A Practical Guide to All-Atom and Coarse-Grained Molecular Dynamics Simulations Using Amber and Gromacs: A Case Study of Disulfide-Bond Impact on the Intrinsically Disordered Amyloid Beta
Abstract
1. Introduction
2. Results
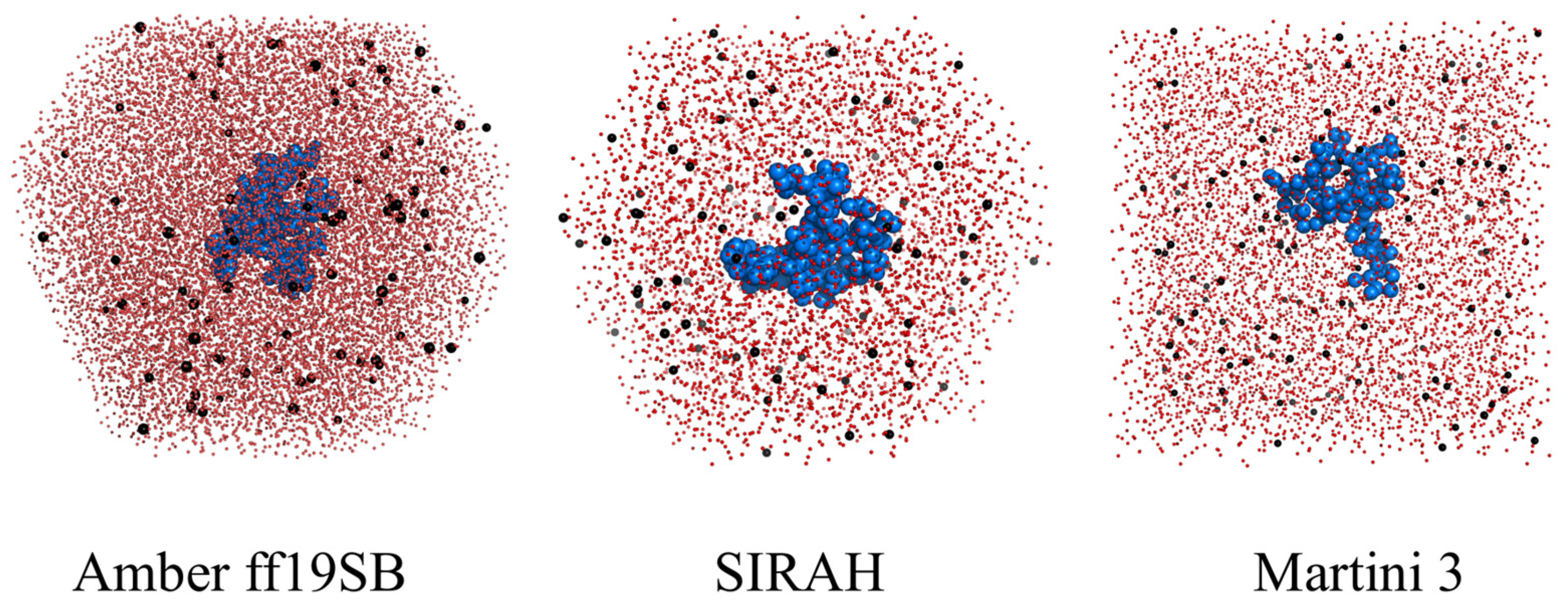
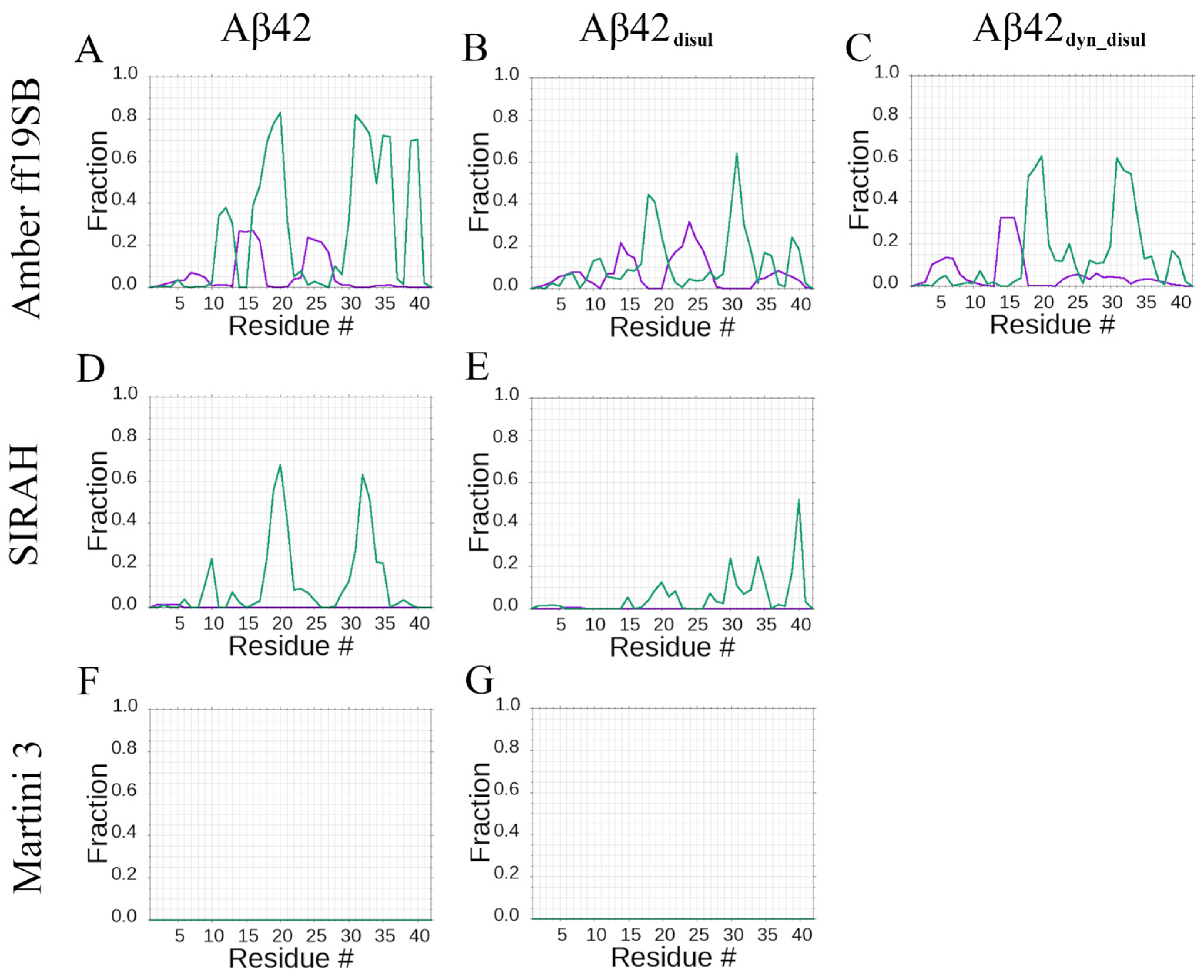
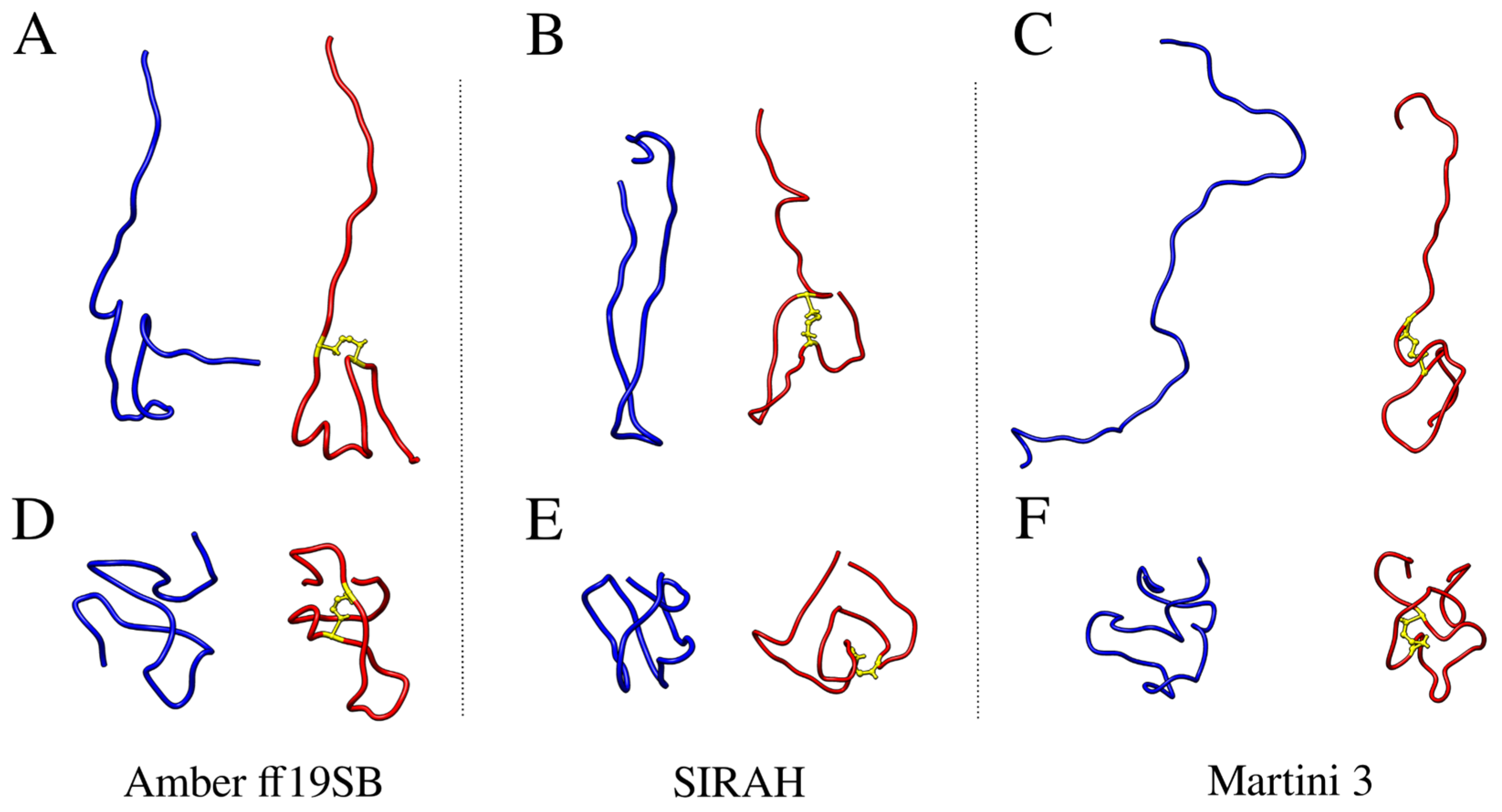
2.1. Structural Properties of the Monomeric Aβ42 Captured by the Force Fields
2.2. Impact of Solute-Solvent Interaction Scaling on the Monomeric Aβ42 in Martini 3 Force Field
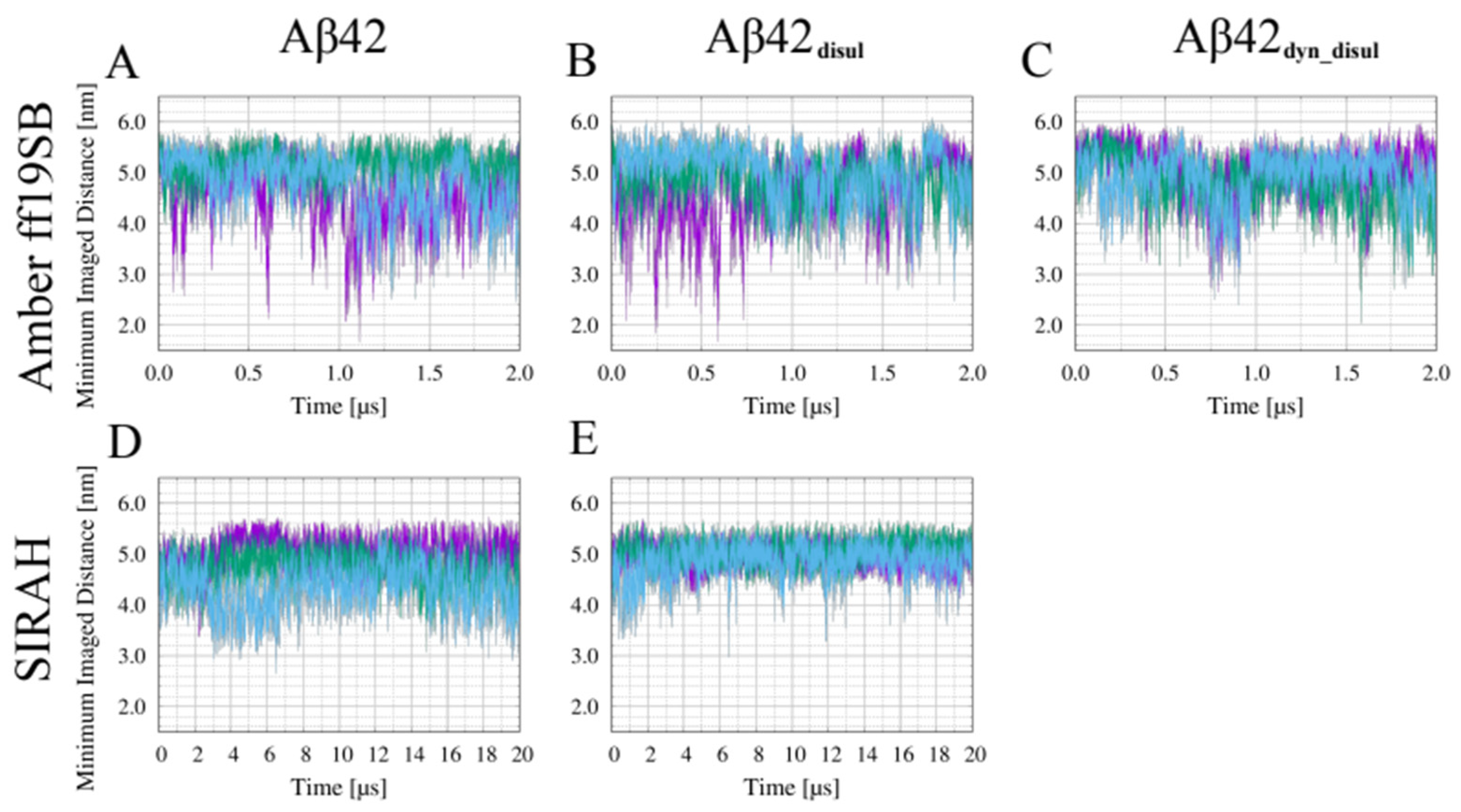
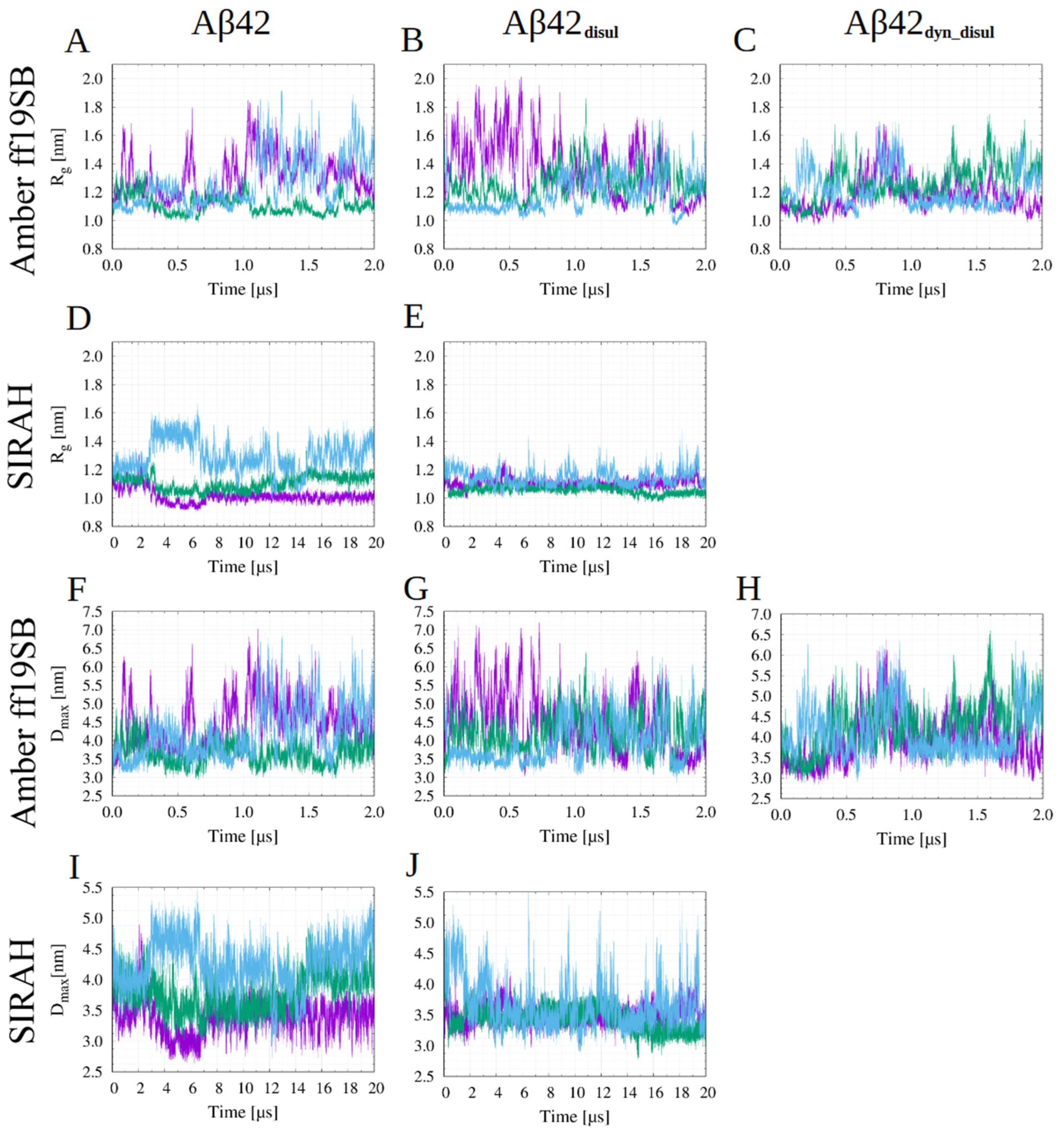
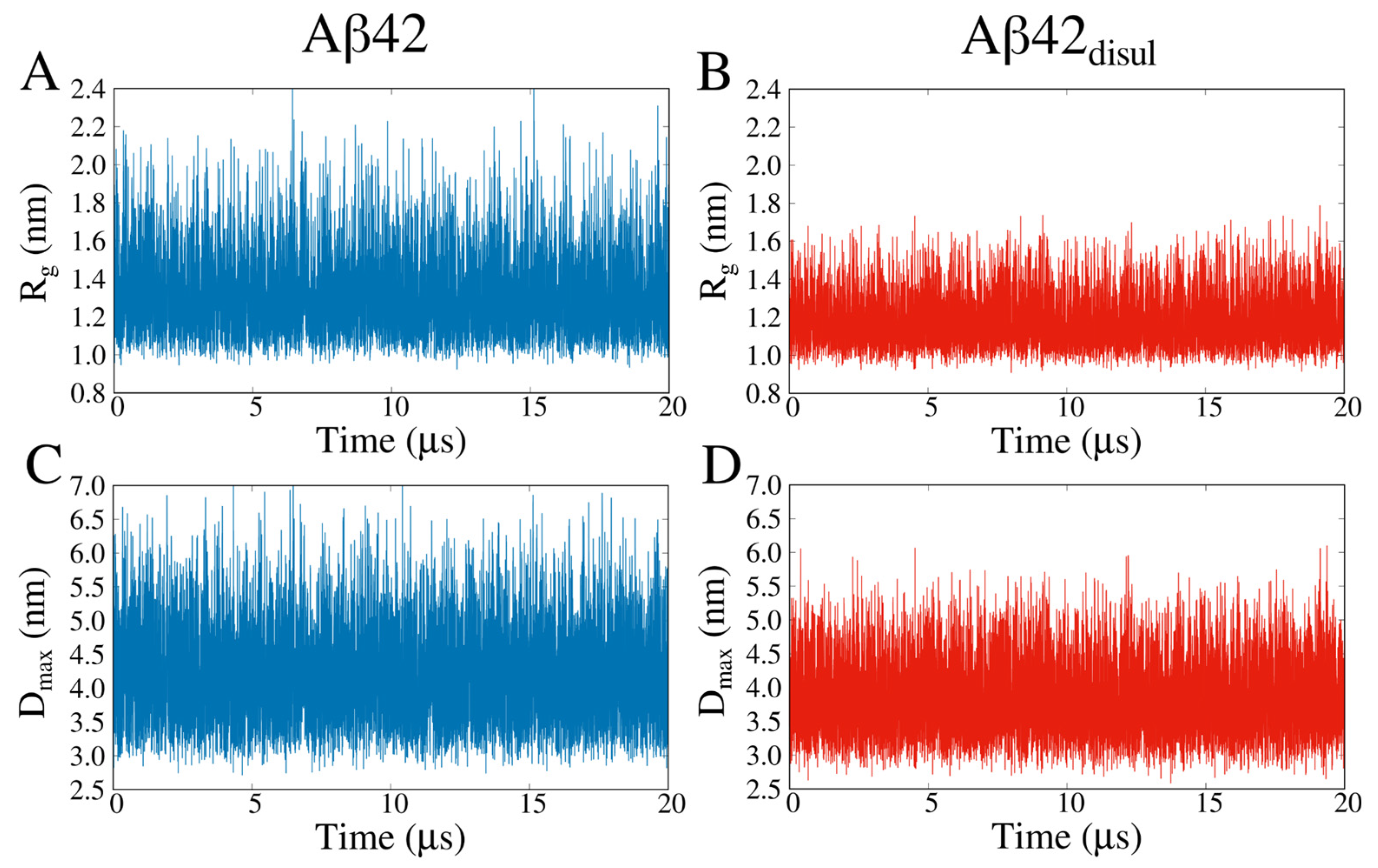
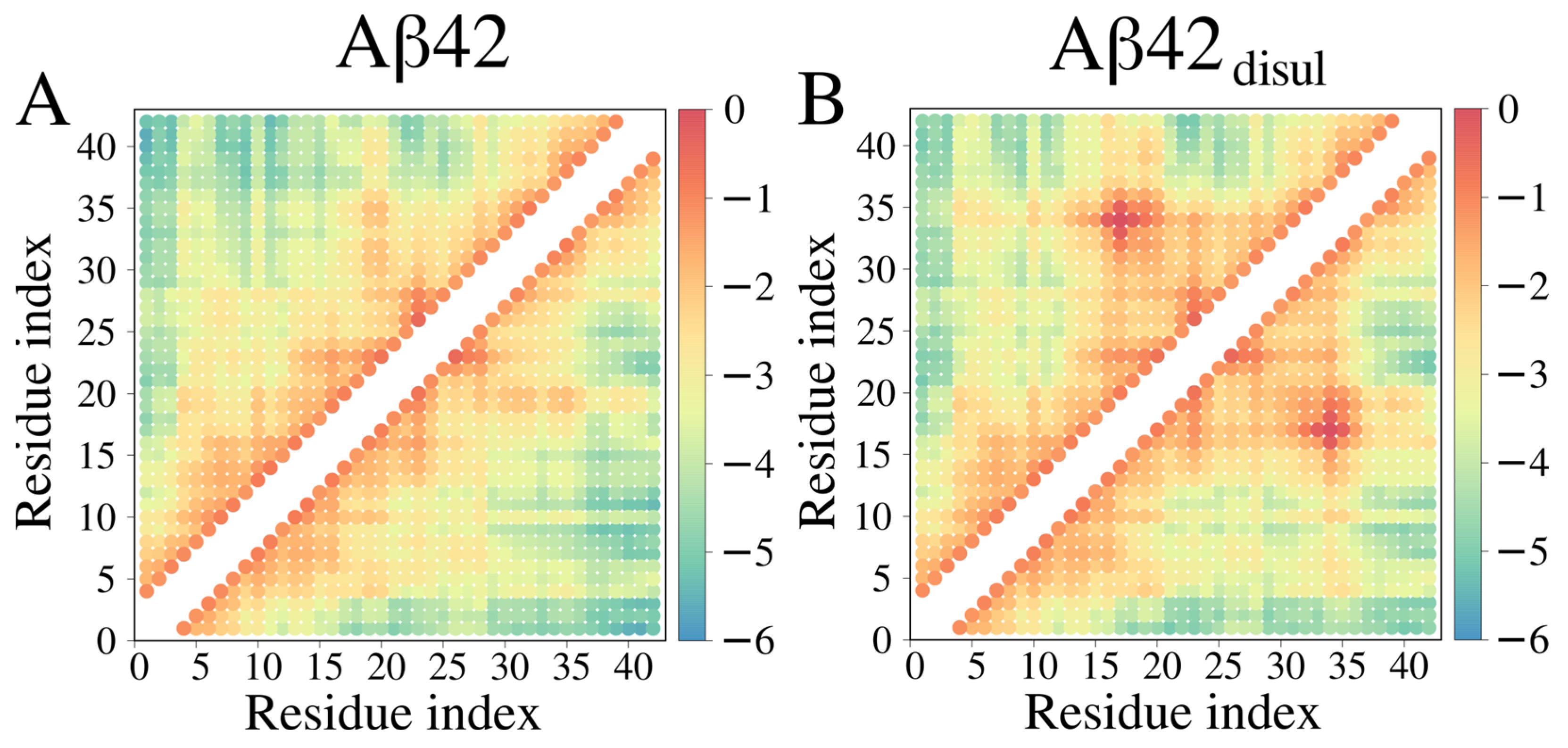
2.3. Impact of the Disulfide Bond Addition to the Monomeric Aβ42
3. Discussion
| Force Field | Variant | Cα | Cβ | N |
|---|---|---|---|---|
| Amber ff19SB | wt | 0.982 (0.007) | 0.990 (0.004) | 0.838 (0.036) |
| CC mutant static | 0.984 (0.003) | 0.978 (0.004) | 0.813 (0.029) | |
| CC mutant dynamic | 0.981 (0.006) | 0.978 (0.001) | 0.864 (0.038) | |
| SIRAH | wt | 0.981 (0.001) | 0.991 (0.002) | 0.780 (0.023) |
| CC mutant | 0.972 (0.004) | 0.979 (0.003) | 0.806 (0.037) | |
| Martini 3 | wt | 0.992 | 0.995 | 0.896 |
| CC mutant | 0.990 | 0.979 | 0.853 |
| Force Field | Cα | Cβ | N | |||
|---|---|---|---|---|---|---|
| ΔH | RMSE | ΔH | RMSE | ΔH | RMSE | |
| Amber ff19SB | 0.22 | 1.06 | −0.58 | 1.52 | −2.17 | 4.03 |
| SIRAH | 0.03 | 0.67 | 0.18 | 0.96 | −1.01 | 2.60 |
| Martini 3 | 0.36 | 0.75 | 0.23 | 0.93 | −6.29 | 6.65 |
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- Simulation Protocol
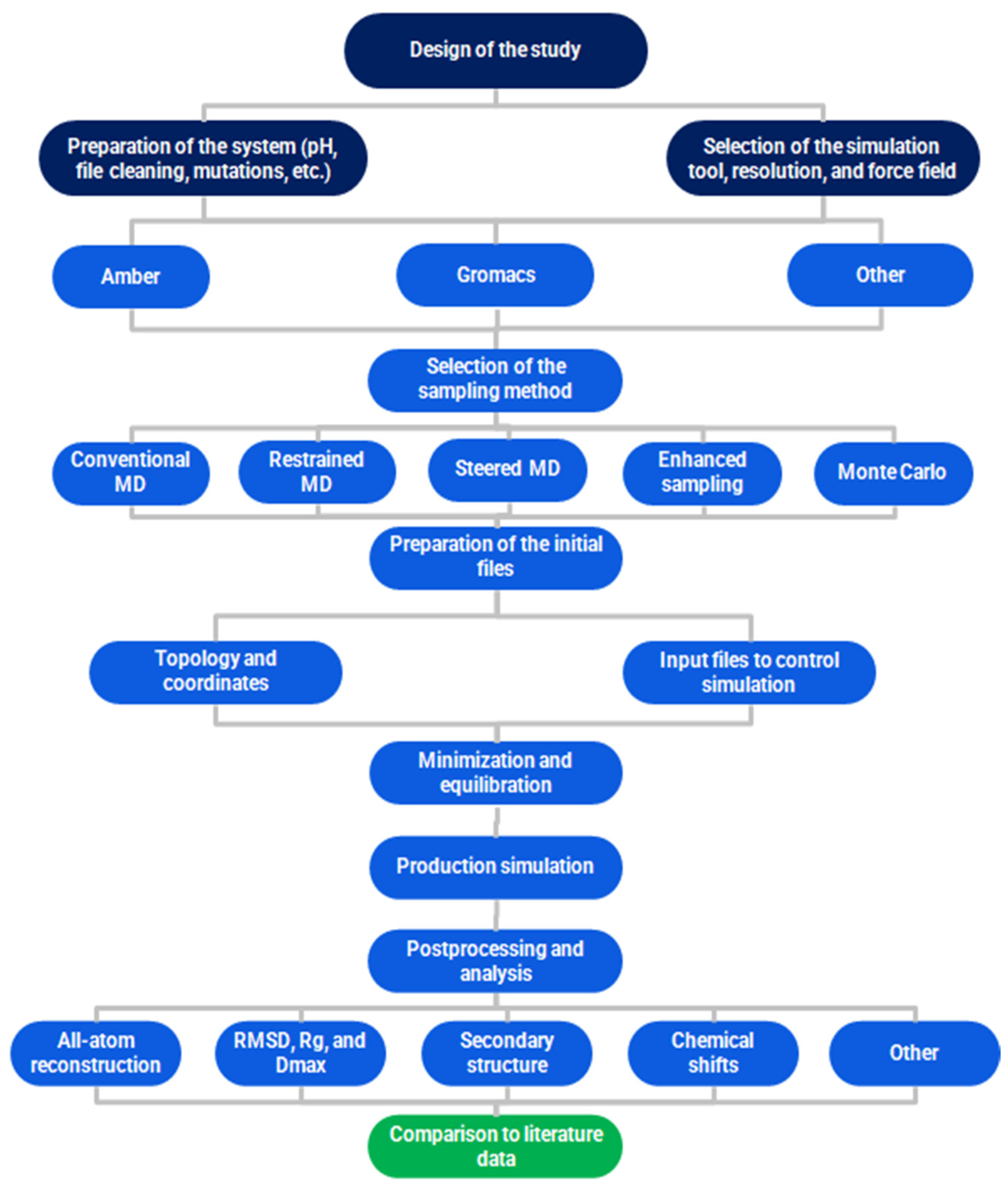
- Prerequisites
- Download the archive file from the Supplementary Materials and decompress it.
- Ensure that all the required software is installed (in the case of a personal computer) or loaded in the form of modules (in the case of computer clusters or supercomputers). Some details about that are provided in the equipment table attached to this publication.
- Make sure that software and packages are not only properly compiled but also installed, which may require adding some directories and libraries to operating system paths.
- In the case of Amber software, make sure that the “amber.sh” file is exported (by the “export AMBERPATH/bash.sh” command or by adding this line permanently to the .bashrc file) before starting the procedure, as indicated in the Amber installation guide.
- Use of Amber as a Computational Engine
- Option 1
- All-atom simulations of monomeric Aβ42 in the ff19SB force field using Amber software.
- 1.1.
- Prepare the PDB input file to run MD simulations using the PDB2PQR server [106]: https://server.poissonboltzmann.org/pdb2pqr (accessed on 10 March 2024)
- 1.1.1.
- Click “Upload a PDB file” button, then click the “Select File” button, and select the PDB file from the extracted directory.
- 1.1.2.
- Fill out the “pH field” with the requested value; in this case, input “7” and select the default option “Use PROPKA to assign protonation states at provided pH”. Select “AMBER” as a requested force field and “AMBER” as an output naming scheme.
- 1.1.3.
- Select the following additional options to ensure proper file processing: (i) ensure that new atoms are not rebuilt too close to existing atoms; (ii) optimize the hydrogen bonding network; (iii) add/keep chain IDs in the PQR file; and (iv) remove the waters from the output file.
- 1.1.4.
- Click “Start Job” and wait for its completion (it usually takes less than a minute).
- 1.1.5.
- Find a file with a “pqr” extension in “PDB2PQR Output Files” and click the “Download” button next to it to download the PQR file. Then rename the downloaded “pqr” file to “input.pqr” and copy it to amber/tleap.
- 1.2.
- Enter the directory Amber_all-atom_and_SIRAH and execute the bash script amber/setup_amber.sh to perform system setup, energy minimization, and equilibration (it can take up to one hour). Ensure that the script is working properly by checking output files at every step by manual inspection in any text editor.
- 1.3.
- Execute the bash script amber/MD1/dyn1/start_dyn1.sh to start the production phase (it can take a few days).
- 1.4.
- Execute the bash script amber_analysis/run_analysis.sh to perform typical analysis of the trajectories with the use of cpptraj, a part of AmberTools package.
- 1.5.
- Execute the script amber_analysis/run_analysis_rg_snap.sh to find and extract snapshots for the all-atom simulations with the minimum and maximum Rg values.
- 1.6.
- To visualize the snapshots, open PyMOL by typing “pymol” in the terminal or clicking the application’s icon:
- 1.6.1.
- Click File from the top menu, then click Open from the list, and select the PDB file generated during the analysis.
- 1.6.2.
- Click “A” in the side menu on the right hand side of the object name, which, in this case, is the name of the PDB file without the extension and click “remove water” to make the visualization of the protein more clear.
- 1.6.3.
- Click “S” in the side menu on the right hand side of the object name and click “cartoon” to select a cartoon representation of the protein.
- 1.6.4.
- Click “C” in the side menu on the right-hand side of the object name, and then click “spectrum” and select “rainbow” to use a rainbow representation to visualize the N- to C-termini of the polypeptide chain.
- Option 2
- 2.
- All-atom simulations of a monomeric Aβ42 mutant with a disulfide bond in the ff19SB force field using Amber software.
- 2.1.
- Enter the directory Amber_all-atom_and_SIRAH and modify the PDB file using PyMOL 2.5 software to mutate two amino-acid residues to cysteines:
- 2.1.1.
- Open PyMOL by typing “pymol” in the terminal window or click the application’s icon.
- 2.1.2.
- Click “S” button in the bottom right corner of the visualization window to show the protein sequence.
- 2.1.3.
- Click “Wizard” from the top window, then click the “Mutagenesis” button from the list and click on the amino-acid residue L17 from the displayed sequence.
- 2.1.4.
- Click on the “No Mutation” button in the right panel and select “CYS”. Ensure that the “No Mutation” button is changed to “Mutate to CYS”, click “Apply”, and click “Done” to mutate selected amino-acid residue.
- 2.1.5.
- Repeat steps 2.1.3 and 2.1.4, selecting the amino-acid residue L34 to mutate it to cysteine.
- 2.1.6.
- Click “File” from the top list and click “Save Molecule…”. Click “OK”, change the directory and name the file mutated_disul.pdb and then click “Save” to save the file on the computer.
- 2.1.7.
- Close PyMOL
- 2.2.
- Prepare the PDB file by using the PDB2PQR tool (i.e., execute step 1.1), copy it to folder amber_disul/smd_disul/tleap and change its name to input_smd.pqr.
- 2.3.
- Execute the bash script amber_disul/smd_disul/setup_smd_disul.sh to perform energy minimization, equilibration, and steered molecular dynamics simulation to bring the sulfur atoms of the two cysteines close to each other. Then execute the bash script amber_disul/tleap/cys2cyx.sh to convert cysteine residues (CYS to CYX) to include information about the disulfide bond between C17 and C34.
- 2.4.
- Execute step 1.1 to process the file amber_disul/tleap/output_smd_cyx.pdb using the PDB2PQR tool. After the pqr file is generated, copy it to amber_disul/tleap as input_disul.pqr.
- 2.5.
- Execute bash script amber_disul/setup_amber_disul.sh to perform system setup, energy minimization, and equilibration, if one would like to use the static disulfide bond model, or amber_dyn_disul/setup_amber_dyn_disul.sh, if one would like to use the dynamic disulfide model (it can take up to one hour). Ensure that the script is working properly by checking output files at every step by manual inspection in any text editor.
- 2.6.
- Execute the bash script amber/MD1/dyn1/start_dyn1.sh to start the production phase (it can take a few days) and run steps 1.4–1.6 to analyze and visualize the simulation results.
- Option 3
- 3.
- Coarse-grained MD simulations of monomeric Aβ42 in the SIRAH force field using Amber software:
- 3.1.
- Enter the directory Amber_all-atom_and_SIRAH and perform step 1.1 to convert the PDB file to the PQR file.
- 3.2.
- Execute amber_sirah/setup_sirah.sh, which converts the .pqr file using the bash script sirah_convert.sh to generate a coarse-grained structural model from the all-atom structure and then performs system setup, energy minimization, and equilibration.
- 3.3.
- Execute the bash script amber_sirah/MD1/dyn1/start_dyn1.sh to start the production phase (it can take a few days).
- 3.4.
- Execute the bash script amber_sirah_analysis/run_analysis.sh to perform typical analyses of the MD trajectories with the use of cpptraj, a part of AmberTools package. Then execute the script amber_sirah_analysis/rg_min_max_cg_snap.sh to identify and extract simulation snapshots with the minimum and maximum Rg values.
- 3.5.
- Execute the VMD script sirah_vmdtk.tcl to map the simulation structures from the coarse-grained representation back to the all-atom representation. To do that, perform the following operations:
- 3.5.1.
- Enter the directory amber_sirah/MD1/dyn1/ for backmapping the whole trajectory or the directory ptraj_MD1 (created in step 3.5 automatically) for backmapping a trajectory stripped from water and ions, and open VMD with sirah_vmdtk.tcl script using the command vmd -e ../../sirah_x2.2_20-08.amber/tools/sirah_vmdtk.tcl in the terminal.
- 3.5.2.
- Click “File” in the top left corner of the VMD Main window and click “New Molecule”.
- 3.5.3.
- In the pop-up window named Molecule File Browser:
- 3.5.3.1.
- Click “Browse…” and select file system.top, then choose from Determine file type menu: “AMBER7 Parm” and click “Load”.
- 3.5.3.2.
- Click “Browse…” and select file traj.nc, then choose from Determine file type menu: AMBER Coordinates with Periodic Box and click “Load”.
- 3.5.3.3.
- Choose the directory and file name, and click “OK”.
- 3.5.4.
- Click “Extensions” and select “Tk Console” to open a new window with a VMD terminal. and type sirah_backmap in the Tkconsole to perform the backmapping of all snapshots in the trajectory (alternatively, type sirah_backmap now to perform the backmapping of only the currently displayed snapshot).
- 3.5.5.
- Save the backmapped trajectory as a single pdb file.
- 3.5.5.1.
- Click “File” in the top left corner of the VMD Main window and click “Save Coordinates”.
- 3.5.5.2.
- In the newly pop-up window named Save Trajectory, choose from the file type list: pdb, then choose from Selected atoms: all, click “Save” and choose the directory and filename, and click “OK”.
- 3.6.
- Perform step 1.6 to visualize the snapshots in the all-atom representation generated in the above step.
- Option 4
- 4.
- Coarse-grained MD simulations of a monomeric Aβ42 mutant with a disulfide bond in the SIRAH force field using Amber software.
- 4.1.
- Perform steps 2.1 to 2.4 to mutate the selected amino-acid residues to cysteines and to convert the PDB file to the PQR format.
- 4.2.
- Execute amber_sirah_disul/setup_sirah_disul.sh to convert the .pqr file using the sirah_convert.sh script to obtain a coarse-grained structural model from the all-atom structure.
- 4.3.
- Execute the bash script amber_sirah/MD1/dyn1/start_dyn1.sh to start the production phase (it can take a few days).
- 4.4.
- Perform steps 3.4 and 3.6 to analyze the simulation data and to convert coarse-grained structural models to all-atom structures, and then run step 1.6 to visualize the snapshots in the all-atom representation.
- Use of Gromacs as a Computational Engine
- Option 5
- 5.
- Coarse-grained MD simulations of monomeric Aβ42 with solute-solvent interaction scaling in the Martini force field using Gromacs 2023.5 software:
- 5.1.
- Enter the directory Gromacs_Martini/ABeta and execute the bash script setup_scripts/setup_martini_AB.sh to convert the all-atom structure of Aβ42 to the corresponding Martini coarse-grained model, solvate it, and perform energy minimization (it can take up to an hour).
- 5.2.
- Execute the bash script equilibration/run_equilibration.sh to perform equilibration simulations (it can take up to an hour).
- 5.3.
- If using a computer cluster, execute 15 bash scripts production/qsub_096.sh to qsub_110.sh to run production simulations with the solute-solvent interaction rescaling parameter in the range from λ = 0.96 to λ = 1.1 (it can take up to days to complete). Alternatively, to run the simulations on a desktop computer, execute the bash script production/run_on_PC.sh.
- 5.4.
- Execute the bash script production/script_pbc.sh to perform the analysis of Rg. Then execute the bash script production/validation.sh to calculate average Rg values for λ = 0.96, …1.1.
- 5.5.
- Execute the bash script production/dmax.sh which calculates the maximum dimension of the Aβ42 molecule, Dmax, for each of the simulation structures in each of the trajectories. Then execute the bash script production/contactdata.sh which calculates contact frequency maps based on the trajectories with λ = 0.96, … 1.1.
- 5.6.
- Open VMD by typing vmd peptide.gro run_pbc_lambda1.00.xtc in the terminal to visualize the Martini trajectories.
- 5.6.1.
- Type pbc box in the command line of VMD to display the simulation box
- 5.6.2.
- Click “Graphics”. Click “Representations”. Select “VDW” from the Drawing Method to visualize the system (by default, the backbone beads are depicted in pink, whereas the side chain beads are shown in yellow).
- 5.6.3.
- Use the slider in the main window to move between frames. Use the play button to visualize the whole trajectory.
- 5.7.
- Execute the script production/backmapping/backmapping.sh to convert the coarse-grained structures generated in the Martini simulations to all-atom structures.
- 5.8.
- Open VMD by typing vmd Ab42_input.pdb merged.xtc to display the trajectory of Aβ42 in the all-atom representation centered in the simulation box.
- 5.8.1.
- Perform steps analogous to 5.6.1–5.6.3 to visualize the trajectory in the all-atom representation.
- 5.8.2.
- Click “Graphics” to change the background color. Click “Colors”. Choose “Display” and then “Background” from “Categories” and set the color to white. Click File → Render and choose Start Rendering to visualize and save a selected snapshot.
- 5.9.
- Open ChimeraX to visualize the system.
- 5.9.1.
- Click File → Open and choose one of the backmapped structures (e.g., production/backmapping/splitted_traj/lambda_1.03/temp/frame_100.pdb). Go to Favorites → Settings and in the first tab, Background change the color in Background color option.
- 5.9.2.
- Click Select → Chains → chain ? to select a whole polypeptide chain. Click Select → Chains → chain ? and then select Actions → Atoms/Bonds → Show to show side-chain atoms. Click Actions → Atoms/Bonds → Atom Style → Ball & Stick to show atoms of the side chain in a different style. Click Actions → Cartoon → Hide to hide the ribbon representation and display backbone atoms.
- 5.9.3.
- Click File → Save… and in Files of type menu, choose the PNG image (*.png), next specify the file name in File name, and, after setting the resolution in the Size option, use the Save button to save the image with the visualization.
- Option 6
- 6.
- Coarse-grained MD simulations of a monomeric Aβ42 mutant with a disulfide bond in a Martini force field using Gromacs software.
- 6.1.
- Enter the directory Gromacs_Martini/ABeta_CC and execute the bash script setup_scripts/setup_martini_AB.sh to convert the all-atom model of Aβ42 to the Martini coarse-grained model, solvate it, and perform energy minimization with the addition of the disulfide bond (it can take up to an hour).
- 6.2.
- Perform steps from 5.2 to 5.9 to simulate, analyze, and visualize the system.
- Option 7
- 7.
- Coarse-grained MD simulations of monomeric Aβ42 in the SIRAH force field using Gromacs software:
- 7.1.
- Enter the directory Gromacs_Sirah and execute the bash script setup_sirah.sh with parameter Ab42 (by typing “bash setup_sirah.sh Ab42” in the terminal) to protonate the all-atom model of Aβ42, convert it to the SIRAH coarse-grained representation, solvate the protein, and create a topology file of the whole simulation system.
- 7.2.
- Run the bash script run_min_eq.sh with parameter Ab42 (i.e., type “./run_min_eq.sh Ab42” in the terminal) to perform energy minimization and equilibration simulations (it can take a few minutes).
- 7.3.
- Execute the bash script run_md.sh with parameter Ab42 (by typing “./run_md.sh Ab42” in the terminal) to run the production simulation on a desktop computer.
- 7.4.
- Run the script vis_trajectory.sh with parameter Ab42 (i.e., type “./vis_trajectoty.sh Ab42“ in the terminal) to visualize the SIRAH trajectory using the VMD program. NOTE: Steps analogous to those described in points 5.4 and 5.5 above can be performed to analyze the SIRAH simulation results.
- Use of CHARMM-GUI to Generate Input Files for the MD Simulations
- Option 8
- 8.
- Go to the official website of CHARMM-GUI: https://www.charmm-gui.org/ (accessed on 10 March 2024)
- 8.1.
- Click “Input Generator” from the list on the left-hand side of the website. If this is the first time, register as a non-profit user (if applicable) or log in with your credentials if the account is already registered.
- 8.2.
- Click “Solution Builder” from the expanded list on the left-hand side of the website, click “Browse…” in the “Protein Solution System” menu, and choose the file Amber_all-atom_and_SIRAH/amber/tleap/input.pdb.
- 8.2.1.
- Select the option “Check/Correct PDB Format” to ensure that CHARMM-GUI will fully process the PDB file. Write down the “JOB ID” from the top right corner of the website in case the procedure is not completed in a single use. This number can be used with the Job Retriever in the top left corner of the Input Generator menu to recover the job.
- 8.3.
- Click “Next Step: Select Model/Chain” in the bottom right corner of the website to proceed (each step may take a few seconds to a few minutes, depending on the PDB file and the current server load).
- 8.4.
- Check if the system has properly recognized all amino-acid residues and click “Next Step: Manipulate PDB” in the bottom right corner of the website to proceed further.
- 8.5.
- Check the option list and click “Next Step: Generate PDB” in the bottom right corner of the website to proceed further.Note: Mutations or adding disulfide bonds can be performed at this step, but it is not possible to perform both operations simultaneously. To do both, first mutate L17 to C17 and L34 to C34, then generate and download the resulting PDB in the next step, and then repeat the procedure points from 8.4 to 8.7 with the newly generated PDB file, selecting the formation of a disulfide bond.
- 8.6.
- In the “Fit Waterbox Size to Protein Size” box, select the thickness of the water layer around the protein to 23 Å or any other value that would prohibit contacts between periodic images. Click “Next Step: Solvate Molecule’‘ in the bottom right corner of the website to proceed further.
- 8.7.
- Click “Next Step: Setup Periodic Boundary Condition” in the bottom right corner of the website to proceed further. Download the step3_pbcsetup.pdb file and visualize it to check if the protein structure is fine. NOTE: This PDB file can be used in the standard Amber or Gromacs procedure (options 1–7) after removing water molecules or by manually specifying box size.
- 8.8.
- Select “Amber” in the “Force Field Options:” field and ensure that “FF19SB” is selected as the force field for proteins.
- 8.9.
- Select “Amber” in the “Input Generation Options:” field. Alternatively, “Gromacs” or another computational engine may be selected.
- 8.10.
- Select “NVT Ensemble” in the “Dynamics Input Generation Options:” field and ensure that “Temperature:” is set to 300K.
- 8.11.
- Download the package of the generated input files by clicking “download.tgz” in the top right corner of the website.
- 8.12.
- Extract the downloaded file, then copy amber/step3_input.rst7 as system.crd to the Amber_all-atom_and_SIRAH/amber/tleap directory and copy amber/step3_input.parm7 as system.top to the Amber_all-atom_and_SIRAH/amber/tleap directory.
- 8.13.
- Execute the bash script Amber_all-atom_and_SIRAH/amber/setup_amber_ch-gui.sh to perform system setup, energy minimization, and equilibration (it can take up to one hour). Ensure that the script is working properly by checking output files at every step through manual inspection in any text editor.
- 8.14.
- Execute the bash script amber/MD1/dyn1/start_dyn1.sh to start the production phase (it can take a few days) and perform steps 1.5–1.7 to analyze and visualize the simulation results.
Appendix B
- Description, options, tips, and tricks to the protocol presented in Appendix A
- Simulations in Amber Software
- Software Tips:
- Option 1: All-Atom MD Simulations of Aβ42
- Option 2: All-Atom MD Simulations of Aβ42 with an Artificially Added Disulfide Bond
- Manual mutation
- Visualization-tool mutation (PyMol)
- Use of a structure prediction tool
- Insertion of a disulfide bond
- Selection of the conformation with cysteine residues in the proximity
- SMD of a structure to bring cysteines together
- Selection of a disulfide-bond model
- Options 3 and 4: SIRAH Coarse-Grained MD Simulations of Aβ42 without and with Disulfide Bonds
- Simulations in Gromacs Software
- Option 5: Martini Coarse-Grained MD Simulations of Aβ42
- Option 6: Martini Coarse-Grained MD Simulations of Aβ42 with an Artificially Added Disulfide Bond
- Application of Martini Coarse-Grained MD Simulations to Structured and Mix-Folded Proteins
- Option 7: SIRAH Coarse-Grained MD Simulations of Aβ42
- Option 8: Use of CHARMM-GUI for MD Simulations of Aβ42
Appendix C
References
- Whitford, D. Proteins: Structure and Function, 1st ed.; Wiley: Hoboken, NJ, USA, 2005. [Google Scholar]
- Anfinsen, C.B. Principles That Govern the Folding of Protein Chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradović, Z. Intrinsic Disorder and Protein Function. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef] [PubMed]
- Sgourakis, N.G.; Yan, Y.; McCallum, S.A.; Wang, C.; Garcia, A.E. The Alzheimer’s Peptides Abeta40 and 42 Adopt Distinct Conformations in Water: A Combined MD/NMR Study. J. Mol. Biol. 2007, 368, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Alessio, E.; Hansen, L.; Iwamoto, M.; Marzilli, L.G. Dynamic Pathways for Fluxional Molecules Defined Using Exchange-NOE Peaks. J. Am. Chem. Soc. 1996, 118, 7593–7600. [Google Scholar] [CrossRef]
- Iannuzzi, C.; Adrover, M.; Puglisi, R.; Yan, R.; Temussi, P.A.; Pastore, A. The Role of Zinc in the Stability of the Marginally Stable IscU Scaffold Protein. Protein Sci. 2014, 23, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Irvine, G.W.; Duncan, K.E.R.; Gullons, M.; Stillman, M.J. Metalation Kinetics of the Human α-Metallothionein 1a Fragment Is Dependent on the Fluxional Structure of the Apo-Protein. Chemistry 2015, 21, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Bokor, M.; Tantos, Á. Protein-Protein Connections-Oligomer, Amyloid and Protein Complex-By Wide Line 1H NMR. Biomolecules 2021, 11, 757. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically Disordered Proteins and Their Environment: Effects of Strong Denaturants, Temperature, pH, Counter Ions, Membranes, Binding Partners, Osmolytes, and Macromolecular Crowding. Protein J. 2009, 28, 305–325. [Google Scholar] [CrossRef] [PubMed]
- Levine, Z.A.; Larini, L.; LaPointe, N.E.; Feinstein, S.C.; Shea, J.-E. Regulation and Aggregation of Intrinsically Disordered Peptides. Proc. Natl. Acad. Sci. USA 2015, 112, 2758–2763. [Google Scholar] [CrossRef]
- Sengupta, U.; Nilson, A.N.; Kayed, R. The Role of Amyloid-β Oligomers in Toxicity, Propagation, and Immunotherapy. EBioMedicine 2016, 6, 42–49. [Google Scholar] [CrossRef]
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, S.; Wetzel, R. An Intersheet Packing Interaction in A Beta Fibrils Mapped by Disulfide Cross-Linking. Biochemistry 2004, 43, 15310–15317. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.; Kwon, Y.; Nam, E.-J.; Park, H.; Paik, S.R. Disulfide-Mediated Elongation of Amyloid Fibrils of α-Synuclein For Use in Producing Self-Healing Hydrogel and Dye-Absorbing Aerogel. Acta Biomater. 2022, 145, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, C.; Kumar, A.; Lang, A.; Ohlenschläger, O. Cysteines and Disulfide Bonds as Structure-Forming Units: Insights From Different Domains of Life and the Potential for Characterization by NMR. Front. Chem. 2020, 8, 280. [Google Scholar] [CrossRef] [PubMed]
- Bulaj, G. Formation of Disulfide Bonds in Proteins and Peptides. Biotechnol. Adv. 2005, 23, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Smardz, P.; Sieradzan, A.K.; Krupa, P. Mechanical Stability of Ribonuclease A Heavily Depends on the Redox Environment. J. Phys. Chem. B 2022, 126, 6240–6249. [Google Scholar] [CrossRef] [PubMed]
- Iftimie, R.; Minary, P.; Tuckerman, M.E. Ab Initio Molecular Dynamics: Concepts, Recent Developments, and Future Trends. Proc. Natl. Acad. Sci. USA 2005, 102, 6654–6659. [Google Scholar] [CrossRef] [PubMed]
- Marx, D.; Hutter, J. Ab Initio Molecular Dynamics: Basic Theory and Advanced Methods; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Kühne, T.D.; Krack, M.; Mohamed, F.R.; Parrinello, M. Efficient and Accurate Car-Parrinello-like Approach to Born-Oppenheimer Molecular Dynamics. Phys. Rev. Lett. 2007, 98, 066401. [Google Scholar] [CrossRef]
- de la Lande, A.; Alvarez-Ibarra, A.; Hasnaoui, K.; Cailliez, F.; Wu, X.; Mineva, T.; Cuny, J.; Calaminici, P.; López-Sosa, L.; Geudtner, G.; et al. Molecular Simulations with in-deMon2k QM/MM, a Tutorial-Review. Molecules 2019, 24, 1653. [Google Scholar] [CrossRef]
- Carnimeo, I.; Affinito, F.; Baroni, S.; Baseggio, O.; Bellentani, L.; Bertossa, R.; Delugas, P.D.; Ruffino, F.F.; Orlandini, S.; Spiga, F.; et al. Quantum ESPRESSO: One Further Step toward the Exascale. J. Chem. Theory Comput. 2023, 19, 6992–7006. [Google Scholar] [CrossRef]
- Cruzeiro, V.W.D.; Manathunga, M.; Merz, K.M., Jr.; Götz, A.W. Open-Source Multi-GPU-Accelerated QM/MM Simulations with AMBER and QUICK. J. Chem. Inf. Model. 2021, 61, 2109–2115. [Google Scholar] [CrossRef]
- Warshel, A.; Levitt, M. Theoretical Studies of Enzymic Reactions: Dielectric, Electrostatic and Steric Stabilization of the Carbonium Ion in the Reaction of Lysozyme. J. Mol. Biol. 1976, 103, 227–249. [Google Scholar] [CrossRef] [PubMed]
- Senftle, T.P.; Hong, S.; Islam, M.M.; Kylasa, S.B.; Zheng, Y.; Shin, Y.K.; Junkermeier, C.; Engel-Herbert, R.; Janik, M.J.; Aktulga, H.M.; et al. The ReaxFF Reactive Force-Field: Development, Applications and Future Directions. NPJ Comput. Mater. 2016, 2, 15011. [Google Scholar] [CrossRef]
- Salahub, D.R.; Wei, D. (Eds.) Multiscale Dynamics Simulations: Nano and Nano-Bio Systems in Complex Environments; RSC Theoretical and Computational Chemistry Series; Royal Society of Chemistry: Cambridge, UK, 2021. [Google Scholar]
- Krupa, P.; Quoc Huy, P.D.; Li, M.S. Properties of Monomeric Aβ42 Probed by Different Sampling Methods and Force Fields: Role of Energy Components. J. Chem. Phys. 2019, 151, 055101. [Google Scholar] [CrossRef]
- Mu, J.; Liu, H.; Zhang, J.; Luo, R.; Chen, H.-F. Recent Force Field Strategies for Intrinsically Disordered Proteins. J. Chem. Inf. Model. 2021, 61, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Klein, F.; Barrera, E.E.; Pantano, S. Assessing SIRAH’s Capability to Simulate Intrinsically Disordered Proteins and Peptides. J. Chem. Theory Comput. 2021, 17, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Mackerell, A.D., Jr.; Feig, M.; Brooks, C.L., 3rd. Extending the Treatment of Backbone Energetics in Protein Force Fields: Limitations of Gas-Phase Quantum Mechanics in Reproducing Protein Conformational Distributions in Molecular Dynamics Simulations. J. Comput. Chem. 2004, 25, 1400–1415. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Piana, S.; Donchev, A.G.; Robustelli, P.; Shaw, D.E. Water Dispersion Interactions Strongly Influence Simulated Structural Properties of Disordered Protein States. J. Phys. Chem. B 2015, 119, 5113–5123. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Li, Z.; Song, L.F.; Li, P.; Merz, K.M., Jr. Parameterization of Monovalent Ions for the OPC3, OPC, TIP3P-FB, and TIP4P-FB Water Models. J. Chem. Inf. Model. 2021, 61, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Somavarapu, A.K.; Kepp, K.P. The Dependence of Amyloid-β Dynamics on Protein Force Fields and Water Models. Chemphyschem 2015, 16, 3278–3289. [Google Scholar] [CrossRef] [PubMed]
- Sugita, Y.; Okamoto, Y. Replica-Exchange Molecular Dynamics Method for Protein Folding. Chem. Phys. Lett. 1999, 314, 141–151. [Google Scholar] [CrossRef]
- Qi, R.; Wei, G.; Ma, B.; Nussinov, R. Replica Exchange Molecular Dynamics: A Practical Application Protocol with Solutions to Common Problems and a Peptide Aggregation and Self-Assembly Example. In Peptide Self-Assembly: Methods and Protocols; Nilsson, B.L., Doran, T.M., Eds.; Springer: New York, NY, USA, 2018; pp. 101–119. [Google Scholar]
- Lee, T.-S.; Cerutti, D.S.; Mermelstein, D.; Lin, C.; LeGrand, S.; Giese, T.J.; Roitberg, A.; Case, D.A.; Walker, R.C.; York, D.M. GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features. J. Chem. Inf. Model. 2018, 58, 2043–2050. [Google Scholar] [CrossRef]
- Kmiecik, S.; Gront, D.; Kolinski, M.; Wieteska, L.; Dawid, A.E.; Kolinski, A. Coarse-Grained Protein Models and Their Applications. Chem. Rev. 2016, 116, 7898–7936. [Google Scholar] [CrossRef]
- Latham, A.P.; Zhang, B. Unifying Coarse-Grained Force Fields for Folded and Disordered Proteins. Curr. Opin. Struct. Biol. 2022, 72, 63–70. [Google Scholar] [CrossRef]
- Liwo, A.; Baranowski, M.; Czaplewski, C.; Gołaś, E.; He, Y.; Jagieła, D.; Krupa, P.; Maciejczyk, M.; Makowski, M.; Mozolewska, M.A.; et al. A Unified Coarse-Grained Model of Biological Macromolecules Based on Mean-Field Multipole-Multipole Interactions. J. Mol. Model. 2014, 20, 2306. [Google Scholar] [CrossRef]
- Liwo, A.; Czaplewski, C.; Sieradzan, A.K.; Lubecka, E.A.; Lipska, A.G.; Golon, Ł.; Karczyńska, A.; Krupa, P.; Mozolewska, M.A.; Makowski, M.; et al. Chapter Two-Scale-Consistent Approach to the Derivation of Coarse-Grained Force Fields for Simulating Structure, Dynamics, and Thermodynamics of Biopolymers. In Progress in Molecular Biology and Translational Science; Strodel, B., Barz, B., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 170, pp. 73–122. [Google Scholar]
- Wu, H.; Wolynes, P.G.; Papoian, G.A. AWSEM-IDP: A Coarse-Grained Force Field for Intrinsically Disordered Proteins. J. Phys. Chem. B 2018, 122, 11115–11125. [Google Scholar] [CrossRef] [PubMed]
- Souza, P.C.T.; Alessandri, R.; Barnoud, J.; Thallmair, S.; Faustino, I.; Grünewald, F.; Patmanidis, I.; Abdizadeh, H.; Bruininks, B.M.H.; Wassenaar, T.A.; et al. Martini 3: A General Purpose Force Field for Coarse-Grained Molecular Dynamics. Nat. Methods 2021, 18, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Thomasen, F.E.; Pesce, F.; Roesgaard, M.A.; Tesei, G.; Lindorff-Larsen, K. Improving Martini 3 for Disordered and Multidomain Proteins. J. Chem. Theory Comput. 2022, 18, 2033–2041. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Berryman, J.T.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E. AMBER; University of California: San Francisco, CA, USA, 2023. [Google Scholar]
- Bekker, H.; Berendsen, H.; Dijkstra, E.J.; Achterop, S.; Vondrumen, R.; Vanderspoel, D.; Sijbers, A.; Keegstra, H.; Renardus, M. GROMACS-A Parallel Computer For Molecular-Dynamics Simulations. In Proceedings of the 4th International Conference on Computational Physics (PC 92), Prague, Czech Republic, 24–28 August 1992; World Scientific Publishing: Singapore, 1993; pp. 252–256. [Google Scholar]
- Izadi, S.; Anandakrishnan, R.; Onufriev, A.V. Building Water Models: A Different Approach. J. Phys. Chem. Lett. 2014, 5, 3863–3871. [Google Scholar] [CrossRef] [PubMed]
- Chan-Yao-Chong, M.; Chan, J.; Kono, H. Benchmarking of Force Fields to Characterize the Intrinsically Disordered R2-FUS-LC Region. Sci. Rep. 2023, 13, 14226. [Google Scholar] [CrossRef]
- Love, O.; Pacheco Lima, M.C.; Clark, C.; Cornillie, S.; Roalstad, S.; Cheatham, T.E., III. Evaluating the Accuracy of the AMBER Protein Force Fields in Modeling Dihydrofolate Reductase Structures: Misbalance in the Conformational Arrangements of the Flexible Loop Domains. J. Biomol. Struct. Dyn. 2023, 41, 5946–5960. [Google Scholar] [CrossRef]
- Shabane, P.S.; Izadi, S.; Onufriev, A.V. General Purpose Water Model Can Improve Atomistic Simulations of Intrinsically Disordered Proteins. J. Chem. Theory Comput. 2019, 15, 2620–2634. [Google Scholar] [CrossRef] [PubMed]
- Klein, F.; Soñora, M.; Helene Santos, L.; Nazareno Frigini, E.; Ballesteros-Casallas, A.; Rodrigo Machado, M.; Pantano, S. The SIRAH Force Field: A Suite for Simulations of Complex Biological Systems at the Coarse-Grained and Multiscale Levels. J. Struct. Biol. 2023, 215, 107985. [Google Scholar] [CrossRef]
- Krupa, P.; Sieradzan, A.K.; Mozolewska, M.A.; Li, H.; Liwo, A.; Scheraga, H.A. Dynamics of Disulfide-Bond Disruption and Formation in the Thermal Unfolding of Ribonuclease A. J. Chem. Theory Comput. 2017, 13, 5721–5730. [Google Scholar] [CrossRef]
- Best, R.B.; Zheng, W.; Mittal, J. Balanced Protein-Water Interactions Improve Properties of Disordered Proteins and Non-Specific Protein Association. J. Chem. Theory Comput. 2014, 10, 5113–5124. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr 2002, 40, 82–92. [Google Scholar]
- Terry, C.A.; Fernández, M.-J.; Gude, L.; Lorente, A.; Grant, K.B. Physiologically Relevant Concentrations of NaCl and KCl Increase DNA Photocleavage by an N-Substituted 9-Aminomethylanthracene Dye. Biochemistry 2011, 50, 10375–10389. [Google Scholar] [CrossRef] [PubMed]
- Różycki, B.; Boura, E. Conformational Ensemble of the Full-Length SARS-CoV-2 Nucleocapsid (N) Protein Based on Molecular Simulations and SAXS Data. Biophys. Chem. 2022, 288, 106843. [Google Scholar] [CrossRef]
- Dignon, G.L.; Zheng, W.; Kim, Y.C.; Best, R.B.; Mittal, J. Sequence Determinants of Protein Phase Behavior from a Coarse-Grained Model. PLoS Comput. Biol. 2018, 14, e1005941. [Google Scholar] [CrossRef]
- Anila, M.M.; Ghosh, R.; Różycki, B. Membrane Curvature Sensing by Model Biomolecular Condensates. Soft Matter 2023, 19, 3723–3732. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.Y.; Doan, D.; Medrano, M.; Chang, C.-E.A. Modeling Structural Interconversion in Alzheimers’ Amyloid Beta Peptide with Classical and Intrinsically Disordered Protein Force Fields. J. Biomol. Struct. Dyn. 2022, 40, 10005–10022. [Google Scholar] [CrossRef]
- Massi, F.; Peng, J.W.; Lee, J.P.; Straub, J.E. Simulation Study of the Structure and Dynamics of the Alzheimer’s Amyloid Peptide Congener in Solution. Biophys. J. 2001, 80, 31–44. [Google Scholar] [PubMed]
- Li, X.; Mehler, E.L. Simulation of Molecular Crowding Effects on an Alzheimer’s Beta-Amyloid Peptide. Cell Biochem. Biophys. 2006, 46, 123–141. [Google Scholar]
- Nag, S.; Sarkar, B.; Bandyopadhyay, A.; Sahoo, B.; Sreenivasan, V.K.A.; Kombrabail, M.; Muralidharan, C.; Maiti, S. Nature of the Amyloid-Beta Monomer and the Monomer-Oligomer Equilibrium. J. Biol. Chem. 2011, 286, 13827–13833. [Google Scholar]
- Festa, G.; Mallamace, F.; Sancesario, G.M.; Corsaro, C.; Mallamace, D.; Fazio, E.; Arcidiacono, L.; Garcia Sakai, V.; Senesi, R.; Preziosi, E.; et al. Aggregation States of Aβ1-40, Aβ1-42 and Aβp3-42 Amyloid Beta Peptides: A SANS Study. Int. J. Mol. Sci. 2019, 20, 4126. [Google Scholar] [CrossRef] [PubMed]
- Zhang-Haagen, B.; Biehl, R.; Nagel-Steger, L.; Radulescu, A.; Richter, D.; Willbold, D. Monomeric Amyloid Beta Peptide in Hexafluoroisopropanol Detected by Small Angle Neutron Scattering. PLoS ONE 2016, 11, e0150267. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choi, B.; Kim, N.H.; Jin, G.Y.; Kim, Y.S.; Kim, Y.H.; Eom, K. Sequence-Dependent Aggregation-Prone Conformations of Islet Amyloid Polypeptide. Phys. Chem. Chem. Phys. 2021, 23, 22532–22542. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A. Normal Mode Analysis of Protein Dynamics. Curr. Opin. Struct. Biol. 1994, 4, 285–290. [Google Scholar] [CrossRef]
- Amadei, A.; Linssen, A.B.; Berendsen, H.J. Essential Dynamics of Proteins. Proteins 1993, 17, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Barz, B.; Olubiyi, O.O.; Strodel, B. Early Amyloid β-Protein Aggregation Precedes Conformational Change. Chem. Commun. 2014, 50, 5373–5375. [Google Scholar] [CrossRef] [PubMed]
- Aho, A. Design and Analysis of Computer Algorithms, 1st ed.; Addison-Wesley: Boston, MA, USA, 1974. [Google Scholar]
- Nguyen, H.L.; Krupa, P.; Hai, N.M.; Linh, H.Q.; Li, M.S. Structure and Physicochemical Properties of the Aβ42 Tetramer: Multiscale Molecular Dynamics Simulations. J. Phys. Chem. B 2019, 123, 7253–7269. [Google Scholar] [CrossRef] [PubMed]
- Crescenzi, O.; Tomaselli, S.; Guerrini, R.; Salvadori, S.; D’Ursi, A.M.; Temussi, P.A.; Picone, D. Solution Structure of the Alzheimer Amyloid Beta-Peptide (1-42) in an Apolar Microenvironment. Similarity with a Virus Fusion Domain. Eur. J. Biochem. 2002, 269, 5642–5648. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, S.; Esposito, V.; Vangone, P.; van Nuland, N.A.J.; Bonvin, A.M.J.J.; Guerrini, R.; Tancredi, T.; Temussi, P.A.; Picone, D. The Alpha-to-Beta Conformational Transition of Alzheimer’s Abeta-(1-42) Peptide in Aqueous Media Is Reversible: A Step by Step Conformational Analysis Suggests the Location of Beta Conformation Seeding. Chembiochem 2006, 7, 257–267. [Google Scholar] [CrossRef]
- Santoro, A.; Grimaldi, M.; Buonocore, M.; Stillitano, I.; D’Ursi, A.M. Exploring the Early Stages of the Amyloid Aβ(1-42) Peptide Aggregation Process: An NMR Study. Pharmaceuticals 2021, 14, 732. [Google Scholar] [CrossRef]
- Hou, L.; Shao, H.; Zhang, Y.; Li, H.; Menon, N.K.; Neuhaus, E.B.; Brewer, J.M.; Byeon, I.-J.L.; Ray, D.G.; Vitek, M.P.; et al. Solution NMR Studies of the A beta(1-40) and A beta(1-42) Peptides Establish That the Met35 Oxidation State Affects the Mechanism of Amyloid Formation. J. Am. Chem. Soc. 2004, 126, 1992–2005. [Google Scholar] [CrossRef] [PubMed]
- Carballo-Pacheco, M.; Strodel, B. Comparison of Force Fields for Alzheimer’s A β42: A Case Study for Intrinsically Disordered Proteins. Protein Sci. 2017, 26, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Coskuner-Weber, O.; Mirzanli, O.; Uversky, V.N. Intrinsically Disordered Proteins and Proteins with Intrinsically Disordered Regions in Neurodegenerative Diseases. Biophys. Rev. 2022, 14, 679–707. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-S.; Pande, V.S. Effects of Familial Mutations on the Monomer Structure of Aβ42. Biophys. J. 2012, 103, L47–L49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, J.; Bennett, K.C.; Liu, Y.; Martin, M.V.; Head-Gordon, T. Accurate Prediction of Chemical Shifts for Aqueous Protein Structure on “Real World” Data. Chem. Sci. 2020, 11, 3180–3191. [Google Scholar] [CrossRef] [PubMed]
- Ball, K.A.; Phillips, A.H.; Nerenberg, P.S.; Fawzi, N.L.; Wemmer, D.E.; Head-Gordon, T. Homogeneous and Heterogeneous Tertiary Structure Ensembles of Amyloid-β Peptides. Biochemistry 2011, 50, 7612–7628. [Google Scholar] [CrossRef] [PubMed]
- Osapay, K.; Case, D.A. A New Analysis of Proton Chemical Shifts in Proteins. J. Am. Chem. Soc. 1991, 113, 9436–9444. [Google Scholar] [CrossRef]
- Kirkitadze, M.D.; Condron, M.M.; Teplow, D.B. Identification and Characterization of Key Kinetic Intermediates in Amyloid Beta-Protein Fibrillogenesis. J. Mol. Biol. 2001, 312, 1103–1119. [Google Scholar] [CrossRef] [PubMed]
- Baumketner, A.; Bernstein, S.L.; Wyttenbach, T.; Bitan, G.; Teplow, D.B.; Bowers, M.T.; Shea, J.-E. Amyloid Beta-Protein Monomer Structure: A Computational and Experimental Study. Protein Sci. 2006, 15, 420–428. [Google Scholar] [CrossRef]
- Itoh, S.G.; Okumura, H. Oligomer Formation of Amyloid-β(29-42) from Its Monomers Using the Hamiltonian Replica-Permutation Molecular Dynamics Simulation. J. Phys. Chem. B 2016, 120, 6555–6561. [Google Scholar] [CrossRef]
- Itoh, S.G.; Yagi-Utsumi, M.; Kato, K.; Okumura, H. Key Residue for Aggregation of Amyloid-β Peptides. ACS Chem. Neurosci. 2022, 13, 3139–3151. [Google Scholar] [CrossRef] [PubMed]
- Li, M.S.; Co, N.T.; Reddy, G.; Hu, C.-K.; Straub, J.E.; Thirumalai, D. Factors Governing Fibrillogenesis of Polypeptide Chains Revealed by Lattice Models. Phys. Rev. Lett. 2010, 105, 218101. [Google Scholar] [CrossRef] [PubMed]
- Kurouski, D.; Lednev, I.K. The Impact of Protein Disulfide Bonds on the Amyloid Fibril Morphology. Int. J. Biomed Nanosci. Nanotechnol. 2011, 2, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Mossuto, M.F.; Bolognesi, B.; Guixer, B.; Dhulesia, A.; Agostini, F.; Kumita, J.R.; Tartaglia, G.G.; Dumoulin, M.; Dobson, C.M.; Salvatella, X. Disulfide Bonds Reduce the Toxicity of the Amyloid Fibrils Formed by an Extracellular Protein. Angew. Chem. Int. Ed. Engl. 2011, 50, 7048–7051. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, A.; Luheshi, L.M.; Söllvander, S.; Pereira de Barros, T.; Macao, B.; Knowles, T.P.J.; Biverstål, H.; Lendel, C.; Ekholm-Petterson, F.; Dubnovitsky, A.; et al. Stabilization of Neurotoxic Alzheimer Amyloid-Beta Oligomers by Protein Engineering. Proc. Natl. Acad. Sci. USA 2010, 107, 15595–15600. [Google Scholar] [CrossRef] [PubMed]
- Reif, M.M.; Hünenberger, P.H.; Oostenbrink, C. New Interaction Parameters for Charged Amino Acid Side Chains in the GROMOS Force Field. J. Chem. Theory Comput. 2012, 8, 3705–3723. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D., Jr.; Brooks, B.; Brooks, C.L., III; Nilsson, L.; Roux, B.; Won, Y.; Karplus, M. CHARMM: The Energy Function and Its Parameterization. In Encyclopedia of Computational Chemistry; John Wiley & Sons, Ltd.: Chichester, UK, 2002. [Google Scholar]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Alessandri, R.; Thallmair, S.; Herrero, C.G.; Mera-Adasme, R.; Marrink, S.J.; Souza, P.C.T. A Practical Introduction to Martini 3 and Its Application to Protein-Ligand Binding Simulations. In A Practical Guide to Recent Advances in Multiscale Modeling and Simulation of Biomolecules; AIP Publishing: Melville, NY, USA, 2023; pp. 1–34. [Google Scholar]
- Borges-Araújo, L.; Patmanidis, I.; Singh, A.P.; Santos, L.H.S.; Sieradzan, A.K.; Vanni, S.; Czaplewski, C.; Pantano, S.; Shinoda, W.; Monticelli, L.; et al. Pragmatic Coarse-Graining of Proteins: Models and Applications. J. Chem. Theory Comput. 2023, 19, 7112–7135. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; Monticelli, L.; Melo, M.N.; Alessandri, R.; Tieleman, D.P.; Souza, P.C.T. Two Decades of Martini: Better Beads, Broader Scope. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2023, 13, e1620. [Google Scholar] [CrossRef]
- Periole, X.; Cavalli, M.; Marrink, S.-J.; Ceruso, M.A. Combining an Elastic Network With a Coarse-Grained Molecular Force Field: Structure, Dynamics, and Intermolecular Recognition. J. Chem. Theory Comput. 2009, 5, 2531–2543. [Google Scholar] [CrossRef]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.-P.; Lane, T.J.; Pande, V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef] [PubMed]
- Weiser, J.; Shenkin, P.S.; Still, W.C. Approximate Atomic Surfaces from Linear Combinations of Pairwise Overlaps (LCPO). J. Comput. Chem. 1999, 20, 217–230. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Cerutti, D.S.; Cisneros, G.A.; Cruzeiro, V.W.D.; Forouzesh, N.; Giese, T.J.; Götz, A.W.; Gohlke, H.; et al. AmberTools. J. Chem. Inf. Model. 2023, 63, 6183–6191. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Brooks, B.R. A Protocol for Preparing Explicitly Solvated Systems for Stable Molecular Dynamics Simulations. J. Chem. Phys. 2020, 153, 054123. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. Protein Structure and Function Prediction Using I-TASSER. Curr. Protoc. Bioinform. 2015, 52, 5.8.1–5.8.15. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.-Y.R.E.D. Server: A Web Service for Deriving RESP and ESP Charges and Building Force Field Libraries for New Molecules and Molecular Fragments. Nucleic Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B.; Frantal, S.; Cibena, M.; Schreiner, W.; Bauer, P. Is an Intuitive Convergence Definition of Molecular Dynamics Simulations Solely Based on the Root Mean Square Deviation Possible? J. Comput. Biol. 2011, 18, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Sawle, L.; Ghosh, K. Convergence of Molecular Dynamics Simulation of Protein Native States: Feasibility vs Self-Consistency Dilemma. J. Chem. Theory Comput. 2016, 12, 861–869. [Google Scholar] [CrossRef]
- Ormeño, F.; General, I.J. Convergence and Equilibrium in Molecular Dynamics Simulations. Commun. Chem. 2024, 7, 26. [Google Scholar] [CrossRef]
- Nielsen, J.E.; Baker, N.A.; Czodrowski, P.; Klebe, G. pdb2pqr: PDB2PQR-Determining Titration States, Adding Missing Atoms, and Assigning Charges/Radii to Biomolecules; Github: San Francisco, CA, USA.
- Calculating Salt Molarity in an Explicit Water System. Available online: https://ambermd.org/tutorials/basic/tutorial8/index.php (accessed on 15 February 2024).
- Song, D.; Wang, W.; Ye, W.; Ji, D.; Luo, R.; Chen, H.-F. ff14IDPs Force Field Improving the Conformation Sampling of Intrinsically Disordered Proteins. Chem. Biol. Drug Des. 2017, 89, 5–15. [Google Scholar] [CrossRef]
- The pmemd.cuda GPU Implementation. Available online: https://ambermd.org/GPULogistics.php (accessed on 15 February 2024).
- Mutagenesis-PyMOLWiki. Available online: https://pymolwiki.org/index.php/Mutagenesis (accessed on 15 February 2024).
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Im, D.; Liu, Y.; Fang, J.; Tian, X.; Kim, M.; Zhang, W.-B.; Seo, J. Distinguishing Protein Chemical Topologies Using Supercharging Ion Mobility Spectrometry-mass Spectrometry. Angew. Chem. 2023, 135, e202314980. [Google Scholar] [CrossRef]
- Qin, M.; Wang, W.; Thirumalai, D. Protein Folding Guides Disulfide Bond Formation. Proc. Natl. Acad. Sci. USA 2015, 112, 11241–11246. [Google Scholar] [CrossRef]
- Wassenaar, T.A.; Pluhackova, K.; Böckmann, R.A.; Marrink, S.J.; Tieleman, D.P. Going Backward: A Flexible Geometric Approach to Reverse Transformation from Coarse Grained to Atomistic Models. J. Chem. Theory Comput. 2014, 10, 676–690. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Herzog, F.A.; Braun, L.; Schoen, I.; Vogel, V. Improved Side Chain Dynamics in MARTINI Simulations of Protein-Lipid Interfaces. J. Chem. Theory Comput. 2016, 12, 2446–2458. [Google Scholar] [CrossRef] [PubMed]
- CHARMM-GUI. Available online: https://www.charmm-gui.org/ (accessed on 15 February 2024).







| Amber ff19SB Aβ42 | Amber ff19SB Aβ42disul | Amber ff19SB Aβ42dyn-disul | SIRAH Aβ42 | SIRAH Aβ42disul | Martini Aβ42 with λ = 1 | Martini Aβ42disul with λ = 1 | |
|---|---|---|---|---|---|---|---|
| Rg (nm) | 1.29 ± 0.14 | 1.27 ± 0.01 | 1.22 ± 0.07 | 1.13 ± 0.11 | 1.10 ± 0.04 | 1.32 ± 0.01 | 1.19 ± 0.01 |
| Dmax (nm) | 4.22 ± 0.42 | 4.16 ± 0.06 | 4.09 ± 0.21 | 3.84 ± 0.32 | 3.48 ± 0.09 | 4.20 ± 0.04 | 3.83 ± 0.03 |
| SASA (nm2) | 37.67 ± 2.78 | 38.57 ± 0.92 | 37.73 ± 1.89 | 36.47 ± 2.63 | 38.08 ± 0.74 | 42.90 ± 3.45 | 39.54 ± 3.87 |
| Martini Aβ42 with λ = 0.98 | Martini Aβ42 with λ = 1.00 | Martini Aβ42 with λ = 1.02 | Martini Aβ42disul with λ = 0.98 | Martini Aβ42disul with λ = 1.00 | Martini Aβ42disul with λ = 1.02 | |
|---|---|---|---|---|---|---|
| Rg (nm) | 1.20 ± 0.16 | 1.32 ± 0.19 | 1.44 ± 0.22 | 1.11 ± 0.10 | 1.18 ± 0.12 | 1.26 ± 0.13 |
| Dmax (nm) | 3.85 ± 0.59 | 4.20 ± 0.67 | 4.52 ± 0.71 | 3.59 ± 0.47 | 3.83 ± 0.52 | 4.06 ± 0.55 |
| End-to-end distance (nm) | 2.65 ± 0.94 | 3.00 ± 1.05 | 3.32 ± 1.11 | 2.47 ± 0.83 | 2.70 ± 0.90 | 2.93 ± 0.94 |
| Amber ff19SB Aβ42 | Amber ff19SB Aβ42disul | Amber ff19SB Aβ42dyn-disul | SIRAH Aβ42 | SIRAH Aβ42disul | Martini Aβ42 | Martini Aβ42disul | |
|---|---|---|---|---|---|---|---|
| Q15-V36 | 12.54 ± 6.40 | 9.09 ± 9.94 | 1.55 ± 1.67 | 3.45 ± 2.65 | 0.00 ± 0.00 | 1.63 ± 0.35 | 7.11 ± 0.63 |
| L17-L34 (C17-C34) | 59.10 ± 29.75 | 100.00 ± 0.00 | 100.00 ± 0.00 | 43.35 ± 7.25 | 100.00 ± 0.00 | 5.39 ± 0.75 | 100.00 ± 0.00 |
| F19-I32 | 82.34 ± 10.27 | 58.20 ± 41.84 | 70.05 ± 36.89 | 25.72 ± 22.65 | 26.88 ± 36.38 | 12.02 ± 0.92 | 24.72 ± 1.09 |
| Force Field | Variant | α-Helices | β-Strand | Other |
|---|---|---|---|---|
| Amber ff19SB | wt | 5.4 (7.3) | 23.9 (11.3) | 70.7 (7.0) |
| CC mutant static | 7.7 (3.4) | 9.8 (4.6) | 82.5 (7.9) | |
| CC mutant dynamic | 5.6 (2.0) | 13.1 (8.6) | 81.2 (9.3) | |
| SIRAH | wt | 0.1 (0.2) | 11.4 (3.5) | 88.4 (3.7) |
| CC mutant | 0.0 (0.1) | 5.4 (2.8) | 94.5 (2.8) | |
| Martini 3 | wt | 0.8 (2.5) | 0.5 (1.6) | 98.7 (3.0) |
| CC mutant | 0.8 (2.5) | 0.6 (1.8) | 98.6 (3.1) |
| Force Field | Variant | 2BEG | 2LMN | 2M4J | 2MXU | 2NAO | 5KK3 | 5OQV |
|---|---|---|---|---|---|---|---|---|
| Amber ff19SB | wt | 0.0 (0.0) | 6.3 (10.2) | 43.2 (17.6) | 2.6 (2.2) | 5.9 (4.1) | 6.4 (4.9) | 2.2 (1.5) |
| CC mutant static | 0.1 (0.1) | 2.6 (3.7) | 20.2 (13.8) | 34.4 (12.7) | 53.7 (20.2) | 53.0 (21.2) | 31.9 (12.0) | |
| CC mutant dynamic | 1.6 (2.7) | 2.3 (2.6) | 43.8 (28.7) | 12.3 (11.7) | 29.4 (13.9) | 21.1 (4.5) | 7.7 (6.7) | |
| SIRAH | wt | 0.0 (0.0) | 0.0 (0.0) | 10.8 (10.3) | 3.6 (6.2) | 29.7 (46.5) | 8.8 (14.4) | 0.3 (0.6) |
| CC mutant | 0.0 (0.0) | 0.0 (0.0) | 64.7 (56.0) | 26.1 (45.3) | 93.1 (5.1) | 75.2 (16.4) | 20.6 (30.6) | |
| Martini 3 | wt | 2.5 | 2.8 | 13.2 | 8.5 | 11.4 | 8.7 | 5.5 |
| CC mutant | 1.3 | 2.3 | 25.8 | 7.5 | 18.1 | 13.5 | 5.0 |
| Force Field | Variant | 2BEG | 2LMN | 2M4J | 2MXU | 2NAO | 5KK3 | 5OQV |
|---|---|---|---|---|---|---|---|---|
| Amber ff19SB | wt | 6.39 | 5.20 | 4.24 | 4.23 | 4.07 | 4.06 | 4.66 |
| CC mutant static | 5.74 | 4.51 | 3.46 | 3.78 | 3.31 | 3.28 | 4.03 | |
| CC mutant dynamic | 4.87 | 4.89 | 3.68 | 3.69 | 3.61 | 3.60 | 4.04 | |
| SIRAH | wt | 6.27 | 6.22 | 5.46 | 5.55 | 4.69 | 5.34 | 5.90 |
| CC mutant | 7.02 | 6.94 | 4.48 | 4.18 | 3.52 | 3.86 | 4.84 | |
| Martini 3 | wt | 3.95 | 4.40 | 3.38 | 3.31 | 3.82 | 3.80 | 3.62 |
| CC mutant | 5.12 | 4.68 | 3.52 | 3.96 | 3.18 | 3.34 | 3.97 |
| Software | Force Field | Disulfide Bond | Number of Interaction Centers: | Timestep, Cutoff Values (Method to Calculate van der Waals/Coulomb Interactions) | Real Time for 1,000,000 Steps | Hardware | ||
|---|---|---|---|---|---|---|---|---|
| Peptide | Total | CPU/GPU Model | Core No. and Frequency | |||||
| Amber 22 | All-atom Amber ff19SB | none | 627 | 60,718 | 2 fs 0.9 nm (PME/PME) | 580 min 0 s | 2*Intel(R) Xeon(R) E5-2670 v2 | 20 cores @ 2500 MHz |
| 8 min 50 s | NVIDIA A100-SXM4-40GB | 6912 cores @ 1410 MHz | ||||||
| 38 min 21 s | GeForce GTX 1660 SUPER | 3584 cores @ 1800 MHz | ||||||
| static | 609 | 62,396 | 2 fs 0.9 nm (PME/PME) | 6 min 50 s | NVIDIA GeForce RTX 4090 | 16,384 cores @ 2520 MHz | ||
| 6 min | NVIDIA A100-SXM4-40GB | 6912 cores @ 1410 MHz | ||||||
| 40 min 13 s | GeForce GTX 1660 SUPER | 3584 cores @ 1800 MHz | ||||||
| dynamic | 611 | 62,378 | 2 fs 0.9 nm (PME/PME) | 10 min | NVIDIA A100-SXM4-40GB | 6912 cores @ 1410 MHz | ||
| 40 min 20 s | GeForce GTX 1660 SUPER | 3584 cores @ 1800 MHz | ||||||
| Coarse-grained SIRAH | none | 195 | 4774 | 20 fs 1.2 nm (PME/PME) | 50 min 20 s | 2*Intel(R) Xeon(R) E5-2670 v2 | 20 cores @ 2500 MHz | |
| 4 min 35 s | NVIDIA GeForce RTX 4090 | 16,384 cores @ 2520 MHz | ||||||
| 3 min 40 s | NVIDIA A100-SXM4-40GB | 6912 cores @ 1410 MHz | ||||||
| 9 min 45 s | GeForce GTX 1660 SUPER | 3584 cores @ 1800 MHz | ||||||
| static | 195 | 4926 | 20 fs 1.2 nm (PME/PME) | 3 min 40 s | NVIDIA A100-SXM4-40GB | 6912 cores @ 1410 MHz | ||
| 7 min 10 s | GeForce GTX 1660 SUPER | 3584 cores @ 1800 MHz | ||||||
| Gromacs 2023.5 | Coarse-grained MARTINI 3 | none | 96 | 6722 | 20 fs 1.1 nm (cutoff/reaction-field) | 1 min 07 s | AMD EPYC 7763 | 64 cores @ 2450 MHz |
| 4 min 20 s | Intel(R) Core(TM) i7-5820K | 6 cores @ 3300 MHz | ||||||
| 1 min 22 s | GeForce GTX 1660 SUPER | 3584 cores @ 1800 MHz | ||||||
| static | 96 | 6770 | 20 fs 1.1 nm (cutoff/reaction-field) | 5 min 5 s | Intel(R) Core(TM) i7-5820K | 6 cores @ 3300 MHz | ||
| 1 min 19 s | GeForce GTX 1660 SUPER | 3584 cores @ 1800 MHz | ||||||
| SIRAH | none | 195 | 4644 | 20 fs 1.2 nm (cutoff/PME) | 1 min 43 s | AMD EPYC 7763 | 64 cores @ 2450 MHz | |
| 4 min 37 s | AMD Ryzen 7 5700G | 8 cores @ 3800 MHz | ||||||
| 1 min 59 s | GeForce GTX 1660 SUPER | 3584 cores @ 1800 MHz | ||||||
| static | 195 | 4644 | 20 fs 1.2 nm (PME/PME) | 2 min 53 s | AMD EPYC 7763 | 64 cores @ 2450 MHz | ||
| 7 min 38 s | AMD Ryzen 7 5700G | 8 cores @ 3800 MHz | ||||||
| 9 min 38 s | GeForce GTX 1660 SUPER | 3584 cores @ 1800 MHz | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smardz, P.; Anila, M.M.; Rogowski, P.; Li, M.S.; Różycki, B.; Krupa, P. A Practical Guide to All-Atom and Coarse-Grained Molecular Dynamics Simulations Using Amber and Gromacs: A Case Study of Disulfide-Bond Impact on the Intrinsically Disordered Amyloid Beta. Int. J. Mol. Sci. 2024, 25, 6698. https://doi.org/10.3390/ijms25126698
Smardz P, Anila MM, Rogowski P, Li MS, Różycki B, Krupa P. A Practical Guide to All-Atom and Coarse-Grained Molecular Dynamics Simulations Using Amber and Gromacs: A Case Study of Disulfide-Bond Impact on the Intrinsically Disordered Amyloid Beta. International Journal of Molecular Sciences. 2024; 25(12):6698. https://doi.org/10.3390/ijms25126698
Chicago/Turabian StyleSmardz, Pamela, Midhun Mohan Anila, Paweł Rogowski, Mai Suan Li, Bartosz Różycki, and Pawel Krupa. 2024. "A Practical Guide to All-Atom and Coarse-Grained Molecular Dynamics Simulations Using Amber and Gromacs: A Case Study of Disulfide-Bond Impact on the Intrinsically Disordered Amyloid Beta" International Journal of Molecular Sciences 25, no. 12: 6698. https://doi.org/10.3390/ijms25126698
APA StyleSmardz, P., Anila, M. M., Rogowski, P., Li, M. S., Różycki, B., & Krupa, P. (2024). A Practical Guide to All-Atom and Coarse-Grained Molecular Dynamics Simulations Using Amber and Gromacs: A Case Study of Disulfide-Bond Impact on the Intrinsically Disordered Amyloid Beta. International Journal of Molecular Sciences, 25(12), 6698. https://doi.org/10.3390/ijms25126698







