Exploring the Metabolic Differences between Cisplatin- and UV Light-Induced Apoptotic Bodies in HK-2 Cells by an Untargeted Metabolomics Approach
Abstract
1. Introduction
2. Results and Discussion
2.1. Non-Targeted Metabolomics Analysis of ABs Induced by Cisplatin and UV Light Exposure
2.2. Selection of Variables Based on Variable Importance
2.3. Identification of the Selected Molecular Features
2.4. Biological Interpretation
3. Materials and Methods
3.1. Reagents and Solvents
3.2. Cell Culture
3.3. Isolation of ABs from Apoptotic HK-2 Cells
3.4. Optimized Sample Preparation Protocol
3.5. Liquid Chromatography-Mass spectrometry Metabolomics Analysis
3.6. Data Treatment and Analysis
3.7. Identification of Metabolites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Battistelli, M.; Falciri, E. Apoptotic bodies: Particular extracellular vesicles involved in intercellular communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Deng, J.; Zhou, H.; Tan, W.; Lin, L.; Yang, J. Programmed cell death in sepsis associated acute kidney injury. Front. Med. 2022, 9, 883028. [Google Scholar] [CrossRef] [PubMed]
- Wickman, G.; Julian, L.; Olson, M.F. How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ. 2012, 19, 735–742. [Google Scholar] [CrossRef]
- Xu, X.; Lai, Y.; Hua, Z.C. Apoptosis and apoptotic body: Disease message and therapeutic target potentials. Biosci. Rep. 2019, 39, BSR20180992. [Google Scholar] [CrossRef]
- Garcia-Pastor, C.; Blazquez-Serra, R.; Bosch, R.J.; Lucio Cazana, F.J. Fernandez-Martinez, A.B. Apoptosis and cell proliferation in proximal tubular cells exposed to apoptotic bodies. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2504–2515. [Google Scholar] [CrossRef] [PubMed]
- Giera, M.; Yanes, O.; Siuzdak, G. Metabolite discovery: Biochemistry’s scientific. Cell Metab. 2022, 34, 21–34. [Google Scholar] [CrossRef]
- Yan, M.; Xu, G. Current and future perspectives of functional metabolomics indisease studies—A review. Anal. Chim. Acta 2018, 1037, 41–54. [Google Scholar] [CrossRef]
- Dudzik, D.; Macioszeka, S.; Struck-Lewicka, W.; Kordalewska, M.; Buszewska-Forajta, M.; Waszczuk-Jankowska, M.; Wawrzyniak, R.; Artymowicz, M.; Raczak-Gutknecht, J.; Siluk, D.; et al. Perspectives and challenges in extracellular vesicles untargeted metabolomics analysis. TrAC Trends Anal. Chem. 2021, 143, 116382. [Google Scholar] [CrossRef]
- Bernardo-Bermejo, S.; Sánchez-López, E.; Castro-Puyana, M.; Benito, S.; Lucio-Cazaña, F.J.; Marina, M.L. An untargeted metabolomic strategy based on liquid chromatography-mass spectrometry to study high glucose-induced changes in HK-2 cells. J. Chromatogr. A 2019, 1596, 124–133. [Google Scholar] [CrossRef]
- Bernardo-Bermejo, S.; Sánchez-López, E.; Tan, L.; Benito-Martínez, S.; Jiang, Z.; Castro-Puyana, M.; Lucio-Cazaña, F.J.; Marina, M.L. Exploratory metabolomic analysis based on reverse-phase liquid chromatography-mass spectrometry to study an in vitro model of hypoxia-induced metabolic alterations in HK-2 cells. Int. J. Mol. Sci. 2021, 22, 7399. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Phil. Trans. R. Soc. A 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016, 5, e10250. [Google Scholar] [CrossRef]
- Tadokoro, H.; Hirayama, A.; Kudo, R.; Hasebe, M.; Yoshioka, Y.; Matsuzaki, J.; Yamamoto, Y.; Sugimoto, M.; Soga, T.; Ochiya, T. Adenosine leakage from perforinburst extracellular vesicles inhibits perforin secretion by cytotoxic T-lymphocytes. PLoS ONE 2020, 15, e0231430. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Sumner, W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemicalanalysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Bergsmedh, A.; Szeles, A.; Henriksson, M.; Bratt, A.; Folkman, M.J.; Spetz, A.-L.-L.; Holmgren, L. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc. Natl. Acad. Sci. USA 2001, 98, 6407–6411. [Google Scholar] [CrossRef]
- Schiller, M.; Bekeredjian-Ding, I.; Heyder, P.; Blank, N.; Ho, A.D.; Lorenz, H.M. Autoantigens are translocated into small apoptotic bodies during early stages of apoptosis. Cell Death Differ. 2008, 15, 183–191. [Google Scholar] [CrossRef]
- Cocca, B.A.; Cline, A.M.; Radic, M.Z. Blebs and Apoptotic Bodies Are B Cell Autoantigens. J. Immunol. 2002, 169, 159–166. [Google Scholar] [CrossRef]
- Tran, H.B.; Ohlsson, M.; Beroukas, D.; Hiscock, J.; Bradley, J.; Buyon, J.P.; Gordon, T.P. Subcellular redistribution of La/SSB autoantigen during physiologic apoptosis in the fetal mouse heart and conduction system: A clue to the pathogenesis of congenital heart block. Arthritis Rheum. 2002, 46, 202–208. [Google Scholar] [CrossRef]
- Berda-Haddad, Y.; Robert, S.; Salers, P.; Zekraoui, L.; Farnarier, C.; Dinarello, C.A.; Dignat-George, F.; Kaplanski, G. Sterile inflammation of endothelial cell-derived apoptotic bodies is mediated by interleukin-1. Proc. Natl. Acad. Sci. USA 2011, 108, 20684–20689. [Google Scholar] [CrossRef] [PubMed]
- Atkin-Smith, G.K.; Duan, M.; Zanker, D.J.; Loh, L.; Nguyen, T.H.O.; Koutsakos, M.; Nguyen, T.; Jiang, X.; Carrera, J.; Phan, T.K.; et al. Monocyte apoptotic bodies are vehicles for influenza A virus propagation. Commun. Biol. 2020, 3, 223. [Google Scholar] [CrossRef] [PubMed]
- Stach, K.; Stach, W.; Augoff, K. Vitamin B6 in health and disease. Nutrients 2021, 13, 3229. [Google Scholar] [CrossRef] [PubMed]
- Ralto, K.M.; Rhee, E.P.; Parikh, S.M. NAD+ homeostasis in renal health and disease. Nat. Rev. Nephrol. 2020, 16, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Marsili, S.; Vitale, I.; Senovilla, L.; Michels, J.; Garcia, P.; Vacchelli, E.; Chatelut, E.; Castedo, M.; Kroemer, G. Vitamin B6 metabolism influences the intracellular accumulation of cisplatin. Cell Cycle. 2013, 12, 417–421. [Google Scholar] [CrossRef]
- Sato, K.; Taguchi, H.; Maeda, T.; Yoshikawa, K. Pyridoxine toxicity to cultured fibroblasts caused by near-ultraviolet light. J. Investig. Dermatol. 1993, 100, 266–270. [Google Scholar] [CrossRef]
- Ren, R.; Fang, Y.; Sherchan, P.; Lu, Q.; Lenahan, C.; Zhang, J.H.; Zhang, J.; Tang, J. Kynurenine/aryl hydrocarbon receptor modulates mitochondria-mediated oxidative stress and neuronal apoptosis in experimental intracerebral hemorrhage. Antioxid. Redox. Signal. 2022, 37, 1111–1129. [Google Scholar] [CrossRef]
- Mohib, K.; Guan, Q.; Diao, H.; Du, C.; Jevnikar, A.M. Proapoptotic activity of indoleamine 2,3-dioxygenase expressed in renal tubular epithelial cells. Am. J. Physiol. Renal. Physiol. 2007, 293, F801–F812. [Google Scholar] [CrossRef]
- Bonventre, J.V.; Yang, L. Cellular pathophysiology of ischemic acute kidney injury. J. Clin. Investig. 2011, 121, 4210–4221. [Google Scholar] [CrossRef]
- Pabla, N.; Dong, Z. Cisplatin nephrotoxicity: Mechanisms and renoprotective strategies. Kidney Int. 2008, 73, 994–1007. [Google Scholar] [CrossRef]
- Maiese, K. New insights for nicotinamide: Metabolic disease, autophagy, and mTOR. Front. Biosci. 2020, 25, 1925–1973. [Google Scholar] [CrossRef] [PubMed]
- Gopal, E.; Fei, Y.J.; Miyauchi, S.; Zhuang, L.; Prasad, P.D.; Ganapathy, V. Sodium-coupled and electrogenic transport of B-complex vitamin nicotinic acid by slc5a8, a member of the Na/glucose co-transporter gene family. Biochem. J. 2005, 388, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, K.A.; Martin, A.S.; Hirschey, M.D. Role of NAD. Nat. Rev. Nephrol. 2017, 13, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Thaysen, J.H.; Lassen, N.A.; Munck, O. Sodium transport and oxygen consumption in the mammalian kidney. Nature 1961, 190, 919–921. [Google Scholar] [CrossRef]
- Zahedi, K.; Barone, S.; Destefano-Shields, C.; Brooks, M.; Murray-Stewart, T.; Dunworth, M.; Li, W.; Doherty, J.R.; Hall, M.A.; Smith, R.D.; et al. Activation of endoplasmic reticulum stress response by enhanced polyamine catabolism is important in the mediation of cisplatin-induced acute kidney injury. PLoS ONE 2017, 12, e0184570. [Google Scholar] [CrossRef]
- Avila, M.A.; García-Trevijano, E.R.; Lu, S.C.; Corrales, F.J.; Mato, J.M. Methylthioadenosine. Int. J. Biochem. Cell Biol. 2004, 36, 2125–2130. [Google Scholar] [CrossRef] [PubMed]
- Tomasi, M.L.; Cossu, C.; Spissu, Y.; Floris, A.; Ryoo, M.; Iglesias-Ara, A.; Wang, Q.; Pandol, S.J.; Bhowmick, N.A.; Seki, E.; et al. S-adenosylmethionine and methylthioadenosine inhibit cancer metastasis by targeting microRNA 34a/b-methionine adenosyltransferase 2A/2B axis. Oncotarget 2017, 8, 78851–78869. [Google Scholar] [CrossRef]
- Li, T.W.; Zhang, Q.; Oh, P.; Xia, M.; Chen, H.; Bemanian, S.; Lastra, N.; Circ, M.; Moyer, M.P.; Mato, J.M.; et al. S-Adenosylmethionine and methylthioadenosine inhibit cellular FLICE inhibitory protein expression and induce apoptosis in colon cancer cells. Mol. Pharmacol. 2009, 76, 192–200. [Google Scholar] [CrossRef]
- Ansorena, E.; Garcia-Trevijano, E.R.; Martinez-Chantar, M.L.; Huang, Z.Z.; Chen, L.; Mato, J.M.; Iraburu, M.; Lu, S.C.; Avila, M.A. S-adenosylmethionine and methylthioadenosine are antiapoptotic in cultured rat hepatocytes but proapoptotic in human hepatoma cells. Hepatology 2002, 35, 274–280. [Google Scholar] [CrossRef]
- Makrides, V.; Camargo, S.M.R.; Verrey, F. Transport of amino acids in the kidney. Compr. Physiol. 2014, 4, 367–403. [Google Scholar]
- Gerrard, J.A. New aspects of an ageing chemistry-recent developments concerning the Maillard reaction. Austral. J. Chem. 2002, 55, 299–310. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, W.; Guo, M.; Tan, Q.; Zhou, E.; Deng, J.; Li, M.; Chen, J.; Yang, Z.; Jin, Y. Metabolomics of extracellular vesicles: A future promise of multiple clinical applications. Int. J. Nanomed. 2022, 17, 6113–6129. [Google Scholar] [CrossRef] [PubMed]

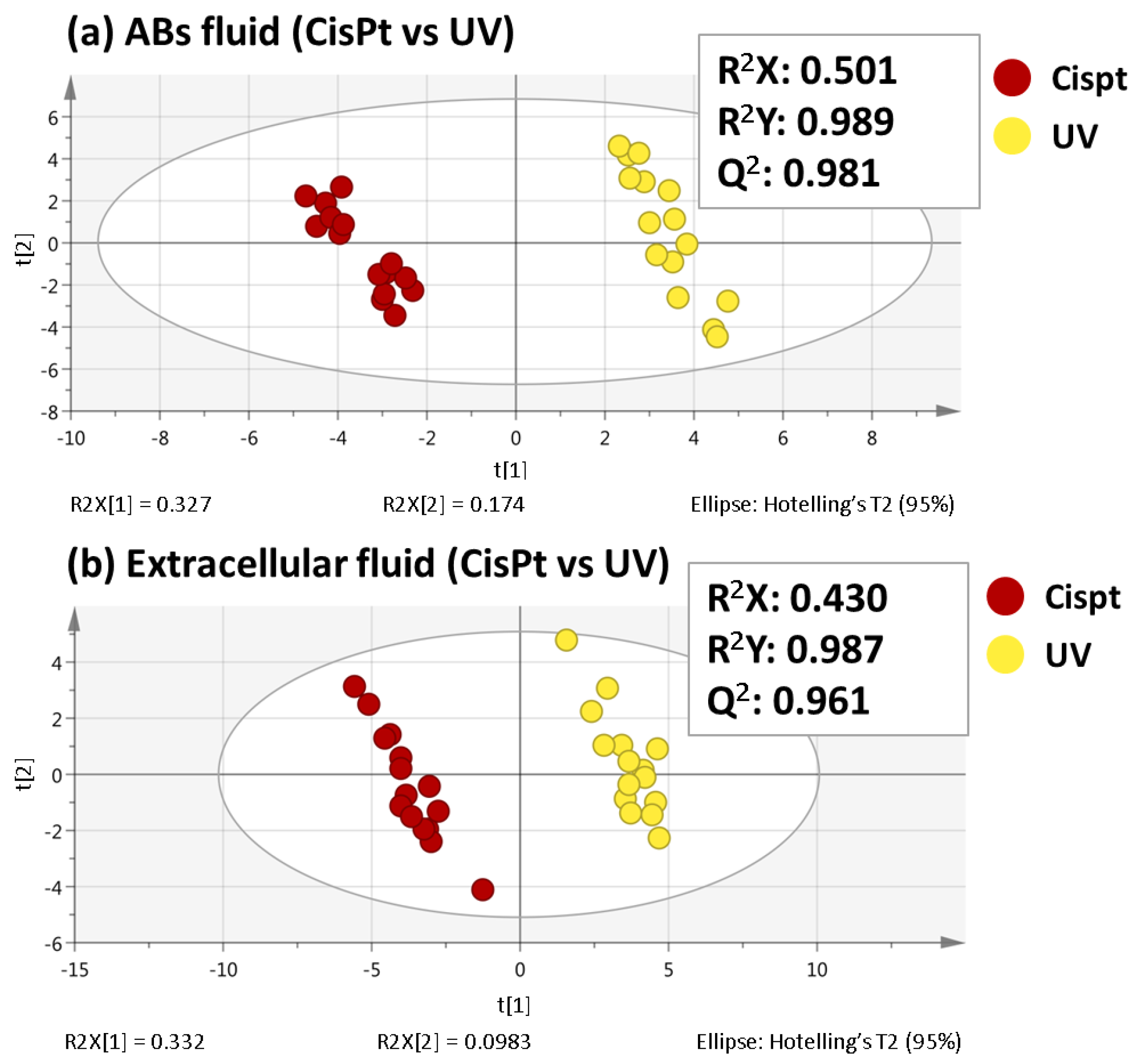
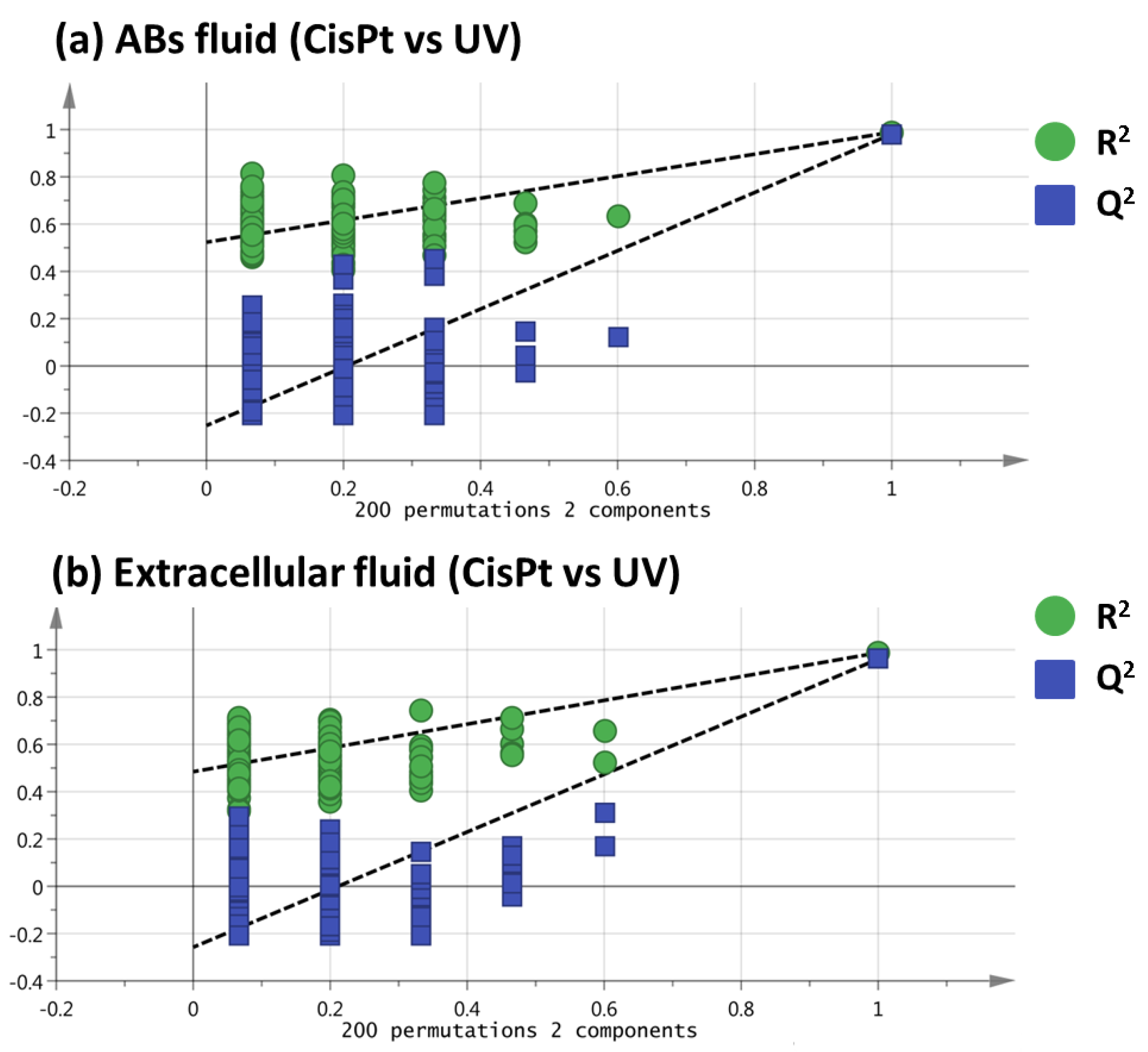
| # | RT (min) | Molecular Formula | Metabolite | Identification Level * | Monoisotopic Mass (Da) | Mass Error (ppm) | Main Fragments (MS/MS) | VIP (Trend CisPt vs. UV) ** |
|---|---|---|---|---|---|---|---|---|
| Intracellular fluid | ||||||||
| 1 | 0.5 | C10H26N4 | Spermine | 1 | 202.2156 | 1 | 112.1105, 84.0804, 129.1387 | 1.3 (↑) |
| 2 | 0.7 | C4H9N3O2 | Creatine | 1 | 131.0663 | 24 | 90.0546, 87.0584 | 1.2 (↓) |
| 3 | 0.8 | C5H11NO2 | Valine | 1 | 117.0767 | 20 | 72.0794, 55.0533 | 1.2 (↓) |
| 4 | 0.8 | C8H11NO3 | Pyridoxine | 1 | 169.0727 | 7 | 134.0592, 152.0687 | 2.2 (↑) |
| 5 | 0.8 | C15H21NO8 | N-(1-deoxy-1-fructosyl)tyrosine | 2 | 343.1250 | 5 | 308.1066, 280.1201, 326.1201 | 3.5 (↓) |
| 6 | 0.9 | C12H23NO7 | N-(1-deoxy-1-fructosyl)-(iso)leucine | 2 | 293.1457 | 6 | 230.1371, 258.1314, 132.0993 | 3.7 (↓) |
| 7 | 0.9 | C6H13NO2 | Leucine | 1 | 131.0940 | 5 | 86.0948 | 2.0 (↓) |
| 8 | 1.2 | C10H12N2O3 | Kynurenine | 1 | 208.0843 | 2 | 146.0589, 94.0619, 118.0618, 74.0236, 174.0538, 120.0408 | 1.3 (↓) |
| 9 | 1.3 | C15H21NO7 | N-(1-deoxy-1-fructosyl)phenylalanine | 2 | 327.1310 | 2 | 292.1183, 310.1264, 166.0868, 178.0849 | 3.4 (↓) |
| 10 | 2.5 | C11H15N5O3S | 5’-Methylthioadenosine | 1 | 297.0889 | 2 | 136.0613 | 3.0 (↑) |
| Extracellular fluid | ||||||||
| 11 | 0.8 | C6H6N2O | Nicotinamide | 1 | 122.0463 | 14 | 80.0486, 78.0337 | 1.5 (↓) |
| 12 | 0.8 | C5H11NO2 | Valine | 1 | 117.0764 | 22 | 72.0808, 58.0648, 59.0730 | 1.2 (↓) |
| 13 | 0.8 | C8H11NO3 | Pyridoxine | 1 | 169.0727 | 7 | 134.0587, 152.0695 | 2.0 (↓) |
| 14 | 0.8 | C15H21NO8 | N-(1-deoxy-1-fructosyl)tyrosine | 2 | 343.1225 | 12 | 308.1172, 280.0972, 326.1222 | 2.2 (↓) |
| 15 | 0.9 | C12H23NO7 | N-(1-deoxy-1-fructosyl)-(iso)leucine | 2 | 293.1463 | 4 | 230.1353, 258.1304, 276.1415 | 2.4 (↓) |
| 16 | 0.9 | C6H13NO2 | Leucine | 1 | 131.0937 | 7 | 86.0950 | 1.7 (↓) |
| 17 | 1.2 | C10H12N2O3 | Kynurenine | 1 | 208.0849 | 1 | 146.0600, 94.0651, 118.0642, 74.0233, 174.0503, 120.0429 | 1.3 (↓) |
| 18 | 1.3 | C15H21NO7 | N-(1-deoxy-1-fructosyl)phenylalanine | 2 | 327.1312 | 2 | 292.1177, 310.1231, 166.0857, 178.0860 | 1.8 (↓) |
| 19 | 2.5 | C11H15N5O3S | 5’-Methylthioadenosine | 1 | 297.0891 | 2 | 136.0620 | 2.3 (↑) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardo-Bermejo, S.; Sánchez-López, E.; Castro-Puyana, M.; Fernández-Martínez, A.B.; Lucio-Cazaña, F.J.; Marina, M.L. Exploring the Metabolic Differences between Cisplatin- and UV Light-Induced Apoptotic Bodies in HK-2 Cells by an Untargeted Metabolomics Approach. Int. J. Mol. Sci. 2023, 24, 7237. https://doi.org/10.3390/ijms24087237
Bernardo-Bermejo S, Sánchez-López E, Castro-Puyana M, Fernández-Martínez AB, Lucio-Cazaña FJ, Marina ML. Exploring the Metabolic Differences between Cisplatin- and UV Light-Induced Apoptotic Bodies in HK-2 Cells by an Untargeted Metabolomics Approach. International Journal of Molecular Sciences. 2023; 24(8):7237. https://doi.org/10.3390/ijms24087237
Chicago/Turabian StyleBernardo-Bermejo, Samuel, Elena Sánchez-López, María Castro-Puyana, Ana B. Fernández-Martínez, Francisco Javier Lucio-Cazaña, and María Luisa Marina. 2023. "Exploring the Metabolic Differences between Cisplatin- and UV Light-Induced Apoptotic Bodies in HK-2 Cells by an Untargeted Metabolomics Approach" International Journal of Molecular Sciences 24, no. 8: 7237. https://doi.org/10.3390/ijms24087237
APA StyleBernardo-Bermejo, S., Sánchez-López, E., Castro-Puyana, M., Fernández-Martínez, A. B., Lucio-Cazaña, F. J., & Marina, M. L. (2023). Exploring the Metabolic Differences between Cisplatin- and UV Light-Induced Apoptotic Bodies in HK-2 Cells by an Untargeted Metabolomics Approach. International Journal of Molecular Sciences, 24(8), 7237. https://doi.org/10.3390/ijms24087237







