought stress and may play a potential role in drought-stress tolerance of sorghum.
2.2. SbNAC9 Functions as a Transcriptional Activator
SbNAC9 encodes a 385-amino-acid protein with a 163-amino-acid conserved NAC domain (1-163 aa) harboring five conserved subdomains (a–e) (Figure S2). To test if SbNAC9 had transcription activity, we constructed three vectors as pBD-SbNAC9-FL (containing the full length of SbNAC9), pBD-SbNAC9-N (containing N-terminal of SbNAC9), and pBD-SbNAC9-C (containing C terminal of SbNAC9). In these three vectors, full length or partial coding sequence of SbNAC9 were fused with GAL4 DNA-binding domain (Figure 2A). Yeast cells transformed with pBD-SbNAC9-FL and pBD-SbNAC9-C, but not with pBD-SbNAC9-N, grew well on the SD/-His medium (Figure 2B), indicating that SbNAC9 may function as a transcriptional activator and its transcriptional activation domain was in the C terminal.
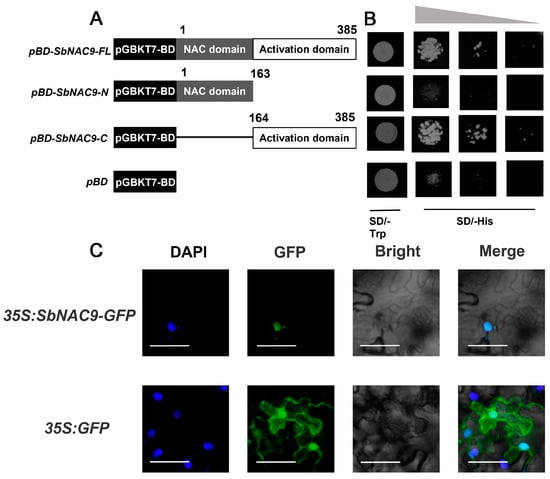
Figure 2.
SbNAC9 functions as a transcriptional activator. (A) Schematic diagrams of constructs in SbNAC9 transcriptional activation assay. (B) Transcriptional activation of SbNAC9. Yeast cells were diluted to 1, 10−1, 10−2 from the left panel to the right panel. pBD-SbNAC9-FL, full-length CDS of SbNAC9; pBD-SbNAC9-N, N-terminal of SbNAC9; pBD-SbNAC9-C, C-terminal of SbNAC9; and pBD, pGBKT7-BD vector. pGBKT7-BD was used as a negative control. (C) Subcellular localization of SbNAC9 in tobacco leaves. DAPI (2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride) channel represents the signal of nucleus colored in blue. GFP channel represents the signal of SbNAC9-GFP or GFP colored in green. Merge indicates combination of GFP (green fluorescent protein), DAPI, and Bright together colored in cyan. Bars indicate 50 μm.
We also analyzed the SbNAC9 protein sequence using an online webtool (https://psort.hgc.jp/) and noticed a nuclear localization signal of PRDRKYP inside the SbNAC9 protein. To examine the subcellular localization of SbNAC9, we checked the protein localization of 35S: SbNAC9-GFP in tobacco leaf cells. A strong GFP signal can be detected in the nucleus (Figure 2C), suggesting that SbNAC9 is a transcriptional activator localized in the nucleus.
2.3. Overexpression of SbNAC9 Can Enhance Drought-Stress Tolerance of Sorghum
To investigate the role of SbNAC9 in sorghum under drought stress, we overexpressed SbNAC9 in sorghum and chose three independent lines with various SbNAC9 expression levels (OE-1, OE-9, OE-14) for further study (Figure S3). At the six-leaf stage, the selected transgenic lines and wild-type plants were treated with water deprivation for 21 days. The transgenic lines displayed less withered leaves and stronger root architecture than the wild-type plants after drought treatment (Figure 3A,B). Additionally, the transgenic lines showed a lower water loss rate than the wild-type plants (Figure S4).
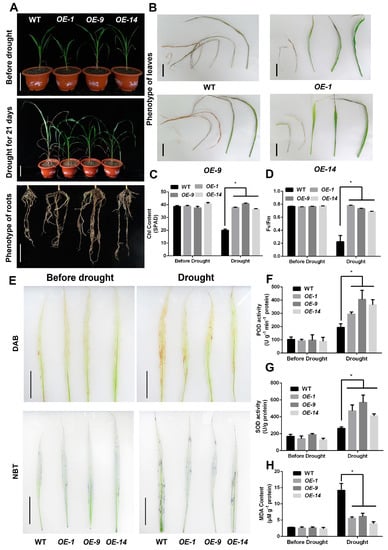
Figure 3.
Overexpression of SbNAC9 enhanced drought-stress tolerance of sorghum. (A) Phenotype of sorghum plants under drought-stress treatment. Three-week-old WT and transgenic lines (OE-1, OE-9, OE-14) were treated with water deprivation for 21 days. Bars indicate 10 cm. (B) Phenotype of first five leaves of WT and transgenic lines after 21-day drought-stress treatment. Bars indicate 5 cm. (C,D) Chlorophyll content (C) and chlorophyll fluorescence Fv/Fm (D) in the fifth leaf of WT and transgenic lines after drought treatment. Error bars indicate SD of three independent experiments. * p < 0.05 by Student’s t-test. (E) DAB and NBT staining of leaves of WT and transgenic sorghum seedlings treated with drought stress for 5 days. Bars indicate 5 cm. (F–H) POD activity (F), SOD activity (G), and MDA content (H) of WT and transgenic lines after drought-stress treatment. Error bars indicate SD of three independent experiments. * p < 0.05 by Student’s t-test.
Chlorophyll content is a vital indicator to assess drought-stress tolerance of plants [30]. Therefore, we measured chlorophyll contents in SbNAC9 overexpression lines and wild-type plants under normal or continuous drought conditions. The results showed that the transgenic lines had significantly higher chlorophyll content than the wild-type plants under drought treatments (Figure 3C). Consistently, the transgenic lines exhibited an obviously higher Fv/Fm (variable fluorescence/maximal fluorescence) ratio than the wild-type plants (Figure 3D). These results suggested that photosynthesis in sorghum transgenic lines with SbNAC9 overexpression were less affected by drought stress.
Drought stress triggered oxidative damage to plant cells. Thus, the ability to eliminate the excess accumulation of ROS was also considered a critical index for plant response to drought stress. We compared ROS scavenging activities of transgenic lines with SbNAC9 overexpression and wild-type plants under drought stress. The DAB and NBT staining assays showed that transgenic lines accumulated less H2O2 and O2− compared with wild-type (Figure 3E). Meanwhile, transgenic lines had significantly higher POD and SOD activities than the wild-type plants (Figure 3F,G), and the MDA content of transgenic lines was noticeably lower than in wild-type plants (Figure 3H). Taken together, these results showed that overexpression of SbNAC9 enhanced drought-stress tolerance of sorghum through maintaining relative high photosynthesis, strengthening root architecture, and increasing ROS scavenging ability.
2.4. Silencing of SbNAC9 Weakens Drought-Stress Tolerance of Sorghum Seedlings
To further test the role of SbNAC9 in sorghum under drought stress, we silenced SbNAC9 in sorghum seedlings by VIGS. We observed phenotypes of plants inoculated with empty virus (BSMV:00) or with BSMV:SbNAC9 under normal conditions or continuous drought treatments. Sorghum seedlings inoculated with BSMV:SbNAC9 showed obviously weakened drought stress tolerance with more wilting and chlorosis of leaves than the plants inoculated with empty virus (Figure 4A,B). Consistently, chlorophyll content and Fv/Fm ratio in plants inoculated with BSMV:SbNAC9 were significantly lower than those in plants inoculated with empty virus (Figure 4C,D). Moreover, the activities of antioxidative enzymes including SOD, POD, and CAT in plants inoculated with BSMV:SbNAC9 were significantly lower than those in plants inoculated with empty virus under drought-stress treatment (Figure 4E–G). DAB and NBT staining assays also showed that plants inoculated with BSMV:SbNAC9 had more H2O2 and O2− than plants inoculated with empty virus (Figure 4H). Together with the traits of SbNAC9 overexpression lines under drought-stress treatments (Figure 3), all these results suggest that SbNAC9 may play a vital function in drought-stress tolerance of sorghum.
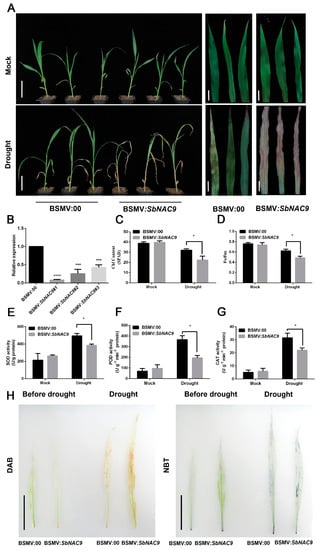
Figure 4.
Silenced SbNAC9 weakened drought-stress tolerance of sorghum. (A) Phenotype of sorghum seedlings inoculated with BSMV:00 and BSMV:SbNAC9 under mock and drought-stress treatments. Bars indicate 6 cm for the whole plants in the left panels and 2 cm for the leaves in the middle and right panels. (B) Relative transcript level of SbNAC9 in sorghum seedlings silenced by VIGS. SbEIF4A was used as the internal reference. Error bars indicate SD of three independent experiments. **** p < 0.0001 and *** p < 0.001 by Student’s t-test. (C,D) Chlorophyll content (C) and chlorophyll fluorescence Fv/Fm (D) in the third leaf of sorghum seedlings inoculated with BSMV:00 and BSMV:SbNAC9 under mock or drought-stress treatments. Error bars indicate SD of three independent experiments. * p < 0.05 by Student’s t-test. (E–G) SOD (E), POD (F), and CAT (G) activities in sorghum seedlings inoculated with BSMV:00 and BSMV:SbNAC9 under mock or drought-stress treatments. Error bars indicate SD of three independent experiments. * p < 0.05 by Student’s t-test. (H) DAB and NBT staining of leaves of sorghum seedlings inoculated with BSMV:00 and BSMV:SbNAC9 under drought-stress treatment for 5 days. Bars indicate 5 cm.
2.5. Heterologous Overexpression of SbNAC9 Enhances Drought Tolerance of Arabidopsis
To further confirm the function of SbNAC9 in drought-stress response, we also overexpressed SbNAC9 in Arabidopsis. We treated wild-type Col-0 plants and three independent transgenic lines (#9, #10, and #11 with various expression level of SbNAC9) with water deprivation for 10 days. After 5 continuous days with rewatering, the transgenic lines showed obviously higher survival rate than wild-type plants (Figure S5A–C). Under drought-stress treatment, transgenic plants showed lower H2O2 and O2− level by DAB and NBT staining than wild-type (Figure S5D,E). Meanwhile, the antioxidative enzyme activities were higher and MDA content was lower in transgenic lines than those in wild-type plants under drought-stress treatment (Figure S5I).
Next, we examined relative expression of several drought-responsive genes of Arabidopsis including Dehydration Responsive Element-Binding protein 1A (DREB1A), Dehydration Responsive Element-Binding protein 2A (DREB2A), and KIN1 [31,32,33]. We also examined relative expression of a key enzyme gene 9-Cis-Epoxycarotenoid Dioxygenase 3 (AtNCED3) involved in biosynthesis of ABA [16], which can regulate stomatal aperture and water loss rate [29]. Compared with wild-type plants, relative expression of these four genes in SbNAC9 overexpression Arabidopsis was significantly upregulated under drought stress (Figure S6).
Drought stress retarded various developmental processes including root formation. We observed that the roots of transgenic lines were longer than wild-type plants in 10% PEG6000 MS medium (Figure S7). Taken together, heterologous overexpression of SbNAC9 in Arabidopsis also can enhance plant drought-stress tolerance by reducing water loss, increasing antioxidative activity, and forming elongated roots.
2.6. SbNAC9 Directly Activates Expression of SbC5YQ75 and SbNCED3
Many NAC TFs are involved in abiotic stress by regulating downstream stress-induced genes [34]. In Arabidopsis, researchers identified consensus binding sites for two representative NAC proteins, ANAC019 and ANAC092 [35]. SbNAC9 shows higher similarity to ANAC019 by protein sequence alignment (Figure S8). Therefore, we used the putative binding motif (TTNCGTA) of ANAC019 [35] as the putative binding site of SbNAC9. To investigate how SbNAC9 is involved in drought-stress tolerance of sorghum, we searched for putative downstream targets of SbNAC9 based on the study of differentially expressed genes (DEGs) under sorbitol-simulated osmotic stress in sorghum [27]. We found 16 putative targets of SbNAC9 by searching the 2000-bp promoter sequences (including the proximal promoter and 5′ UTR) upstream of the start codons of these DEGs (Table S2). After prediction of genes’ function and analysis of the positions of SbNAC9-binding motifs, we chose SbC5YQ75, FASCICLIN-LIKE ARABINOGALACTAN 1 (SbFLA1), and SbC5XIY1 as candidates. As AtNCED3 was significantly upregulated in SbNAC9 overexpression Arabidopsis, we, therefore, searched the promoters of SbNCED3 and SbNCED9, both of which are AtNCED3 homologs (Figure S9). We found that both SbNCED3 and SbNCED9 promoters contain SbNAC9 binding motifs, with two different bases (TCTCGTG) at -838 bp and one different base (TTGCGTG) at −1126 bp upstream of their transcriptional start sites, respectively. Hence, we finally chose five genes including SbC5YQ75, SbFLA1, SbC5XIY1, SbNCED3, and SbNCED9 for further analysis.
We tested the transcript level of these five candidate genes in sorghum seedlings under 20% PEG6000-simulated dehydration-stress treatment and found that expression levels of all the five genes peaked later than SbNAC9 (Figure S10A). In addition, the five genes also rapidly responded to ABA treatment (Figure S10B). These indicated that these five candidates were the potential targets of SbNAC9 and possibly involved in ABA signaling for drought-stress tolerance. We then tested relative expression of the five genes in sorghum seedlings with silenced SbNAC9, and found that expression of SbC5YQ75, SbNCED3, and SbNCED9 were significantly downregulated (Figure 5A,B, and Figure S11A). However, due to the obvious upregulation of only SbC5YQ75, SbNCED3, and SbNCED9, but not the other two candidates in sorghum with SbNAC9-overexpression (Figure S11), we, therefore, chose SbC5YQ75, SbNCED3, and SbNCED9 for further analysis.
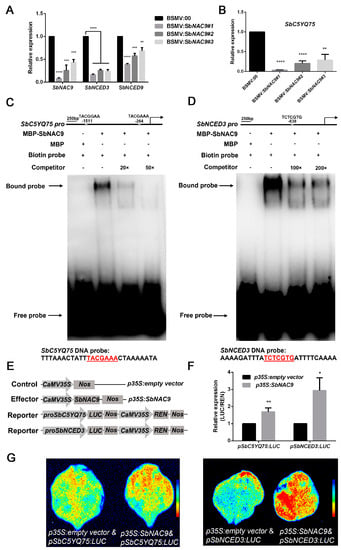
Figure 5.
SbNAC9 directly activated the expression of SbC5YQ75 and SbNCED3. (A,B) Relative transcript level of SbNAC9, SbNCED3, SbNCED9 (A), and C5YQ75 (B) in the third leaf of sorghum seedlings with silenced SbNAC9. SbEIF4A was used as the internal reference. Error bars indicate SD of three independent experiments. **** p < 0.0001, *** p < 0.001, and ** p < 0.01 by Student’s t-test. (C,D) EMSA assays of SbNAC9 bound to the promoters of SbC5YQ75 and SbNCED3 in vitro. Competition experiments were performed with excessive amounts of unlabeled probes (20× and 50× for SbC5YQ75 as well as 100× and 200× for SbNCED3). Schematic diagrams of putative binding motifs of SbNAC9 on the promoters of SbC5YQ75 and SbNCED3 were listed at the top. The motif at −264 bp upstream of transcription start sites of SbC5YQ75 and the motif at −838 bp upstream of transcription start sites of SbNCED3 were used for EMSA assays. The sequences of probes were listed at the bottom. (E) Schematic diagrams of constructs used in luciferase assays. (F) Relative transcript level of LUC/REN activity normalized by REN in luciferase assays. Error bars indicate SD of three independent experiments. ** p < 0.01 and * p < 0.05 by Student’s t-test. (G) Luciferase assays of SbNAC9 binding on SbC5YQ75 and SbNCED3 promoters in tobacco leaves. pSbC5YQ75:LUC or pSbNCED3:LUC with an empty vector were used as negative control.
To test the direct bindings, we first performed EMSA assays, and found that SbNAC9 directly bound the promoter fragments of SbC5YQ75 and SbNCED3, and the bindings were weakened with increased amounts of competitive probes (Figure 5C,D). However, we could not detect the binding of SbNAC9 to the promoter of SbNCED9 (Figure S12). Thus, we focused on SbC5YQ75 and SbNCED3 for further study. We also performed luciferase assays and found that the relative expression level of LUC/REN was higher in the cotransformed leaves with effector and two individual reporters than the control leaves (Figure 5E–G). All these results indicated that SbNAC9 can directly activate SbC5YQ75 and SbNCED3.
2.7. Functions of SbC5YQ75 and SbNCED3 under Drought Stress in Sorghum
To further explore the functions of the two SbNAC9 targets SbC5YQ75 and SbNCED3 under drought stress, we first silenced SbC5YQ75 by VIGS in the sorghum seedlings. Compared with sorghum seedlings inoculated with empty virus (named BSMV:00), the sorghum seedlings inoculated with BSMV:SbC5YQ75 (named BSMV:SbC5YQ75) showed weakened drought-stress tolerance with more wilted leaves (Figure 6A). The DAB and NBT staining indicated that BSMV:SbC5YQ75 accumulated more H2O2 and O2− (Figure 6B,C). Consistently, the activities of antioxidative enzymes including POD and SOD in BSMV:SbC5YQ75 were more weakened than those in BSMV:00 (Figure 6D,E). Moreover, MDA content in BSMV:SbC5YQ75 was obviously higher than those in BSMV:00 under drought-stress treatment (Figure 6F). These results indicated that silenced SbC5YQ75 weakened the plants’ drought-stress tolerance by decreasing ROS scavenging ability in sorghum.
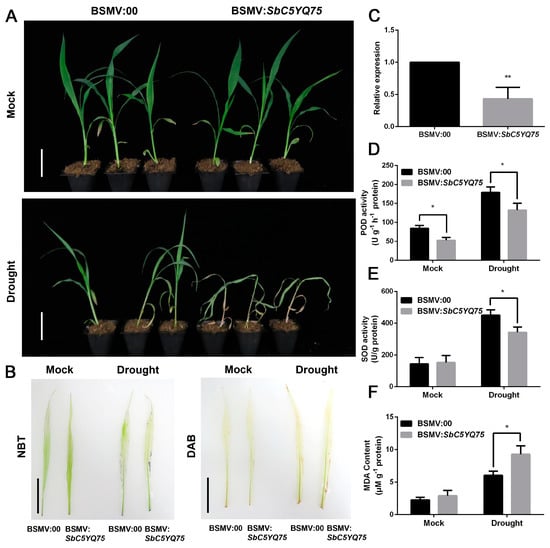
Figure 6.
The function of SbC5YQ75 in response to drought stress in sorghum. (A) Phenotype of sorghum seedlings inoculated with BSMV:00 and BSMV:SbC5YQ75 under mock and drought-stress treatments. Bars indicate 6 cm. (B) DAB and NBT staining of leaves of sorghum seedlings inoculated with BSMV:00 and BSMV:SbC5YQ75 treated with drought stress for 5 days. Bars indicate 5 cm. (C) Relative transcript level of SbC5YQ75 in sorghum seedlings silenced by VIGS. SbEIF4A was used as the internal reference. Error bars indicate SD of three independent experiments. ** p < 0.01 by Student’s t-test. (D–F) POD and (D) SOD (E) activities and MDA content (F) in sorghum seedlings inoculated with BSMV:00 and BSMV:SbC5YQ75 under mock or drought-stress treatments. Error bars indicate SD of three independent experiments. * p < 0.05 by Student’s t-test.
We also silenced SbNCED3 by VIGS in sorghum seedlings to explore its function in response to drought stress. The sorghum seedlings inoculated with BSMV:SbNCED3 (named BSMV:SbNCED3) exhibited weakened drought-stress tolerance with more wilted leaves than those inoculated with empty virus (named BSMV:00) (Figure S13A). In Arabidopsis, researchers found that AtNCED3 is the key enzyme to promote ABA biosynthesis [16]. To further investigate the role of SbNAC9 and SbNCED3 in ABA signaling, we compared relative ABA content in sorghum seedlings with silenced SbNAC9 or silenced SbNCED3 under dehydration treatment. The results showed that the ratio of ABA content of 12 h over that of 0 h after dehydration treatment in sorghum seedlings with silenced SbNAC9 or silenced SbNCED3 were both significantly lower than that in BSMV:00 (Figure S13B,D). These suggested that SbNAC9 and SbNCED3 can function in ABA signaling, thereby affecting drought-stress resistance of sorghum.
3. Discussion
The functions of sorghum NAC TFs which can respond to abiotic stresses remain largely unknown. In this study, we cloned SbNAC9, which has 63.22% similarity to OsNAC5 that can be induced by abiotic stress (Figure S2) [28]. Our results show that SbNAC9 can be induced by abiotic stresses including low temperature, salt, PEG-simulated, and mannitol-simulated dehydration stress, as well as ABA treatments. Overexpression of SbNAC9 enhanced drought tolerance of both Arabidopsis and sorghum, while silencing of SbNAC9 weakened drought tolerance of sorghum. In sorghum, SbNAC9 can directly activate the putative peroxidase gene SbC5YQ75 and the putative ABA biosynthesis gene SbNCED3 to enhance plant drought tolerance.
When sensing drought-stress signals, plants evolve various strategies to adapt to water-deficient conditions. Among these strategies, morphological adaptation such as “stay green” of leaves, strengthening of root architecture, and stomatal closure are considered as typical responses of plants [3]. In this study, sorghum transgenic lines with SbNAC9 overexpression exhibited higher chlorophyll content, higher photosynthetic rate, stronger root architecture, and lower water loss rate than wild-type plants (Figure 3A–D and Figure S4), while seedlings with silenced SbNAC9 showed opposite traits (Figure 4A–D). These findings suggest that SbNAC9 plays an essential role by enhancing plant physical adaptation to drought stress.
Drought stress also induces ROS accumulation. The scavenging ability of ROS is vital for plants to respond to drought stress. Our previous study showed that SbNAC2 overexpression in Arabidopsis has higher antioxidative enzyme activities under multiple abiotic stresses than wild-type plants [26]. In this study, we showed that SbNAC9 is both capable and required for ROS scavenging in sorghum ((Figure 3E–G and (Figure 4E–H). To investigate the molecular function of SbNAC9, we analyzed two putative peroxidases, SbC5YQ75 and SbC5XIY1. Given SbNAC9 as a transcription activator (Figure 2A,B), we tested the transcript level of these two genes in sorghum seedlings with silenced SbNAC9, and only SbC5YQ75 was downregulated ((Figure 5B and Figure S11A). Our EMSA assay and luciferase assays confirmed the direct activation of SbC5YQ75 by SbNAC9 (Figure 5C–F). Moreover, sorghum seedlings with silenced SbC5YQ75 showed weakened drought-stress tolerance with decreased antioxidative enzyme activities under drought stress (Figure 6). Taken together, SbNAC9 may enhance the ROS scavenging ability of sorghum by directly activating SbC5YQ75 expression.
ABA level quickly elevates when plants sense drought-stress signals [36]. Previous studies revealed that many TFs are involved in ABA signaling pathway [16]. In Arabidopsis, the NAC transcription factor ATAF1 can bind to the TTGCGTA motif in the promoter of NCED3 to regulate ABA biosynthesis [37]. In rice, WRKY5 functions as a negative regulator by binding to the abiotic stress-related gene OsMYB2, which can enhance ABA-induced drought tolerance [38]. The mutants oswrky5–2 and oswrky5–3 showed enhanced drought-stress tolerance and higher sensitivity to ABA. In this study, SbNAC9 was rapidly upregulated under ABA treatment (Figure S10B). Additionally, the transcript level of NCED3 involved in ABA biosynthesis was upregulated under drought stress in Arabidopsis transgenic lines overexpressing SbNAC9 (Figure S6). In sorghum, we showed that SbNAC9 can directly induce SbNCED3 (Figure 5C–F), which may be potentially involved in ABA biosynthesis. Moreover, after dehydration treatment, we found that the relative ABA content in sorghum seedlings with silenced SbNCED3 was much lower than that in plants inoculated with empty virus. Sorghum seedlings with silenced SbNAC9 showed similar results (Figure S13B–D). Thus, SbNAC9 can be induced by ABA, and in turn, SbNAC9 may possibly promote ABA biosynthesis by directly activating the putative ABA biosynthesis gene SbNCED3, thereby forming a positive feedback loop for drought-stress response in sorghum.
Recent reports provided details on the molecular and structural study about NAC TFs. For example, transgenic tobacco overexpressing the NAC domain of GhNAC4 exhibits higher tolerance to stress and higher sensitivity to ABA compared with lines overexpressing GhNAC4 transactivation domain [39]. In maize, ZmMPK5 can phosphate ZmNAC49 on the Thr-26 by increasing the binding capability of ZmNAC49 to ZmSOD3 and, thus, enhances the oxidative stress tolerance of maize [40]. Natural antisense transcript also participates in drought stress as well. Cis-NATZmNAC48 represses the expression of ZmNAC48 and affects stomatal movement to alter drought-stress tolerance of maize [41]. Whether SbNAC9 can be similarly regulated like these NAC TFs remains to be uncovered.
In summary, we proposed a model of how SbNAC9 enhances drought-stress tolerance in sorghum (Figure 7). Drought stress can induce SbNAC9 expression. Thereby, SbNAC9 directly activates the expression of SbC5YQ75 and SbNCED3 by binding to their promoters. In addition, SbNCED3 may potentially promote biosynthesis of ABA, which can induce SbNAC9 expression to form a feedback regulation. Additionally, SbNAC9 overexpression enhances drought-stress tolerance of sorghum through strengthening root architecture and increasing ROS scavenging ability.
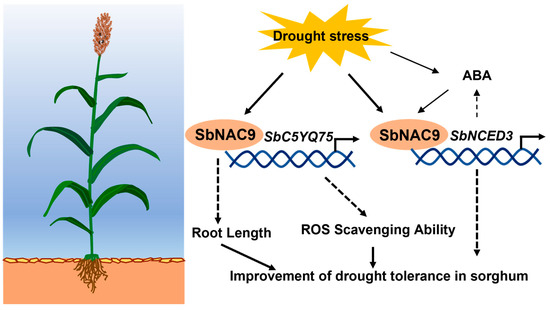
Figure 7.
Model for SbNAC9 function in drought-stress tolerance of sorghum. Drought stress induces SbNAC9 expression. SbNAC9 directly activates the expression of SbC5YQ75 and SbNCED3 by binding to their promoters. In addition, SbNCED3 may potentially promote biosynthesis of ABA, which can induce SbNAC9 expression to form a feedback regulation. Additionally, SbNAC9 overexpression enhances drought-stress tolerance of sorghum through altering root architecture and increasing ROS scavenging ability.
4. Materials and Methods
4.1. Plant Materials and Growth Condition
Tx430, a nontannin genotype, was used for sorghum transformation. The transgenic lines were grown at 28 °C under 12 h light/12 h dark photoperiod at 60% humidity for seed harvest. The sorghum genotype BTx623 was used for the other assays. The sorghum seedlings used in the study were grown at 25 °C under 16 h light/ 8 h dark photoperiod at 60% humidity. Transformation of Arabidopsis was performed on Columbia wild-type (Col-0). Tobacco (Nicotiana tabacum) was used for the subcellular localization assay and luciferase assay. Arabidopsis and tobacco used in the study were grown at 24 °C under 16 h light/ 8 h dark photoperiod at 60% humidity.
4.2. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Assay
Total RNA extraction, cDNA synthesis, and qRT-PCR were carried out as previously described [42]. Actin2 and SbEIF4A were used as internal controls in Arabidopsis and sorghum, respectively. The primers used in qRT-PCR are listed in Table S3.
4.3. Phytohormone and Abiotic Stress Treatments
The roots of sorghum seedlings at four-leaf stage were subjected to 150 μM GA, 150 μM ABA, 200 mM NaCl, 200 mM mannitol, 20% PEG6000 treatments or kept at 4 °C, and the fourth leaves were collected for qRT-PCR. For drought-stress treatments, sorghum seedlings overexpressing SbNAC9 at six-leaf stage were treated with water deprivation for 21 days. An amount of 15 sorghum plants each of WT and transgenic lines were used for control and drought-stress treatments. The sorghum seedlings with VIGS-mediated silencing of SbNAC9, SbC5YQ75, and SbNCED3 were treated with water deprivation for 7 days on the sixth day after inoculation with virus. Then, 12 sorghum seedlings each with BSMV:00 or silenced SbNAC9, SbC5YQ75, or SbNCED3 were used for control and drought-stress treatments. Three biological replicates were performed in drought-stress treatments. Three-week-old Arabidopsis transgenic plants heterologously overexpressing SbNAC9 and WT plants were treated with water deprivation for 10 days.
4.4. In Situ Hybridization Assay
The sorghum seeds used for the in situ hybridization assay were sterilized by 30% sodium hypochlorite for 10 min, repeated three times, and then the seeds were washed by sterilized water five times. The sterilized seeds were put on the filter paper to absorb water completely. The seeds were grown on the MS medium for three days. The root tips were collected and soaked in 20% PEG solution for 4 h and 6 h, and then embedded into paraffin for in situ hybridization or put into liquid nitrogen for qRT-PCR. The in situ hybridization assay was performed as previously described [42]. The primer sequences used for probes are listed in Table S3.
4.5. Transactivation Assay
The pGBKT7 (Clontech) constructs containing the full-length N-terminal and C-terminal coding sequences of SbNAC9 were transformed into yeast strain AH109. Empty pGBKT7 was used as the negative control. SD/-Trp medium was used for selecting the successful transformants. SD/-His medium was used to test the transactivation activity. The primers are listed in Table S3.
4.6. Subcellular Localization Assay
The coding sequence of SbNAC9 (without stop codon) was amplified and fused with GFP coding sequence within the vector pGreen to generate 35S:SbNAC9-GFP. Agrobacterium strain GV3101 with pSoup containing 35S:SbNAC9-GFP or 35S:GFP was transformed into three-week-old tobacco leaves. After two days of infiltration, the leaves were harvested and stained by 10 μg/mL DAPI for one hour and then GFP fluorescence was observed via Olympus (BX53) microscope. The primers are listed in Table S3.
4.7. Transformation of Arabidopsis and Sorghum
We used the pGreen vector harboring 35S:SbNAC9-GFP for plant transformation. Agrobacterium strain GV3101 with pSoup was used. The method of Arabidopsis transformation was carried out as previously described [43]. Sorghum transformation was performed according to a previous protocol [44].
4.8. Measurement of Chlorophyll Content
Chlorophyll contents in the fifth leaves of WT and transgenic lines overexpressing SbNAC9, as well as in the third leaves of sorghum seedlings with silenced SbNAC9, were measured by a chlorophyll meter SPAD-502 PLUS (Konika Minolta, Tokyo, Japan).
4.9. Measurement of Chlorophyll Fv/Fm
We selected the fifth leaf of sorghum seedlings overexpressing SbNAC9 and WT, as well as the third leaf of sorghum seedlings with silenced SbNAC9 by VIGS, for the measurement of chlorophyll fluorescence Fv/Fm. The measurement was performed as previously described [45].
4.10. Measurement of Antioxidative Enzyme Activities and MDA Content
Rosette leaves from transgenic Arabidopsis, the fifth leaf from transgenic sorghum seedlings overexpressing SbNAC9, and the fourth leaf from sorghum seedlings with silenced SbNAC9 and SbC5YQ75 were used for the measurement of antioxidative enzyme activities and MDA content. The SOD, CAT, and POD activity measurement and MDA content measurement were performed as previously described [12].
4.11. Diaminobenzidine (DAB) and Nitroblue Tetrazolium (NBT) Staining Assays
Rosette leaves of transgenic Arabidopsis and leaves of sorghum seedlings were collected into 1 mg/mL DAB staining solution (prepared in 50 mM phosphate buffer pH3.8). The leaves were placed at 25 °C for 6 h and decolored by 90% ethanol in boiling water bath. The decolored leaves were placed in petri dish filled with water for taking photos. The NBT staining was conducted as previously described [40].
4.12. Virus-Induced Gene Silencing (VIGS) Assay
Barley stripe mosaic virus (BSMV)-based vectors α, β, and γ were used for VIGS. The target fragments of SbNAC9, SbNCED3, and SbC5YQ75 were predicted by SGN VIGS Tool (https://vigs.solgenomics.net/). The predicted target fragments were inserted into γ vector. The primers are listed in Table S3. The α, β, γ, γ-SbNAC9, γ-SbNCED3, and γ-SbC5YQ75 were transformed into EHA105. The VIGS assay was performed as previously described [26]. Sorghum seedlings used for VIGS assays were grown under normal conditions for 5 days after being inoculated with virus, and then treated with water deprivation for 7 days.
4.13. Water Loss Rate Assay
The sixth leaves of sorghum seedlings at six-leaf stage were used for measurement of water loss rate. The assays were performed as previously described [23]. Briefly, the detached leaves were placed in a growth chamber at 25 °C. Fresh weight of detached leaves was recorded every 15 min from when the leaves were collected from sorghum.
4.14. Electrophoretic Mobility Shift (EMSA) Assay
The coding sequence of SbNAC9 was amplified and introduced into the vector pMAL-c5G. The construct was transformed into Transetta competent cells (TransGen Biotech, Beijing, China) to acquire MBP-SbNAC9 fusion protein. The EMSA assays were conducted as previously described [42]. The primers used for this assay are listed in Table S3.
4.15. Luciferase Assay
The 2000-bp promoter fragments of SbC5YQ75 and SbNCED3 were cloned into pGreenII-0800-LUC (luciferase) which contains a separated 35S Renilla (REN) luciferase reporter gene for normalization. Then, 35S:SbNAC9 was used as an effector. Luciferase assays were carried out as described previously [46].
4.16. Measurement of ABA Content in Sorghum
Two-week-old sorghum seedlings were used to measure ABA content under dehydration treatment. Before treatment, roots of sorghum seedlings were soaked in water for 16 h. Then, sorghum seedlings were transferred from water to 50 mL empty tubes for 12 h dehydration treatment. To measure ABA content, 100 mg tissue from the fourth leaves of each group were collected into liquid nitrogen. The frozen ground leaves were then mixed with PBS buffer. After centrifugation, the supernatant was used for ABA measurement according to the instruction of ABA ELISA Kit (MBE21031; MALLBIO Biological Technology, Nanjing, China).
4.17. Statistical Analysis
Statistical analyses were performed by GraphPad Prism (GraphPad Software Inc., San Diego, CA, USA). Experimental data were analyzed by one-way ANOVA test with ** p < 0.01 and * p < 0.05 for Figure S4 and by Student’s t-test with * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001 for the other figures.
4.18. Gene Accession Number
SbNAC9 (SOBIC.005G064600.2) was obtained from Phytozome13 of JGI Genome Portal website (https://genome.jgi.doe.gov/portal/, accessed on 9 January 2018). All gene information in Table S2 as well as SbNCED3 (Sb01g013520) and SbNCED9 (Sb02g003230) were obtained from PlantGDB (http://www.plantgdb.org/, accessed on 2 February 2020). All gene information of Arabidopsis was obtained from TAIR (https://www.arabidopsis.org/, accessed on 3 October 2020) according to the accession numbers as follows: DREB1A (AT4G25480), DREB2A (AT5G05410), KIN1 (AT5G15960), and NCED3 (AT3G14440).
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24032401/s1.
Author Contributions
Conceptualization, B.S.; Methodology, X.J., Y.Z., J.W., Y.C. and Z.Y.; Investigation, X.J., Y.Z., J.W. and Y.C.; Writing—original draft preparation, X.J. and B.S.; Writing—review and editing, W.C., Y.Y., G.L. and B.S.; Supervision, B.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fundamental Research Funds for the Central Universities (020814380180).
Data Availability Statement
The data presented in this study are available in the article or in the supplementary material.
Acknowledgments
We thank Junyi Chen (Fudan University) for providing pGreenII-0800-LUC and the pMal-c5G vector. Additionally, we are also grateful to Xiaofeng Gu and Lifang Niu (Biotechnology Research Institute, CAAS) for providing sorghum genotype BTx623 and to Huiyong Yang for providing sorghum genotype Tx430. Moreover, we thank Aizhong Cao (State Key Laboratory of Crop Genetics and Germplasm Enhancement, Cytogenetics Institute, Nanjing Agricultural University) for providing the vectors related to VIGS. We also thank Lingyan Dai (Heilongjiang Bayi Agricultural University) for help in sorghum transformation. We thank Lijuan Zhao (Nanjing University) for providing chlorophyll meter SPAD-502 PLUS for the measurement of chlorophyll content.
Conflicts of Interest
The authors declare no conflict of interests.
Abbreviations
| ABA | Abscisic acid |
| BSMV | Barley stripe mosaic virus |
| CAT | Catalase |
| DAB | Diaminobenzidine |
| Fv/Fm | Fluorescence/maximal fluorescence |
| GA | Gibberellin |
| GFP | Green fluorescent protein |
| LUC | Luciferase |
| MDA | Malondialdehyde |
| NBT | Nitroblue tetrazolium |
| PEG | Polyethylene glycol |
| POD | Peroxidase |
| REN | Renilla |
| ROS | Reactive oxygen species |
| SOD | Superoxide dismutase |
| TFs | Transcription factors |
| VIGS | Virus-induced gene silencing |
References
- Sehgal, A.; Sita, K.; Siddique, K.H.M.; Kumar, R.; Bhogireddy, S.; Varshney, R.K.; HanumanthaRao, B.; Nair, R.M.; Prasad, P.V.V.; Nayyar, H. Drought or/and Heat-Stress Effects on Seed Filling in Food Crops: Impacts on Functional Biochemistry, Seed Yields, and Nutritional Quality. Front. Plant Sci. 2018, 9, 1705. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, M.S.; Oliveira, C.; Steiner, F.; Zuffo, A.M.; Menis, F.T. Drought Stresses on Seed Germination and Early Growth of Maize and Sorghum. J. Agric. Sci. 2019, 11, 310–318. [Google Scholar] [CrossRef]
- Abreha, K.B.; Enyew, M.; Carlsson, A.S.; Vetukuri, R.R.; Feyissa, T.; Motlhaodi, T.; Ng’uni, D.; Geleta, M. Sorghum in dryland: Morphological, physiological, and molecular responses of sorghum under drought stress. Planta 2021, 255, 20. [Google Scholar] [CrossRef] [PubMed]
- Sanjari, S.; Shirzadian-Khorramabad, R.; Shobbar, Z.S.; Shahbazi, M. Systematic analysis of NAC transcription factors’ gene family and identification of post-flowering drought stress responsive members in sorghum. Plant Cell Rep. 2019, 38, 361–376. [Google Scholar] [CrossRef] [PubMed]
- John, J.; Burke, Y.E.; Hayes, C.; Xin, Z.; Burow, G. Registration of Four Postlowering Drought-Tolerant Grain Sorghum Lines with Early-Season Cold Tolerance. J. Plant Regist. 2018, 12, 386–390. [Google Scholar]
- Qi, J.; Song, C.P.; Wang, B.; Zhou, J.; Kangasjarvi, J.; Zhu, J.K.; Gong, Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Petrov, V.D.; Van Breusegem, F. Hydrogen peroxide-a central hub for information flow in plant cells. AoB Plants 2012, 2012, pls014. [Google Scholar] [CrossRef]
- Asada, K. THE WATER-WATER CYCLE IN CHLOROPLASTS:Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Qiu, Y.; Hu, Y.; Yu, D. Heterologous Expression of AtWRKY57 Confers Drought Tolerance in Oryza sativa. Front. Plant Sci. 2016, 7, 145. [Google Scholar] [CrossRef] [PubMed]
- Sheoran, S.; Thakur, V.; Narwal, S.; Turan, R.; Mamrutha, H.M.; Singh, V.; Tiwari, V.; Sharma, I. Differential Activity and Expression Profile of Antioxidant Enzymes and Physiological Changes in Wheat (Triticum aestivum L.) Under Drought. Appl. Biochem. Biotechnol. 2015, 177, 1282–1298. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Ju, Y.L.; Yue, X.F.; Min, Z.; Wang, X.H.; Fang, Y.L.; Zhang, J.X. VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 146, 98–111. [Google Scholar] [CrossRef]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef]
- Varoquaux, N.; Cole, B.; Gao, C.; Pierroz, G.; Baker, C.R.; Patel, D.; Madera, M.; Jeffers, T.; Hollingsworth, J.; Sievert, J.; et al. Transcriptomic analysis of field-droughted sorghum from seedling to maturity reveals biotic and metabolic responses. Proc. Natl. Acad. Sci. USA 2019, 116, 27124–27132. [Google Scholar] [CrossRef]
- Jian, W.; Zheng, Y.; Yu, T.; Cao, H.; Chen, Y.; Cui, Q.; Xu, C.; Li, Z. SlNAC6, A NAC transcription factor, is involved in drought stress response and reproductive process in tomato. J. Plant Physiol. 2021, 264, 153483. [Google Scholar] [CrossRef]
- Hu, P.; Zhang, K.; Yang, C. BpNAC012 Positively Regulates Abiotic Stress Responses and Secondary Wall Biosynthesis. Plant Physiol. 2019, 179, 700–717. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Sun, X.; Bian, X.; Wei, T.; Han, T.; Yan, J.; Zhang, A. The transcription factor ZmNAC49 reduces stomatal density and improves drought tolerance in maize. J. Exp. Bot. 2021, 72, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, D.-F.; Shi, Y.-S.; Song, Y.-C.; Wang, T.-Y.; Li, Y. Expression of SbSNAC1, a NAC transcription factor from sorghum, confers drought tolerance to transgenic Arabidopsis. Plant Cell. Tissue Organ Cult. 2013, 115, 443–455. [Google Scholar] [CrossRef]
- Jin, X.; Long, Y.; Xiong, S.; Yang, Z.; Chen, W.; Hawar, A.; Chi, X.; Chen, Y.; Luo, H.; Qi, J.; et al. SbNAC2 enhances abiotic stress tolerance by upregulating ROS scavenging activities and inducing stress-response genes in sorghum. Environ. Exp. Bot. 2021, 192, 104664. [Google Scholar] [CrossRef]
- Ngara, R.; Ramulifho, E.; Movahedi, M.; Shargie, N.G.; Brown, A.P.; Chivasa, S. Identifying differentially expressed proteins in sorghum cell cultures exposed to osmotic stress. Sci. Rep. 2018, 8, 8671. [Google Scholar] [CrossRef]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genom. 2010, 284, 173–183. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Chen, D.; Wang, S.; Cao, B.; Cao, D.; Leng, G.; Li, H.; Yin, L.; Shan, L.; Deng, X. Genotypic Variation in Growth and Physiological Response to Drought Stress and Re-Watering Reveals the Critical Role of Recovery in Drought Adaptation in Maize Seedlings. Front Plant Sci. 2015, 6, 1241. [Google Scholar] [CrossRef]
- Kurkela, M.F.S. Cloning and characterization of a cold-and ABA-inducible Arabidopsis gene. Plant Mol. Biol. 1990, 15, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Diao, P.; Chen, C.; Zhang, Y.; Meng, Q.; Lv, W.; Ma, N. The role of NAC transcription factor in plant cold response. Plant Signal. Behav. 2020, 15, 1785668. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 2005, 169, 785–797. [Google Scholar] [CrossRef]
- Zeevaart, J. Changes in the Levels of Abscisic Acid and Its Metabolites in Excised Leaf Blades of Xanthium strumarium during and after Water Stress. Plant Physiol. 1980, 66, 672–678. [Google Scholar] [CrossRef]
- Jensen, M.K.; Lindemose, S.; de Masi, F.; Reimer, J.J.; Nielsen, M.; Perera, V.; Workman, C.T.; Turck, F.; Grant, M.R.; Mundy, J.; et al. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 2013, 3, 321–327. [Google Scholar] [CrossRef]
- Lim, C.; Kang, K.; Shim, Y.; Yoo, S.C.; Paek, N.C. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 2022, 188, 1900–1916. [Google Scholar] [CrossRef]
- Trishla, V.S.; Kirti, P.B. Structure-function relationship of Gossypium hirsutum NAC transcription factor, GhNAC4 with regard to ABA and abiotic stress responses. Plant Sci. 2021, 302, 110718. [Google Scholar] [CrossRef]
- Xiang, Y.; Bian, X.; Wei, T.; Yan, J.; Sun, X.; Han, T.; Dong, B.; Zhang, G.; Li, J.; Zhang, A. ZmMPK5 phosphorylates ZmNAC49 to enhance oxidative stress tolerance in maize. New Phytol. 2021, 232, 2400–2417. [Google Scholar] [CrossRef]
- Mao, Y.; Xu, J.; Wang, Q.; Li, G.; Tang, X.; Liu, T.; Feng, X.; Wu, F.; Li, M.; Xie, W.; et al. A natural antisense transcript acts as a negative regulator for the maize drought stress response gene ZmNAC48. J. Exp. Bot. 2021, 72, 2790–2806. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhou, Y.; Cai, J.; Shang, E.; Yamaguchi, N.; Xiao, J.; Looi, L.S.; Wee, W.Y.; Gao, X.; Wagner, D.; et al. Integration of Transcriptional Repression and Polycomb-Mediated Silencing of WUSCHEL in Floral Meristems. Plant Cell 2019, 31, 1488–1505. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Wu, E.; Lenderts, B.; Glassman, K.; Berezowska-Kaniewska, M.; Christensen, H.; Asmus, T.; Zhen, S.; Chu, U.; Cho, M.J.; Zhao, Z.Y. Optimized Agrobacterium-mediated sorghum transformation protocol and molecular data of transgenic sorghum plants. Vitr. Cell. Dev. Biol. Plant 2014, 50, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Hantzis, L.J.; Kroh, G.E.; Jahn, C.E.; Cantrell, M.; Peers, G.; Pilon, M.; Ravet, K. A Program for Iron Economy during Deficiency Targets Specific Fe Proteins. Plant Physiol. 2018, 176, 596–610. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chi, X.; Guo, F.; Jin, X.; Luo, H.; Hawar, A.; Chen, Y.; Feng, K.; Wang, B.; Qi, J.; et al. SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 2020, 246, 153142. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).