Novel Insights into Changes in Gene Expression within the Hypothalamus in Two Asthma Mouse Models: A Transcriptomic Lung–Brain Axis Study
Abstract
1. Introduction
2. Results
2.1. Confirmation of Lung Inflammation in OVA- and LPS-Treated Mice
2.2. Validation of the Microdissection
2.3. Bioinformatics Analysis
2.3.1. Single Genes Differentially Expressed in Asthma Groups vs. Control
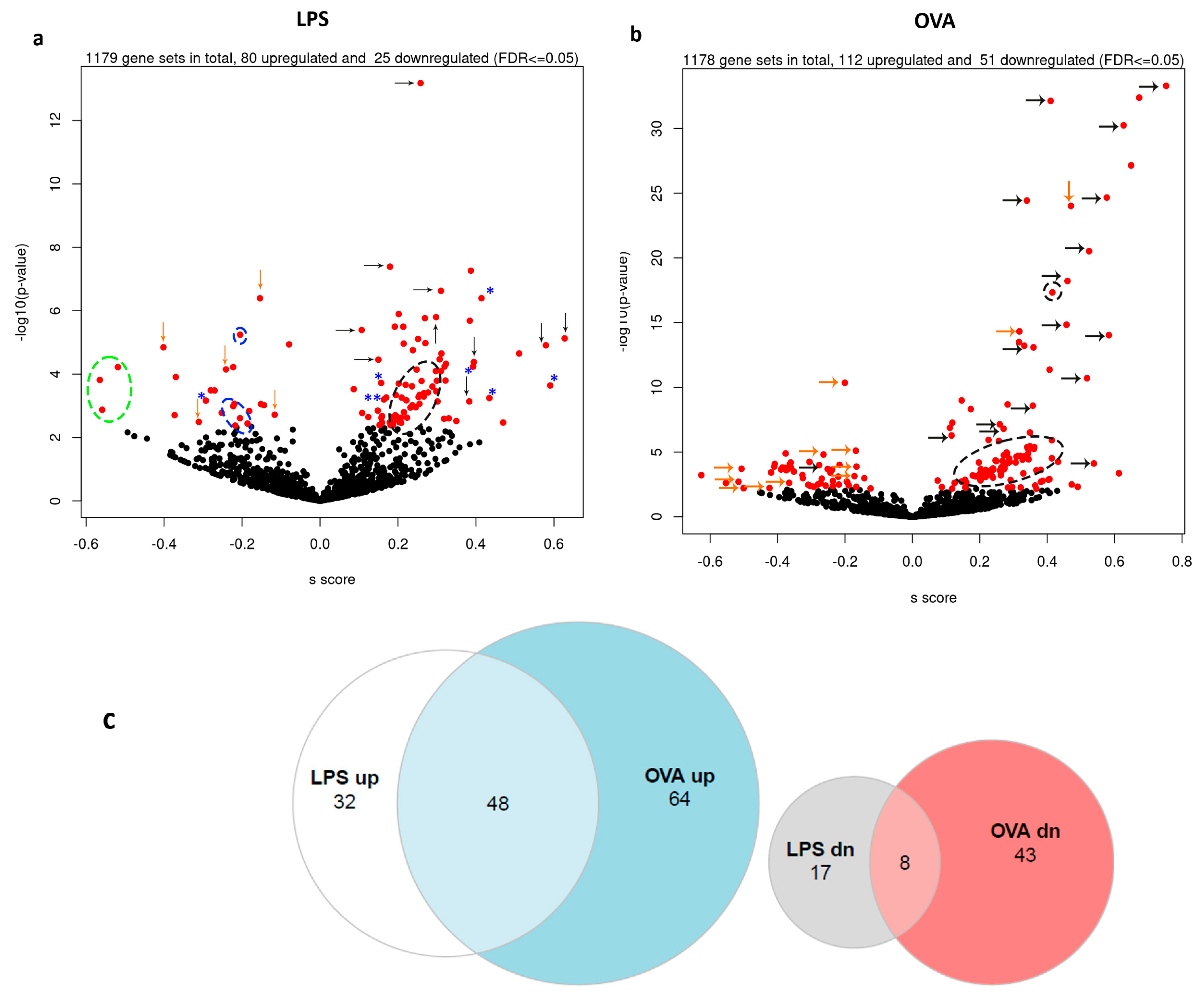
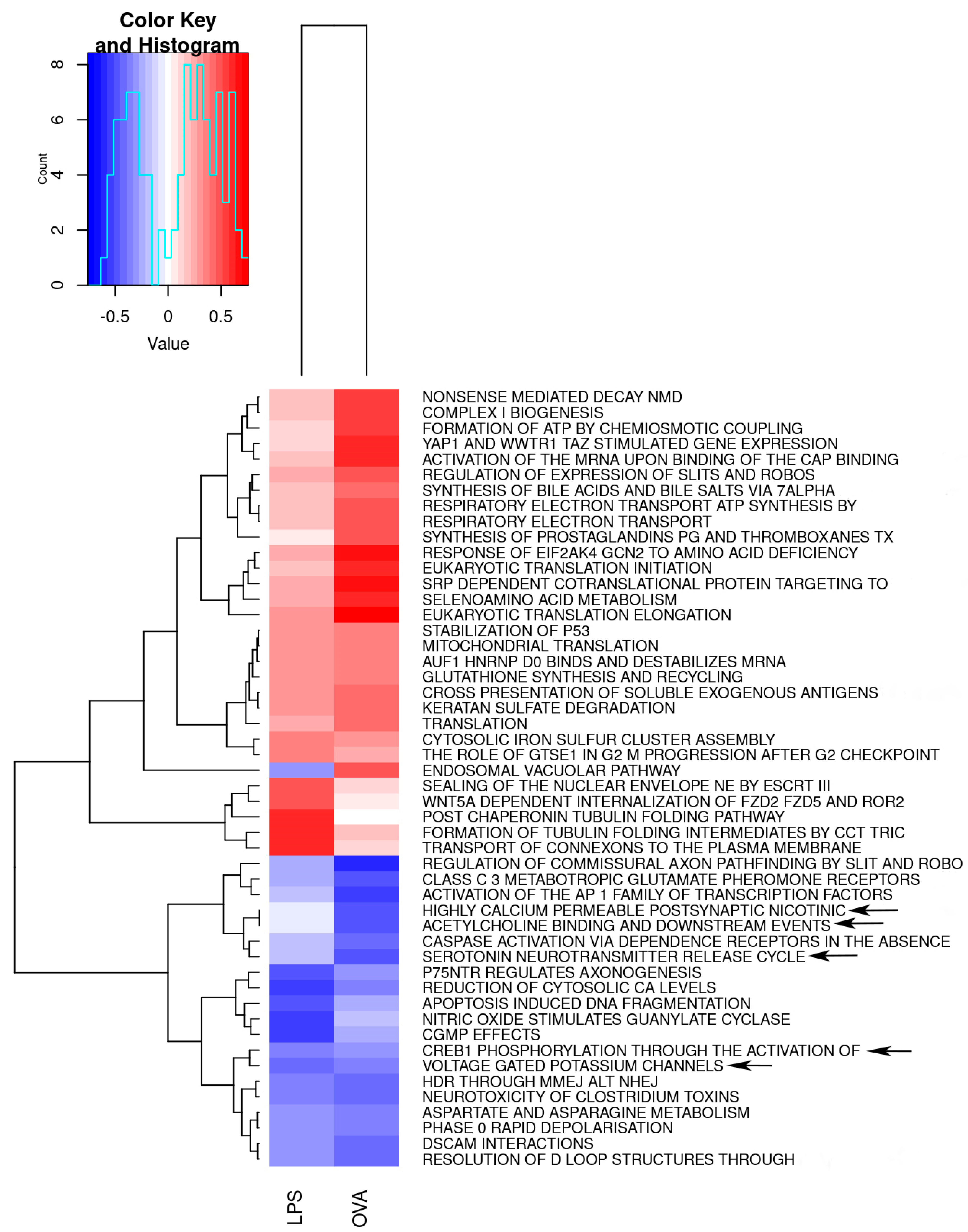
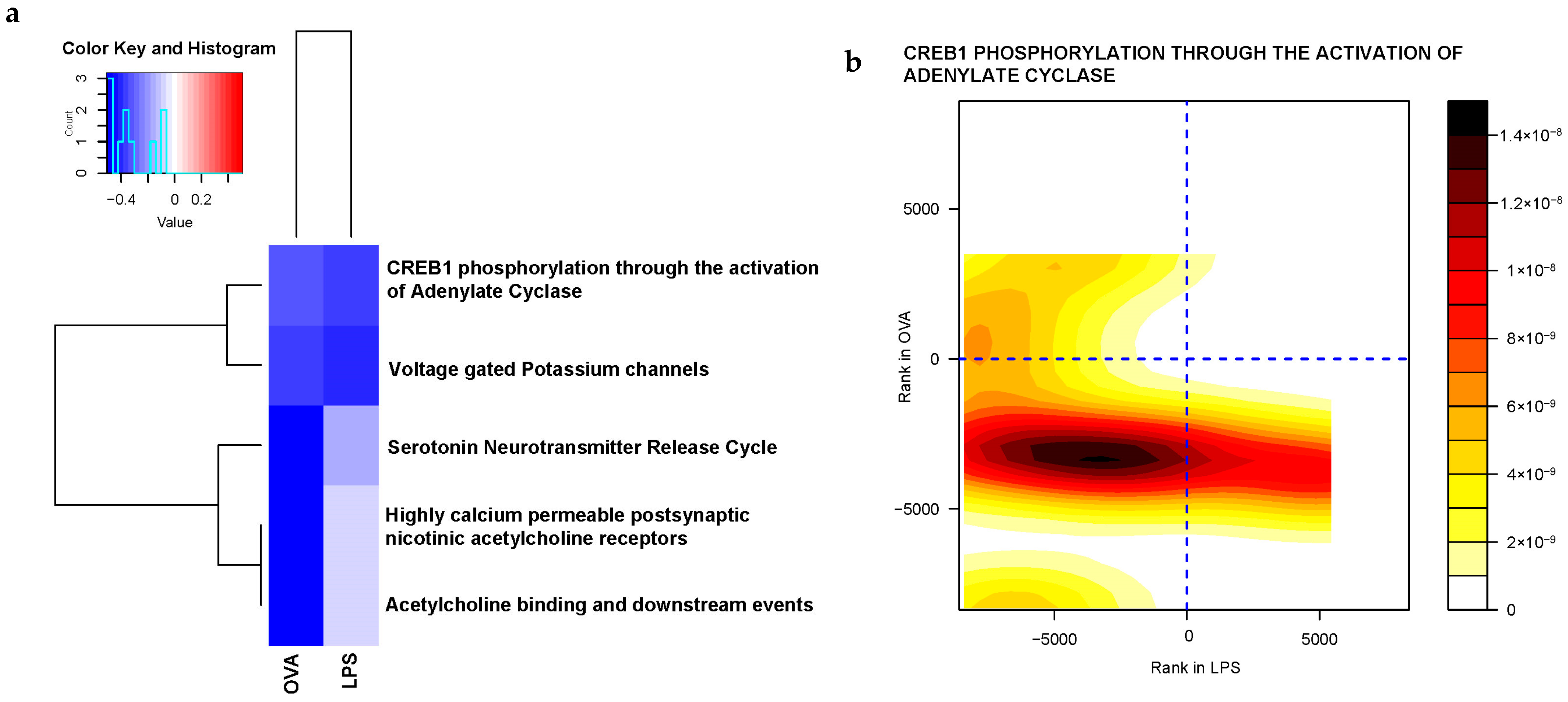
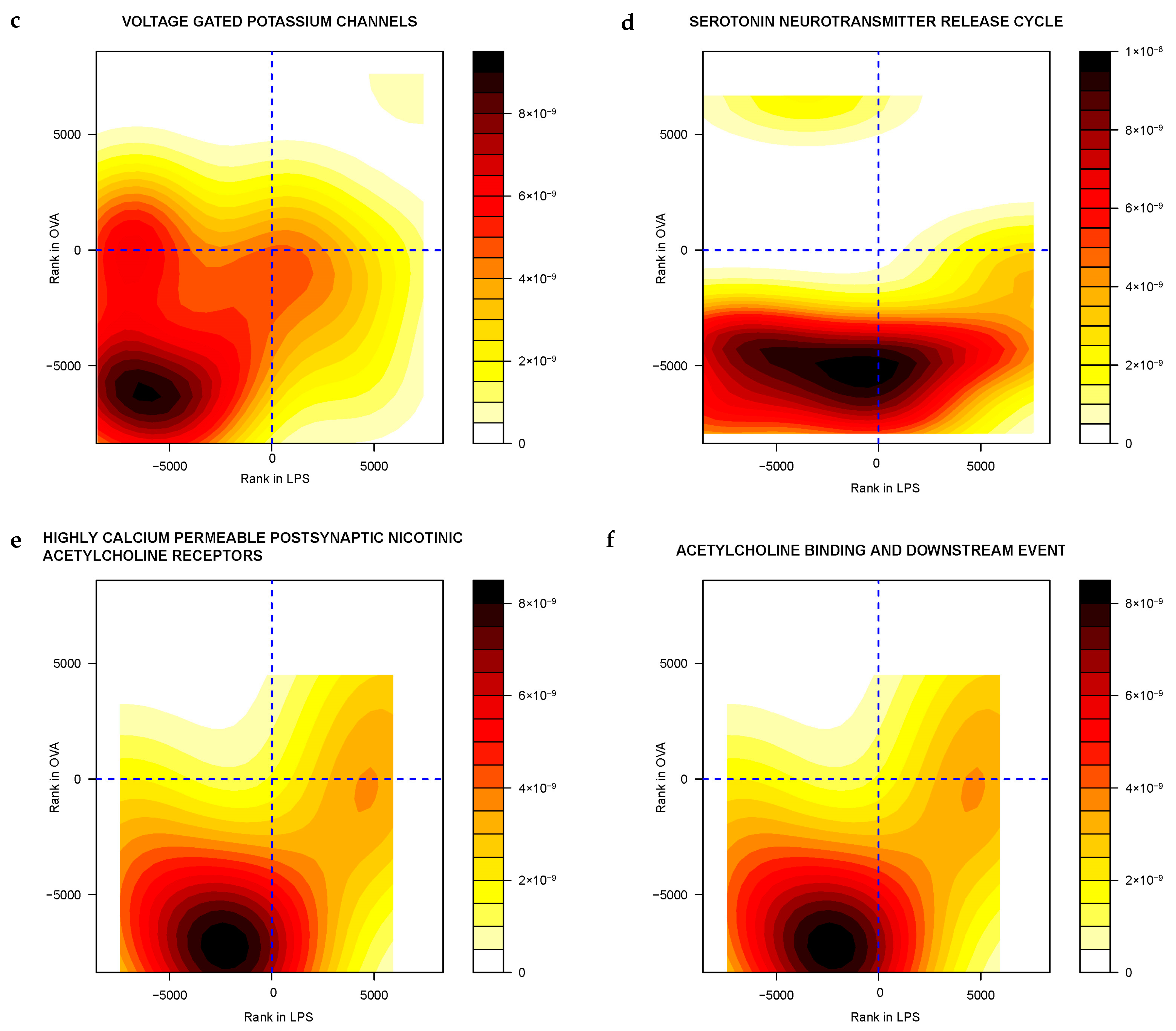
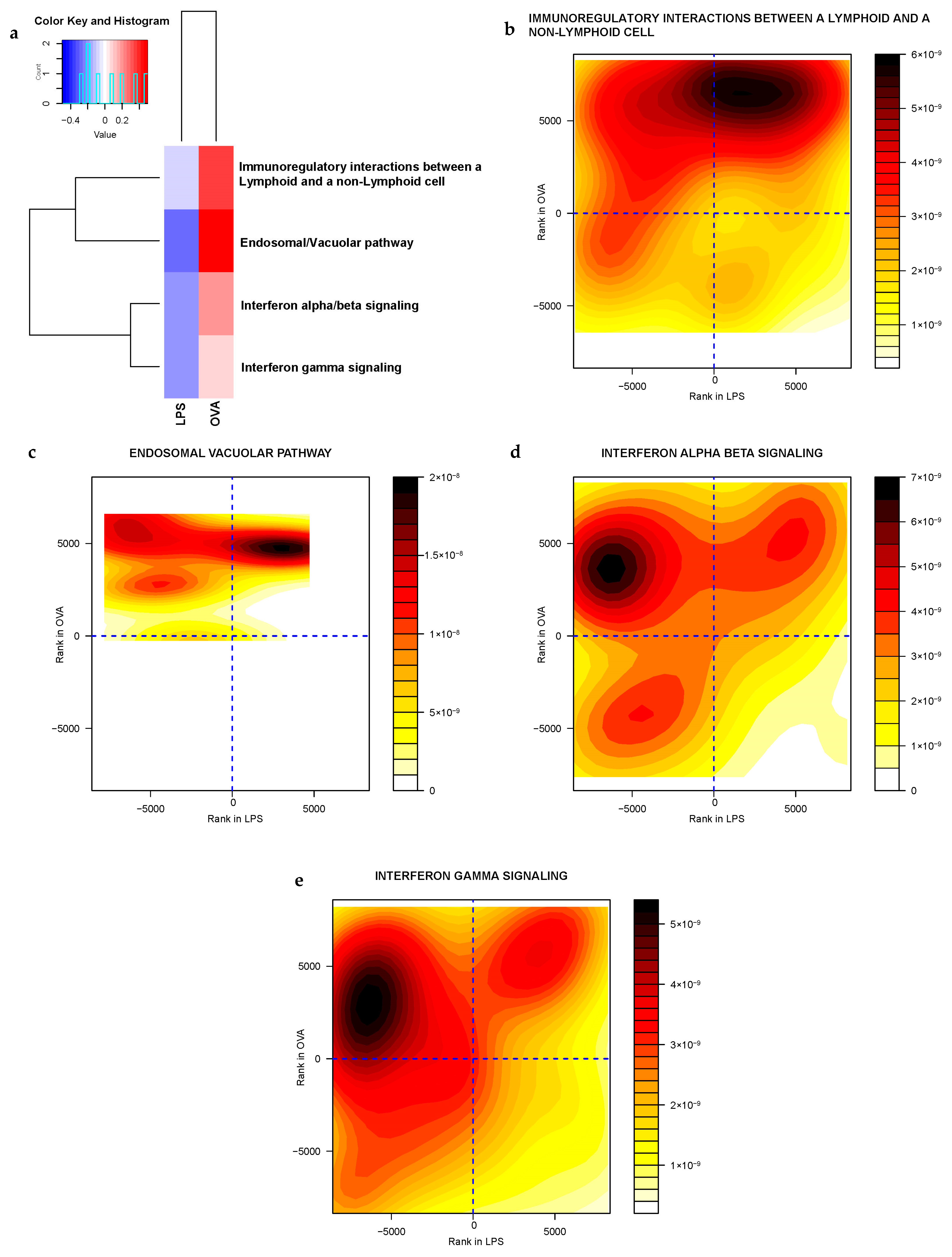
2.3.2. Gene Sets Differentially Expressed in Asthma Groups vs. Control
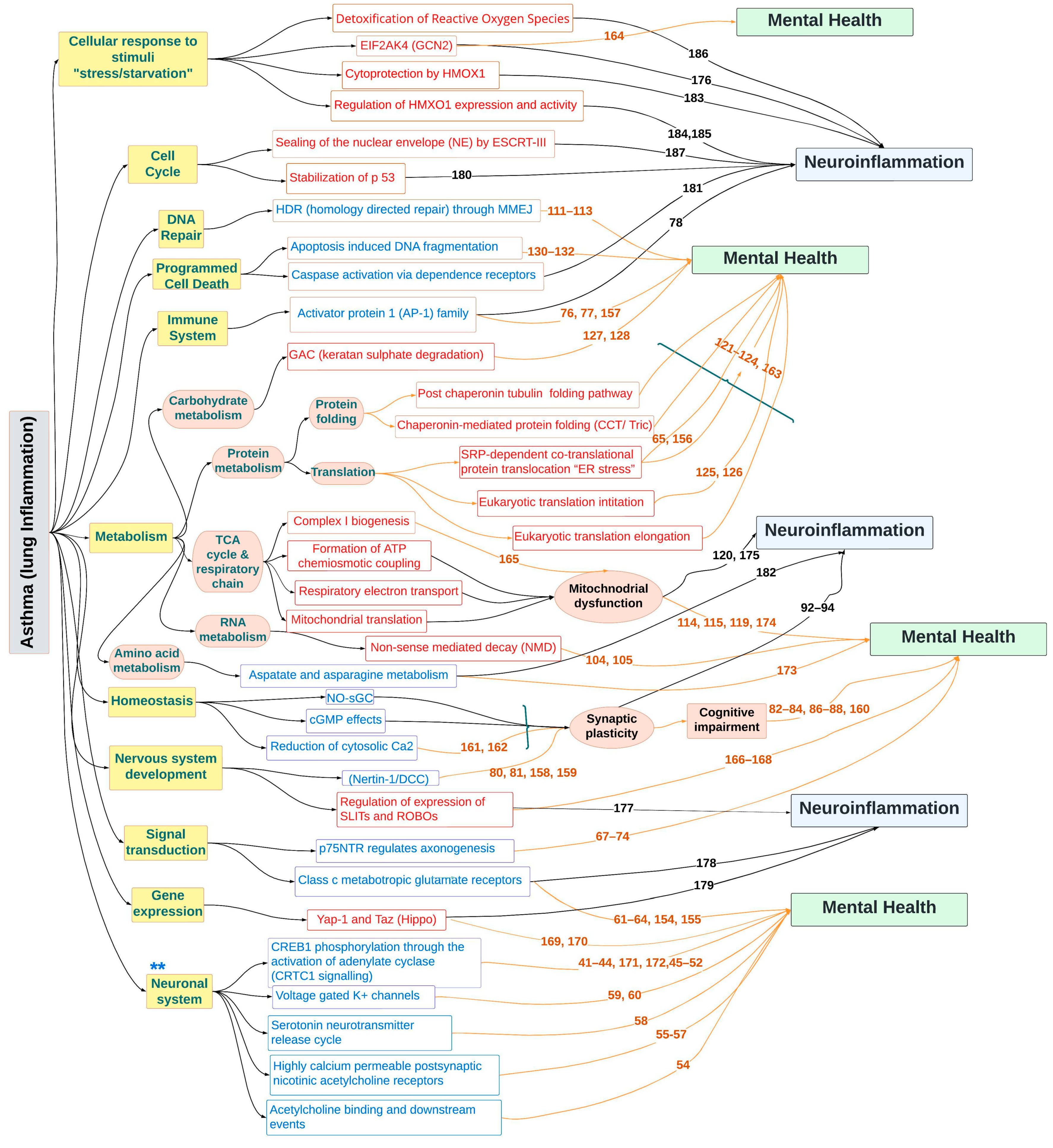
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Experimental Timeline
4.3. Pulmonary Histopathology
4.4. Brain Microdissection
4.5. RNA Extraction and Quality Control
4.6. RNA Transcriptome Sequencing
4.7. Bioinformatics Analysis
4.8. Literature Search
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goldsmith, D.R.; Rapaport, M.H.; Miller, B.J. A Meta-Analysis of Blood Cytokine Network Alterations in Psychiatric Patients: Comparisons between Schizophrenia, Bipolar Disorder and Depression. Mol. Psychiatry 2016, 21, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Y.; Ren, L.; Shao, Y.; Tao, W.; Dai, X.-J. Preexisting Mental Disorders Increase the Risk of COVID-19 Infection and Associated Mortality. Front. Public. Health 2021, 9, 684112. [Google Scholar] [CrossRef]
- Santoft, F.; Hedman-Lagerlöf, E.; Salomonsson, S.; Lindsäter, E.; Ljótsson, B.; Kecklund, G.; Lekander, M.; Andreasson, A. Inflammatory Cytokines in Patients with Common Mental Disorders Treated with Cognitive Behavior Therapy. Brain Behav. Immun. Health 2020, 3, 100045. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, L.; Sheng, C.; Cheng, Z.; Cui, L.; Li, M.; Zhao, Y.; Shi, T.; Yau, T.O.; Li, F.; et al. Increased Serum Levels of Cortisol and Inflammatory Cytokines in People with Depression. J. Nerv. Ment. Dis. 2019, 207, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Garner, M.; Holmes, C.; Osmond, C.; Teeling, J.; Lau, L.; Baldwin, D.S. Peripheral Inflammatory Cytokines and Immune Balance in Generalised Anxiety Disorder: Case-Controlled Study. Brain. Behav. Immun. 2017, 62, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.K.; Miller, B.J. Meta-Analysis of Cerebrospinal Fluid Cytokine and Tryptophan Catabolite Alterations in Psychiatric Patients: Comparisons between Schizophrenia, Bipolar Disorder, and Depression. Schizophr. Bull. 2018, 44, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.E.; Teixeira, A.L. Inflammation in Psychiatric Disorders: What Comes First? Ann. N. Y. Acad. Sci. 2019, 1437, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Kappelmann, N.; Lewis, G.; Dantzer, R.; Jones, P.B.; Khandaker, G.M. Antidepressant Activity of Anti-Cytokine Treatment: A Systematic Review and Meta-Analysis of Clinical Trials of Chronic Inflammatory Conditions. Mol. Psychiatry 2018, 23, 335–343. [Google Scholar] [CrossRef]
- Lee, C.-H.; Giuliani, F. The Role of Inflammation in Depression and Fatigue. Front. Immunol. 2019, 10, 1696. [Google Scholar] [CrossRef]
- Michopoulos, V.; Powers, A.; Gillespie, C.F.; Ressler, K.J.; Jovanovic, T. Inflammation in Fear-and Anxiety-Based Disorders: PTSD, GAD, and Beyond. Neuropsychopharmacology 2017, 42, 254–270. [Google Scholar] [CrossRef]
- Müller, N. Inflammation in Schizophrenia: Pathogenetic Aspects and Therapeutic Considerations. Schizophr. Bull. 2018, 44, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Muneer, A. Bipolar Disorder: Role of Inflammation and the Development of Disease Biomarkers. Psychiatry Investig. 2016, 13, 18–33. [Google Scholar] [CrossRef]
- Sun, Y.; Koyama, Y.; Shimada, S. Inflammation From Peripheral Organs to the Brain: How Does Systemic Inflammation Cause Neuroinflammation? Front. Aging Neurosci. 2022, 14, 903455. [Google Scholar] [CrossRef] [PubMed]
- Atlantis, E.; Fahey, P.; Cochrane, B.; Smith, S. Bidirectional Associations between Clinically Relevant Depression or Anxiety and COPD: A Systematic Review and Meta-Analysis. Chest 2013, 144, 766–777. [Google Scholar] [CrossRef] [PubMed]
- Dalal, A.A.; Shah, M.; Lunacsek, O.; Hanania, N.A. Clinical and Economic Burden of Depression/Anxiety in Chronic Obstructive Pulmonary Disease Patients within a Managed Care Population. COPD J. Chronic Obstr. Pulm. Dis. 2011, 8, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Yohannes, A.M.; Willgoss, T.G.; Baldwin, R.C.; Connolly, M.J. Depression and Anxiety in Chronic Heart Failure and Chronic Obstructive Pulmonary Disease: Prevalence, Relevance, Clinical Implications and Management Principles. Int. J. Geriatr. Psychiatry 2010, 25, 1209–1221. [Google Scholar] [CrossRef]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The Gut-Brain Axis: Interactions between Enteric Microbiota, Central and Enteric Nervous Systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar]
- Appleton, J. The Gut-Brain Axis: Influence of Microbiota on Mood and Mental Health. Integr. Med. (Encinitas) 2018, 17, 28–32. [Google Scholar]
- Gracie, D.J.; Hamlin, P.J.; Ford, A.C. The Influence of the Brain-Gut Axis in Inflammatory Bowel Disease and Possible Implications for Treatment. lancet. Gastroenterol. Hepatol. 2019, 4, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Vos, T.; Lim, S.S.; Abbafati, C.; Abbas, K.M.; Abbasi, M.; Abbasifard, M.; Abbasi-Kangevari, M.; Abbastabar, H.; Abd-Allah, F.; Abdelalim, A. Global Burden of 369 Diseases and Injuries in 204 Countries and Territories, 1990–2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Kharaba, Z.; Feghali, E.; El Husseini, F.; Sacre, H.; Abou Selwan, C.; Saadeh, S.; Hallit, S.; Jirjees, F.; AlObaidi, H.; Salameh, P.; et al. An Assessment of Quality of Life in Patients with Asthma Through Physical, Emotional, Social, and Occupational Aspects. A Cross-Sectional Study. Front. Public. Health 2022, 10, 883784. [Google Scholar] [CrossRef]
- Ali, R.; Ahmed, N.; Salman, M.; Daudpota, S.; Masroor, M.; Nasir, M. Assessment of Quality of Life in Bronchial Asthma Patients. Cureus 2020, 12, e10845. [Google Scholar] [CrossRef] [PubMed]
- Schatz, M.; Rosenwasser, L. The Allergic Asthma Phenotype. J. Allergy Clin. Immunol. Pract. 2014, 2, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Busse, W.W.; Lemanske, R.F.J. Asthma. N. Engl. J. Med. 2001, 344, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Peters, S.P. Airway Remodeling and Persistent Airway Obstruction in Asthma. J. Allergy Clin. Immunol. 1999, 104, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Burfeind, K.G.; Michaelis, K.A.; Marks, D.L. The Central Role of Hypothalamic Inflammation in the Acute Illness Response and Cachexia. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 54, pp. 42–52. [Google Scholar]
- Ambach, G.; Palkovits, M.; Szentagothai, J. Blood Supply of the Rat Hypothalamus. IV. Retrochiasmatic Area, Median Eminence, Arcuate Nucleus. Acta Morphol. Acad. Sci. Hung. 1976, 24, 93–119. [Google Scholar]
- Goldstein, D.S.; Kopin, I.J. Homeostatic Systems, Biocybernetics, and Autonomic Neuroscience. Auton. Neurosci. 2017, 208, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Bao, A.M.; Swaab, D.F. The Human Hypothalamus in Mood Disorders: The HPA Axis in the Center. IBRO Rep. 2019, 6, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.G.; Dobrowolny, H.; Bogerts, B.; Keilhoff, G.; Steiner, J. The Hypothalamus and Neuropsychiatric Disorders: Psychiatry Meets Microscopy. Cell Tissue Res. 2019, 375, 243–258. [Google Scholar] [CrossRef]
- Schindler, S.; Schmidt, L.; Stroske, M.; Storch, M.; Anwander, A.; Trampel, R.; Strauß, M.; Hegerl, U.; Geyer, S.; Schönknecht, P. Hypothalamus Enlargement in Mood Disorders. Acta Psychiatr. Scand. 2019, 139, 56–67. [Google Scholar] [CrossRef]
- Cernackova, A.; Durackova, Z.; Trebaticka, J.; Mravec, B. Neuroinflammation and Depressive Disorder: The Role of the Hypothalamus. J. Clin. Neurosci. 2020, 75, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Tafet, G.E.; Nemeroff, C.B. The Links between Stress and Depression: Psychoneuroendocrinological, Genetic, and Environmental Interactions. J. Neuropsychiatry Clin. Neurosci. 2016, 28, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Casaro, M.; Souza, V.R.; Oliveira, F.A.; Ferreira, C.M. OVA-Induced Allergic Airway Inflammation Mouse Model. Methods Mol. Biol. 2019, 1916, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Nials, A.T.; Uddin, S. Mouse Models of Allergic Asthma: Acute and Chronic Allergen Challenge. Dis. Model. Mech. 2008, 1, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.R.; Khuman, V.; Beladiya, J.V.; Chaudagar, K.K.; Mehta, A.A. An Experimental Model of Asthma in Rats Using Ovalbumin and Lipopolysaccharide Allergens. Heliyon 2019, 5, e02864. [Google Scholar] [CrossRef]
- Murphy, M.; Brown, G.; Wallin, C.; Tatusova, T.; Pruitt, K.; Murphy, T.; Maglott, D. Gene Help: Integrated Access to Genes of Genomes in the Reference Sequence Collection. In Gene Help [Internet]; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2021. [Google Scholar]
- Schou, T.M.; Joca, S.; Wegener, G.; Bay-Richter, C. Psychiatric and Neuropsychiatric Sequelae of COVID-19–A Systematic Review. Brain. Behav. Immun. 2021, 97, 328–348. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Lipsitz, O.; Nasri, F.; Lui, L.M.W.; Gill, H.; Phan, L.; Chen-Li, D.; Iacobucci, M.; Ho, R.; Majeed, A.; et al. Impact of COVID-19 Pandemic on Mental Health in the General Population: A Systematic Review. J. Affect. Disord. 2020, 277, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.G.; Kim, J.-H.; Park, J.-Y.; Hwang, Y.I.; Jang, S.H.; Jung, K.-S. Association Between Asthma and Depression: A National Cohort Study. J. Allergy Clin. Immunol. Pract. 2019, 7, 1239–1245.e1. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, Y.; Pandey, G.N. Adenylyl Cyclase-CyclicAMP Signaling in Mood Disorders: Role of the Crucial Phosphorylating Enzyme Protein Kinase A. Neuropsychiatr. Dis. Treat. 2008, 4, 161–176. [Google Scholar] [CrossRef]
- Breuillaud, L.; Rossetti, C.; Meylan, E.M.; Mérinat, C.; Halfon, O.; Magistretti, P.J.; Cardinaux, J.-R. Deletion of CREB-Regulated Transcription Coactivator 1 Induces Pathological Aggression, Depression-Related Behaviors, and Neuroplasticity Genes Dysregulation in Mice. Biol. Psychiatry 2012, 72, 528–536. [Google Scholar] [CrossRef]
- Sulser, F. The Role of CREB and Other Transcription Factors in the Pharmacotherapy and Etiology of Depression. Ann. Med. 2002, 34, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Meylan, E.M.; Halfon, O.; Magistretti, P.J.; Cardinaux, J.-R. The HDAC Inhibitor SAHA Improves Depressive-like Behavior of CRTC1-Deficient Mice: Possible Relevance for Treatment-Resistant Depression. Neuropharmacology 2016, 107, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, C.; Sciarra, D.; Petit, J.-M.; Eap, C.B.; Halfon, O.; Magistretti, P.J.; Boutrel, B.; Cardinaux, J.-R. Gender-Specific Alteration of Energy Balance and Circadian Locomotor Activity in the Crtc1 Knockout Mouse Model of Depression. Transl. Psychiatry 2017, 7, 1269. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Xue, X.; Liu, S.; Feng, C.; Zhang, H.; Zhang, S.; Ren, Y.; Ma, H.; Dong, Y.; Li, H. CRTC1 Signaling Involvement in Depression-like Behavior of Prenatally Stressed Offspring Rat. Behav. Brain Res. 2021, 399, 113000. [Google Scholar] [CrossRef]
- Koch, J.M.; Kell, S.; Hinze-Selch, D.; Aldenhoff, J.B. Changes in CREB-Phosphorylation during Recovery from Major Depression. J. Psychiatr. Res. 2002, 36, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Cherix, A.; Poitry-Yamate, C.; Lanz, B.; Zanoletti, O.; Grosse, J.; Sandi, C.; Gruetter, R.; Cardinaux, J.-R. Deletion of Crtc1 Leads to Hippocampal Neuroenergetic Impairments Associated with Depressive-like Behavior. Mol. Psychiatry 2022, 27, 4485–4501. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, C.; Cherix, A.; Guiraud, L.F.; Cardinaux, J.-R. New Insights Into the Pivotal Role of CREB-Regulated Transcription Coactivator 1 in Depression and Comorbid Obesity. Front. Mol. Neurosci. 2022, 15, 810641. Available online: https://www.frontiersin.org/articles/10.3389/fnmol.2022.810641 (accessed on 23 February 2022). [CrossRef] [PubMed]
- Young, L.T.; Dowlatshahi, D.; MacQueen, G.M.; Wang, J.F. Increased Temporal Cortex CREB Concentrations and Antidepressant Treatment in Major Depression. Lancet 1998, 352, 1754–1755. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Yamamoto, M.; Ozawa, H.; Riederer, P.; Saito, T. Reduced Phosphorylation of Cyclic AMP-Responsive Element Binding Protein in the Postmortem Orbitofrontal Cortex of Patients with Major Depressive Disorder. J. Neural Transm. 2003, 110, 671–680. [Google Scholar] [CrossRef]
- Dwivedi, Y.; Rizavi, H.S.; Shukla, P.K.; Lyons, J.; Faludi, G.; Palkovits, M.; Sarosi, A.; Conley, R.R.; Roberts, R.C.; Tamminga, C.A. Protein Kinase A in Postmortem Brain of Depressed Suicide Victims: Altered Expression of Specific Regulatory and Catalytic Subunits. Biol. Psychiatry 2004, 55, 234–243. [Google Scholar] [CrossRef]
- Dulawa, S.C.; Janowsky, D.S. Cholinergic Regulation of Mood: From Basic and Clinical Studies to Emerging Therapeutics. Mol. Psychiatry 2019, 24, 694–709. [Google Scholar] [CrossRef]
- Higley, M.J.; Picciotto, M.R. Neuromodulation by Acetylcholine: Examples from Schizophrenia and Depression. Curr. Opin. Neurobiol. 2014, 29, 88–95. [Google Scholar] [CrossRef]
- Bliss, T.V.P.; Cooke, S.F. Long-Term Potentiation and Long-Term Depression: A Clinical Perspective. Clinics 2011, 66 (Suppl. S1), 3–17. [Google Scholar] [CrossRef]
- Duman, R.S.; Aghajanian, G.K. Synaptic Dysfunction in Depression: Potential Therapeutic Targets. Science 2012, 338, 68–72. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, X.; Shi, P.; Yuan, J.; Jia, Q.; Pi, C.; Chen, T.; Xiong, L.; Chen, J.; Tang, J.; et al. A7 Nicotinic Acetylcholine Receptor: A Key Receptor in the Cholinergic Anti-Inflammatory Pathway Exerting an Antidepressant Effect. J. Neuroinflamm. 2023, 20, 84. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Feng, M.; Long, Z.; Ma, J.; Peng, X.; He, G. Allergic Asthma-Induced Cognitive Impairment Is Alleviated by Dexamethasone. Front. Pharmacol. 2021, 12, 680815. [Google Scholar] [CrossRef] [PubMed]
- Della Vecchia, S.; Marchese, M.; Santorelli, F.M.; Sicca, F. Kir4.1 Dysfunction in the Pathophysiology of Depression: A Systematic Review. Cells 2021, 10, 2628. [Google Scholar] [CrossRef]
- Imbrici, P.; Camerino, D.C.; Tricarico, D. Major Channels Involved in Neuropsychiatric Disorders and Therapeutic Perspectives. Front. Genet. 2013, 4, 76. [Google Scholar] [CrossRef]
- Pantazatos, S.P.; Huang, Y.Y.; Rosoklija, G.B.; Dwork, A.J.; Arango, V.; Mann, J.J. Whole-Transcriptome Brain Expression and Exon-Usage Profiling in Major Depression and Suicide: Evidence for Altered Glial, Endothelial and ATPase Activity. Mol. Psychiatry 2017, 22, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Sequeira, A.; Mamdani, F.; Ernst, C.; Vawter, M.P.; Bunney, W.E.; Lebel, V.; Rehal, S.; Klempan, T.; Gratton, A.; Benkelfat, C. Global Brain Gene Expression Analysis Links Glutamatergic and GABAergic Alterations to Suicide and Major Depression. PLoS ONE 2009, 4, e6585. [Google Scholar] [CrossRef]
- Deschwanden, A. Reduced Metabotropic Glutamate Receptor 5 Density in Major Depression Determined by [11 C]ABP688 PET and Postmortem Study. Am. J. Psychiatry 2011, 168, 727–734. [Google Scholar] [CrossRef]
- Moriguchi, S.; Takamiya, A.; Noda, Y.; Horita, N.; Wada, M.; Tsugawa, S.; Plitman, E.; Sano, Y.; Tarumi, R.; ElSalhy, M.; et al. Glutamatergic Neurometabolite Levels in Major Depressive Disorder: A Systematic Review and Meta-Analysis of Proton Magnetic Resonance Spectroscopy Studies. Mol. Psychiatry 2019, 24, 952–964. [Google Scholar] [CrossRef]
- Patel, S.; Howard, D.; Man, A.; Schwartz, D.; Jee, J.; Felsky, D.; Pausova, Z.; Paus, T.; French, L. Donor-Specific Transcriptomic Analysis of Alzheimer’s Disease-Associated Hypometabolism Highlights a Unique Donor, Ribosomal Proteins and Microglia. eNeuro 2020, 7. [Google Scholar] [CrossRef]
- Becker, K.; Cana, A.; Baumgärtner, W.; Spitzbarth, I. P75 Neurotrophin Receptor: A Double-Edged Sword in Pathology and Regeneration of the Central Nervous System. Vet. Pathol. 2018, 55, 786–801. [Google Scholar] [CrossRef]
- Kunugi, H.; Hashimoto, R.; Yoshida, M.; Tatsumi, M.; Kamijima, K. A Missense Polymorphism (S205L) of the Low-Affinity Neurotrophin Receptor P75NTR Gene Is Associated with Depressive Disorder and Attempted Suicide. Am. J. Med. Genet. Neuropsychiatr. Genet. 2004, 129, 44–46. [Google Scholar] [CrossRef]
- Kunugi, H.; Hori, H.; Adachi, N.; Numakawa, T. Interface between Hypothalamic-Pituitary-Adrenal Axis and Brain-Derived Neurotrophic Factor in Depression. Psychiatry Clin. Neurosci. 2010, 64, 447–459. [Google Scholar] [CrossRef]
- Catts, V.S.; Al-Menhali, N.; Burne, T.H.J.; Colditz, M.J.; Coulson, E.J. The P75 Neurotrophin Receptor Regulates Hippocampal Neurogenesis and Related Behaviours. Eur. J. Neurosci. 2008, 28, 883–892. [Google Scholar] [CrossRef]
- Martinowich, K.; Schloesser, R.J.; Lu, Y.; Jimenez, D.V.; Paredes, D.; Greene, J.S.; Greig, N.H.; Manji, H.K.; Lu, B. Roles of P75NTR, Long-Term Depression, and Cholinergic Transmission in Anxiety and Acute Stress Coping. Biol. Psychiatry 2012, 71, 75–83. [Google Scholar] [CrossRef]
- Zanin, J.P.; Montroull, L.E.; Volosin, M.; Friedman, W.J. The P75 Neurotrophin Receptor Facilitates TrkB Signaling and Function in Rat Hippocampal Neurons. Front. Cell. Neurosci. 2019, 13, 485. [Google Scholar] [CrossRef]
- Pérez, V.; Bermedo-Garcia, F.; Zelada, D.; Court, F.A.; Pérez, M.Á.; Fuenzalida, M.; Ábrigo, J.; Cabello-Verrugio, C.; Moya-Alvarado, G.; Tapia, J.C.; et al. The P75NTR Neurotrophin Receptor Is Required to Organize the Mature Neuromuscular Synapse by Regulating Synaptic Vesicle Availability. Acta Neuropathol. Commun. 2019, 7, 147. [Google Scholar] [CrossRef]
- Price, R.B.; Duman, R. Neuroplasticity in Cognitive and Psychological Mechanisms of Depression: An Integrative Model. Mol. Psychiatry 2020, 25, 530–543. [Google Scholar] [CrossRef]
- Yuan, N.; Chen, Y.; Xia, Y.; Dai, J.; Liu, C. Inflammation-Related Biomarkers in Major Psychiatric Disorders: A Cross-Disorder Assessment of Reproducibility and Specificity in 43 Meta-Analyses. Transl. Psychiatry 2019, 9, 233. [Google Scholar] [CrossRef] [PubMed]
- Schulman, H. Intracellular Signaling. In Fundamental Neuroscience; Squire, L.R., Berg, D., Bloom, F.E., du Lac, S., Ghosh, A., Spitzer, N.C.B.T.-F.N., Fourth, E., Eds.; Elsevier: San Diego, CA, USA, 2013; pp. 189–209. Available online: https://www.sciencedirect.com/science/article/pii/B978012397179100004X (accessed on 23 February 2022).
- Appelbaum, L.G.; Shenasa, M.A.; Stolz, L.; Daskalakis, Z. Synaptic Plasticity and Mental Health: Methods, Challenges and Opportunities. Neuropsychopharmacology 2023, 48, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Chottekalapanda, R.U.; Kalik, S.; Gresack, J.; Ayala, A.; Gao, M.; Wang, W.; Meller, S.; Aly, A.; Schaefer, A.; Greengard, P. AP-1 Controls the P11-Dependent Antidepressant Response. Mol. Psychiatry 2020, 25, 1364–1381. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-J.; Li, Y.-L.; Zhu, Z.-L.; Jia, D.-M.; Fan, M.-L.; Li, T.; Wang, X.-J.; Li, Z.-G.; Ma, H.-S. Inhibition of Activator Protein 1 Attenuates Neuroinflammation and Brain Injury after Experimental Intracerebral Hemorrhage. CNS Neurosci. Ther. 2019, 25, 1182–1188. [Google Scholar] [CrossRef] [PubMed]
- Boyer, N.P.; Gupton, S.L. Revisiting Netrin-1: One Who Guides (Axons). Front. Cell. Neurosci. 2018, 12, 221. [Google Scholar] [CrossRef] [PubMed]
- Torres-Berrío, A.; Hernandez, G.; Nestler, E.J.; Flores, C. The Netrin-1/DCC Guidance Cue Pathway as a Molecular Target in Depression: Translational Evidence. Biol. Psychiatry 2020, 88, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Vosberg, D.E.; Leyton, M.; Flores, C. The Netrin-1/DCC Guidance System: Dopamine Pathway Maturation and Psychiatric Disorders Emerging in Adolescence. Mol. Psychiatry 2020, 25, 297–307. [Google Scholar] [CrossRef]
- Arancio, O.; Antonova, I.; Gambaryan, S.; Lohmann, S.M.; Wood, J.S.; Lawrence, D.S.; Hawkins, R.D. Presynaptic Role of CGMP-Dependent Protein Kinase during Long-Lasting Potentiation. J. Neurosci. Off. J. Soc. Neurosci. 2001, 21, 143–149. [Google Scholar] [CrossRef]
- Bernstein, H.G.; Stanarius, A.; Baumann, B.; Henning, H.; Krell, D.; Danos, P.; Falkai, P.; Bogerts, B. Nitric Oxide Synthase-Containing Neurons in the Human Hypothalamus: Reduced Number of Immunoreactive Cells in the Paraventricular Nucleus of Depressive Patients and Schizophrenics. Neuroscience 1998, 83, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.-G.; Heinemann, A.; Krell, D.; Mawrin, C.; Bielau, H.; Danos, P.; Diekmann, S.; Keilhoff, G.; Bogerts, B.; Baumann, B. Further Immunohistochemical Evidence for Impaired NO Signaling in the Hypothalamus of Depressed Patients. Ann. N. Y. Acad. Sci. 2002, 973, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Domek-Łopacińska, K.; Strosznajder, J.B. Cyclic GMP Metabolism and Its Role in Brain Physiology. J. Physiol. Pharmacol. 2005, 56 (Suppl. 2), 15–34. [Google Scholar] [PubMed]
- Joca, S.R.L.; Sartim, A.G.; Roncalho, A.L.; Diniz, C.F.A.; Wegener, G. Nitric Oxide Signalling and Antidepressant Action Revisited. Cell Tissue Res. 2019, 377, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Walia, V.; Garg, C.; Garg, M. NO-SGC-CGMP Signaling Influence the Anxiolytic like Effect of Lithium in Mice in Light and Dark Box and Elevated plus Maze. Brain Res. 2019, 1704, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.; Shuman, M.; Duncan, E. An Emerging Role of CGMP in the Treatment of Schizophrenia: A Review. Schizophr. Res. 2016, 170, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Borghans, L.G.J.M.; Sambeth, A.; Prickaerts, J.; Ramaekers, J.G.; Blokland, A. The Effects of the Soluble Guanylate Cyclase Stimulator Riociguat on Memory Performance in Healthy Volunteers with a Biperiden-Induced Memory Impairment. Psychopharmacology 2018, 235, 2407–2416. [Google Scholar] [CrossRef]
- Nelissen, E.; Argyrousi, E.K.; Van Goethem, N.P.; Zhao, F.; Hines, C.D.G.; Swaminath, G.; Gerisch, M.; Hueser, J.; Sandner, P.; Prickaerts, J. Soluble Guanylate Cyclase Stimulator Vericiguat Enhances Long-Term Memory in Rats without Altering Cerebral Blood Volume. Biomedicines 2021, 9, 1047. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, E.; Possemis, N.; Van Goethem, N.P.; Schepers, M.; Mulder-Jongen, D.A.J.; Dietz, L.; Janssen, W.; Gerisch, M.; Hüser, J.; Sandner, P.; et al. The SGC Stimulator BAY-747 and Activator Runcaciguat Can Enhance Memory In Vivo via Differential Hippocampal Plasticity Mechanisms. Sci. Rep. 2022, 12, 3589. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.S.; Liu, G.; Jacobson, S.; Bernier, S.G.; Tobin, J.V.; Schwartzkopf, C.D.; Atwater, E.; Lonie, E.; Rivers, S.; Carvalho, A.; et al. The CNS-Penetrant Soluble Guanylate Cyclase Stimulator CYR119 Attenuates Markers of Inflammation in the Central Nervous System. J. Neuroinflamm. 2021, 18, 213. [Google Scholar] [CrossRef]
- Sama, D.M.; Norris, C.M. Calcium Dysregulation and Neuroinflammation: Discrete and Integrated Mechanisms for Age-Related Synaptic Dysfunction. Ageing Res. Rev. 2013, 12, 982–995. [Google Scholar] [CrossRef]
- Russwurm, M.; Russwurm, C.; Koesling, D.; Mergia, E. NO/CGMP: The Past, the Present, and the Future. Methods Mol. Biol. 2013, 1020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Hwang, C.J.; Lee, H.P.; Kim, C.S.; Son, D.J.; Ham, Y.W.; Hellström, M.; Han, S.B.; Kim, H.S.; Park, E.K.; et al. Inhibitory Effect of Punicalagin on Lipopolysaccharide-Induced Neuroinflammation, Oxidative Stress and Memory Impairment via Inhibition of Nuclear Factor-KappaB. Neuropharmacology 2017, 117, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Thingore, C.; Kshirsagar, V.; Juvekar, A. Amelioration of Oxidative Stress and Neuroinflammation in Lipopolysaccharide-Induced Memory Impairment Using Rosmarinic Acid in Mice. Metab. Brain Dis. 2021, 36, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Shin, I.S.; Seo, C.S.; Ha, H.; Shin, H.K. Antiasthmatic Effects of Gleditsia Sinensis in an Ovalbumin-Induced Murine Model of Asthma. Int. J. Toxicol. 2011, 30, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.C.; Whelan, C.J.; Purcell, W.M. Reactive Oxygen Species Generation and Histamine Release by Activated Mast Cells: Modulation by Nitric Oxide Synthase Inhibition. Br. J. Pharmacol. 1999, 128, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Nickless, A.; Bailis, J.M.; You, Z. Control of Gene Expression through the Nonsense-Mediated RNA Decay Pathway. Cell Biosci. 2017, 7, 26. [Google Scholar] [CrossRef]
- Goetz, A.E.; Wilkinson, M. Stress and the Nonsense-Mediated RNA Decay Pathway. Cell. Mol. Life Sci. 2017, 74, 3509–3531. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gong, J.; Zeng, H.; Liu, R.; Jin, B.; Chen, L.; Wang, Q. Lipopolysaccharide Activates the Unfolded Protein Response in Human Periodontal Ligament Fibroblasts. J. Periodontol. 2016, 87, e75–e81. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Jeong, J.S.; Kim, S.R.; Park, S.Y.; Chae, H.J.; Lee, Y.C. Inhibition of Endoplasmic Reticulum Stress Alleviates Lipopolysaccharide- Induced Lung Inflammation through Modulation of NF-ΚB/HIF-1α Signaling Pathway. Sci. Rep. 2013, 3, 1142. [Google Scholar] [CrossRef]
- Kim, S.R.; Kim, D.I.; Kang, M.R.; Lee, K.S.; Park, S.Y.; Jeong, J.S.; Lee, Y.C. Endoplasmic Reticulum Stress Influences Bronchial Asthma Pathogenesis by Modulating Nuclear Factor ΚB Activation. J. Allergy Clin. Immunol. 2013, 132, 1397–1408.e11. [Google Scholar] [CrossRef]
- Johnson, J.L.; Stoica, L.; Liu, Y.; Zhu, P.J.; Bhattacharya, A.; Buffington, S.A.; Huq, R.; Eissa, N.T.; Larsson, O.; Porse, B.T.; et al. Inhibition of Upf2-Dependent Nonsense-Mediated Decay Leads to Behavioral and Neurophysiological Abnormalities by Activating the Immune Response. Neuron 2019, 104, 665–679.e8. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R.; Wilkinson, M.F. Nonsense-Mediated RNA Decay in the Brain: Emerging Modulator of Neural Development and Disease. Nat. Rev. Neurosci. 2018, 19, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Plumb, R.; Zhang, Z.R.; Appathurai, S.; Mariappan, M. A Functional Link between the Co-Translational Protein Translocation Pathway and the UPR. Elife 2015, 4, 2–27. [Google Scholar] [CrossRef] [PubMed]
- Muneer, A.; Shamsher Khan, R.M. Endoplasmic Reticulum Stress: Implications for Neuropsychiatric Disorders. Chonnam Med. J. 2019, 55, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Somyajit, K.; Spies, J.; Coscia, F.; Kirik, U.; Rask, M.B.; Lee, J.H.; Neelsen, K.J.; Mund, A.; Jensen, L.J.; Paull, T.T.; et al. Homology-Directed Repair Protects the Replicating Genome from Metabolic Assaults. Dev. Cell 2021, 56, 461–477.e7. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Wang, H.; Peng, Y.; Hu, Y.; Wang, H.; Zhang, X.; Chen, Q.; Bedford, J.S.; Dewhirst, M.W.; Li, C.Y. A Unique Role of the DNA Fragmentation Factor in Maintaining Genomic Stability. Proc. Natl. Acad. Sci. USA 2006, 103, 1504–1509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Xu, M. DNA Fragmentation in Apoptosis. Cell Res. 2000, 10, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Raza, M.U.; Tufan, T.; Wang, Y.; Hill, C.; Zhu, M.-Y. DNA Damage in Major Psychiatric Diseases. Neurotox. Res. 2016, 30, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Buttner, N.; Bhattacharyya, S.; Walsh, J.; Benes, F.M. DNA Fragmentation Is Increased in Non-GABAergic Neurons in Bipolar Disorder but Not in Schizophrenia. Schizophr. Res. 2007, 93, 33–41. [Google Scholar] [CrossRef]
- Mustak, M.S.; Hegde, M.L.; Dinesh, A.; Britton, G.B.; Berrocal, R.; Subba Rao, K.; Shamasundar, N.M.; Rao, K.S.J.; Sathyanarayana Rao, T.S. Evidence of Altered DNA Integrity in the Brain Regions of Suicidal Victims of Bipolar Depression. Indian. J. Psychiatry 2010, 52, 220–228. [Google Scholar] [CrossRef]
- Gong, Q.; Yan, X.-J.; Lei, F.; Wang, M.-L.; He, L.-L.; Luo, Y.-Y.; Gao, H.-W.; Feng, Y.-L.; Yang, S.-L.; Li, J. Proteomic Profiling of the Neurons in Mice with Depressive-like Behavior Induced by Corticosterone and the Regulation of Berberine: Pivotal Sites of Oxidative Phosphorylation. Mol. Brain 2019, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Ke, S.; Wang, Q.; Zhuang, T.; Xia, C.; Xu, Y.; Yang, L.; Zhou, M. Energy Metabolism in Major Depressive Disorder: Recent Advances from Omics Technologies and Imaging. Biomed. Pharmacother. 2021, 141, 111869. [Google Scholar] [CrossRef]
- Kuffner, K.; Triebelhorn, J.; Meindl, K.; Benner, C.; Manook, A.; Sudria-Lopez, D.; Siebert, R.; Nothdurfter, C.; Baghai, T.C.; Drexler, K. Major Depressive Disorder Is Associated with Impaired Mitochondrial Function in Skin Fibroblasts. Cells 2020, 9, 884. [Google Scholar] [CrossRef] [PubMed]
- Camandola, S.; Mattson, M.P. Brain Metabolism in Health, Aging, and Neurodegeneration. EMBO J. 2017, 36, 1474–1492. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.C.B.; Dammer, E.B.; Duong, D.M.; Ping, L.; Zhou, M.; Yin, L.; Higginbotham, L.A.; Guajardo, A.; White, B.; Troncoso, J.C. Large-Scale Proteomic Analysis of Alzheimer’s Disease Brain and Cerebrospinal Fluid Reveals Early Changes in Energy Metabolism Associated with Microglia and Astrocyte Activation. Nat. Med. 2020, 26, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Klempan, T.A.; Sequeira, A.; Canetti, L.; Lalovic, A.; Ernst, C.; Ffrench-Mullen, J.; Turecki, G. Altered Expression of Genes Involved in ATP Biosynthesis and GABAergic Neurotransmission in the Ventral Prefrontal Cortex of Suicides with and without Major Depression. Mol. Psychiatry 2009, 14, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-M.; Liu, N.; Qin, Z.-H.; Wang, Y. Mitochondrial-Derived Damage-Associated Molecular Patterns Amplify Neuroinflammation in Neurodegenerative Diseases. Acta Pharmacol. Sin. 2022, 43, 2439–2447. [Google Scholar] [CrossRef]
- Soto, C.; Pritzkow, S. Protein Misfolding, Aggregation, and Conformational Strains in Neurodegenerative Diseases. Nat. Neurosci. 2018, 21, 1332–1340. [Google Scholar] [CrossRef]
- Bosco, D.A. Translation Dysregulation in Neurodegenerative Disorders. Proc. Natl. Acad. Sci. USA 2018, 115, 12842–12844. [Google Scholar] [CrossRef] [PubMed]
- Jishi, A.; Qi, X.; Miranda, H.C. Implications of MRNA Translation Dysregulation for Neurological Disorders. Semin. Cell Dev. Biol. 2021, 114, 11–19. [Google Scholar] [CrossRef]
- Laguesse, S.; Ron, D. Protein Translation and Psychiatric Disorders. Neuroscientist 2020, 26, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Valles, A.; Haji, N.; De Gregorio, D.; Matta-Camacho, E.; Eslamizade, M.J.; Popic, J.; Sharma, V.; Cao, R.; Rummel, C.; Tanti, A.; et al. Translational Control of Depression-like Behavior via Phosphorylation of Eukaryotic Translation Initiation Factor 4E. Nat. Commun. 2018, 9, 2459. [Google Scholar] [CrossRef] [PubMed]
- Amorim, I.S.; Kedia, S.; Kouloulia, S.; Simbriger, K.; Gantois, I.; Jafarnejad, S.M.; Li, Y.; Kampaite, A.; Pooters, T.; Romanò, N.; et al. Loss of EIF4E Phosphorylation Engenders Depression-like Behaviors via Selective MRNA Translation. J. Neurosci. 2018, 38, 2118–2133. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, D.; Sugahara, K.; Kwok, J.C.F. Glycosaminoglycans and Glycomimetics in the Central Nervous System. Molecules 2015, 20, 3527–3548. [Google Scholar] [CrossRef]
- Huynh, M.B.; Ouidja, M.O.; Chantepie, S.; Carpentier, G.; Maïza, A.; Zhang, G.; Vilares, J.; Raisman-Vozari, R.; Papy-Garcia, D. Glycosaminoglycans from Alzheimer’s Disease Hippocampus Have Altered Capacities to Bind and Regulate Growth Factors Activities and to Bind Tau. PLoS ONE 2019, 14, e0209573. [Google Scholar] [CrossRef]
- Mazgaeen, L.; Gurung, P. Recent Advances in Lipopolysaccharide Recognition Systems. Int. J. Mol. Sci. 2020, 21, 379. [Google Scholar] [CrossRef] [PubMed]
- Cohn, L.; Homer, R.J.; Marinov, A.; Rankin, J.; Bottomly, K. Induction of Airway Mucus Production by T Helper 2 (Th2) Cells: A Critical Role for Interleukin 4 in Cell Recruitment but Not Mucus Production. J. Exp. Med. 1997, 186, 1737–1747. [Google Scholar] [CrossRef]
- Harker, J.A.; Lloyd, C.M. T Helper 2 Cells in Asthma. J. Exp. Med. 2023, 220, e20221094. [Google Scholar] [CrossRef]
- Cemerski, S.; Shaw, A. Immune Synapses in T-Cell Activation. Curr. Opin. Immunol. 2006, 18, 298–304. [Google Scholar] [CrossRef]
- Haspeslagh, E.; Debeuf, N.; Hammad, H.; Lambrecht, B.N. Murine Models of Allergic Asthma. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1559, pp. 121–136. [Google Scholar] [CrossRef]
- Shin, H.S.; Chun, H.R.; Na, H.Y.; Sohn, M.; Ryu, S.H.; Choi, W.; In, B.; Park, J.S.; Park, S.; Park, C.G. Distinct Effects of Different Adjuvants in the Mouse Model of Allergic Airway Inflammation. Asian Pac. J. Allergy Immunol. 2022, 40, 111–120. [Google Scholar] [CrossRef]
- Kool, M.; Soullié, T.; van Nimwegen, M.; Willart, M.A.M.; Muskens, F.; Jung, S.; Hoogsteden, H.C.; Hammad, H.; Lambrecht, B.N. Alum Adjuvant Boosts Adaptive Immunity by Inducing Uric Acid and Activating Inflammatory Dendritic Cells. J. Exp. Med. 2008, 205, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.A.; Austen, K.F. No Audible Wheezing: Nuggets and Conundrums from Mouse Asthma Models. J. Exp. Med. 2005, 201, 1869–1873. [Google Scholar] [CrossRef]
- Zhong, W.; Su, W.; Zhang, Y.; Liu, Q.; Wu, J.; Di, C.; Zhang, Z.; Xia, Z. Basophils as a Primary Inducer of the T Helper Type 2 Immunity in Ovalbumin-induced Allergic Airway Inflammation. Immunology 2014, 142, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, T.W.; Sebastian, M.N.; Battaglia, D.M.; Foster, T.P.; Cormier, S.A.; Nichols, C.D. 5-HT2 Receptor Activation Alleviates Airway Inflammation and Structural Remodeling in a Chronic Mouse Asthma Model. Life Sci. 2019, 236, 116790. [Google Scholar] [CrossRef] [PubMed]
- Gomez, C.D.; Read, J.; Acharjee, S.; Pittman, Q.J. Early Life Inflammation Increases CA1 Pyramidal Neuron Excitability in a Sex and Age Dependent Manner through a Chloride Homeostasis Disruption. J. Neurosci. 2019, 39, 7244–7259. [Google Scholar] [CrossRef]
- Bancroft, J.D.; Gamble, M. Theory and Practice of Histological Techniques; Elsevier Health Sciences: Philadelphia, PA, USA, 2008. [Google Scholar]
- Cataldo, D.D.; Tournoy, K.G.; Vermaelen, K.; Munaut, C.; Foidart, J.-M.; Louis, R.; Noël, A.; Pauwels, R.A. Matrix Metalloproteinase-9 Deficiency Impairs Cellular Infiltration and Bronchial Hyperresponsiveness during Allergen-Induced Airway Inflammation. Am. J. Pathol. 2002, 161, 491–498. [Google Scholar] [CrossRef]
- Curtis, J.L.; Byrd, P.K.; Warnock, M.L.; Kaltreider, H.B. Requirement of CD4-Positive T Cells for Cellular Recruitment to the Lungs of Mice in Response to a Particulate Intratracheal Antigen. J. Clin. Invest. 1991, 88, 1244–1254. [Google Scholar] [CrossRef]
- Franklin, K.B.J.; Paxinos, G. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates, Compact: The Coronal Plates and Diagrams; Elsevier Academic Press: San Diego, CA, USA, 2019. [Google Scholar]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I. GENCODE 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- Andrews, S. Babraham Bioinformatics-FastQC a Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 23 February 2022).
- Jiang, H.; Lei, R.; Ding, S.-W.; Zhu, S. Skewer: A Fast and Accurate Adapter Trimmer for next-Generation Sequencing Paired-End Reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Dolgalev, I. MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format [R Package Msigdbr Version 7.4. 1]. Comprehensive R Archive Network (CRAN). 2020. Available online: https://cran.r-project.org/web/packages/msigdbr/index.html (accessed on 23 February 2022).
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Kaspi, A.; Ziemann, M. Mitch: Multi-Contrast Pathway Enrichment for Multi-Omics and Single-Cell Profiling Data. BMC Genom. 2020, 21, 447. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Peterlik, D.; Flor, P.J.; Uschold-Schmidt, N. The Emerging Role of Metabotropic Glutamate Receptors in the Pathophysiology of Chronic Stress-Related Disorders. Curr. Neuropharmacol. 2016, 14, 514–539. [Google Scholar] [CrossRef] [PubMed]
- Maksymetz, J.; Moran, S.P.; Conn, P.J. Targeting Metabotropic Glutamate Receptors for Novel Treatments of Schizophrenia. Mol. Brain 2017, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, M.; Kowalczyk, E.; Kwiatkowski, P.; Łopusiewicz, Ł.; Talarowska, M.; Sienkiewicz, M. Cellular Response to Unfolded Proteins in Depression. Life 2021, 11, 1376. [Google Scholar] [CrossRef]
- Malki, K.; Pain, O.; Tosto, M.G.; Du Rietz, E.; Carboni, L.; Schalkwyk, L.C. Identification of Genes and Gene Pathways Associated with Major Depressive Disorder by Integrative Brain Analysis of Rat and Human Prefrontal Cortex Transcriptomes. Transl. Psychiatry 2015, 5, e519. [Google Scholar] [CrossRef]
- Li, H.-J.; Qu, N.; Hui, L.; Cai, X.; Zhang, C.-Y.; Zhong, B.-L.; Zhang, S.-F.; Chen, J.; Xia, B.; Wang, L.; et al. Further Confirmation of Netrin 1 Receptor (DCC) as a Depression Risk Gene via Integrations of Multi-Omics Data. Transl. Psychiatry 2020, 10, 98. [Google Scholar] [CrossRef]
- Cline, M.M.; Juarez, B.; Hunker, A.; Regiarto, E.G.; Hariadi, B.; Soden, M.E.; Zweifel, L.S. Netrin-1 Regulates the Balance of Synaptic Glutamate Signaling in the Adult Ventral Tegmental Area. Elife 2023, 12, e83760. [Google Scholar] [CrossRef]
- Zhou, Q.-G.; Zhu, X.-H.; Nemes, A.D.; Zhu, D.-Y. Neuronal Nitric Oxide Synthase and Affective Disorders. IBRO Rep. 2018, 5, 116–132. [Google Scholar] [CrossRef]
- Harrison, P.J.; Hall, N.; Mould, A.; Al-Juffali, N.; Tunbridge, E.M. Cellular Calcium in Bipolar Disorder: Systematic Review and Meta-Analysis. Mol. Psychiatry 2021, 26, 4106–4116. [Google Scholar] [CrossRef]
- Nakao, A.; Matsunaga, Y.; Hayashida, K.; Takahashi, N. Role of Oxidative Stress and Ca(2+) Signaling in Psychiatric Disorders. Front. Cell Dev. Biol. 2021, 9, 615569. [Google Scholar] [CrossRef]
- Kékesi, K.A.; Juhász, G.; Simor, A.; Gulyássy, P.; Szegő, E.M.; Hunyadi-Gulyás, E.; Darula, Z.; Medzihradszky, K.F.; Palkovits, M.; Penke, B.; et al. Altered Functional Protein Networks in the Prefrontal Cortex and Amygdala of Victims of Suicide. PLoS ONE 2012, 7, e50532. [Google Scholar] [CrossRef]
- Yuan, F.; Wu, S.; Zhou, Z.; Jiao, F.; Yin, H.; Niu, Y.; Jiang, H.; Chen, S.; Guo, F. Leucine Deprivation Results in Antidepressant Effects via GCN2 in AgRP Neurons. Life Metab. 2023, 2, load004. [Google Scholar] [CrossRef]
- Trushina, E.; Trushin, S.; Hasan, M.F. Mitochondrial Complex I as a Therapeutic Target for Alzheimer’s Disease. Acta Pharm. Sin. B 2022, 12, 483–495. [Google Scholar] [CrossRef]
- Huang, G.; Wang, S.; Yan, J.; Li, C.; Feng, J.; Chen, Q.; Zheng, X.; Li, H.; He, Y.; Young, A.J.; et al. Depression-/Anxiety-like Behavior Alterations in Adult Slit2 Transgenic Mice. Front. Behav. Neurosci. 2020, 14, 622257. [Google Scholar] [CrossRef]
- van der Zee, Y.Y.; Lardner, C.K.; Parise, E.M.; Mews, P.; Ramakrishnan, A.; Patel, V.; Teague, C.D.; Salery, M.; Walker, D.M.; Browne, C.J.; et al. Sex-Specific Role for SLIT1 in Regulating Stress Susceptibility. Biol. Psychiatry 2022, 91, 81–91. [Google Scholar] [CrossRef]
- Maitra, M.; Mitsuhashi, H.; Rahimian, R.; Chawla, A.; Yang, J.; Fiori, L.M.; Davoli, M.A.; Perlman, K.; Aouabed, Z.; Mash, D.C.; et al. Cell Type Specific Transcriptomic Differences in Depression Show Similar Patterns between Males and Females but Implicate Distinct Cell Types and Genes. Nat. Commun. 2023, 14, 2912. [Google Scholar] [CrossRef]
- Stepan, J.; Anderzhanova, E.; Gassen, N.C. Hippo Signaling: Emerging Pathway in Stress-Related Psychiatric Disorders? Front. Psychiatry 2018, 9, 715. [Google Scholar] [CrossRef]
- Panizzutti, B.; Bortolasci, C.C.; Spolding, B.; Kidnapillai, S.; Connor, T.; Richardson, M.F.; Truong, T.T.T.; Liu, Z.S.J.; Morris, G.; Gray, L.; et al. Transcriptional Modulation of the Hippo Signaling Pathway by Drugs Used to Treat Bipolar Disorder and Schizophrenia. Int. J. Mol. Sci. 2021, 22, 7164. [Google Scholar] [CrossRef]
- Wang, P.; Yang, Y.; Yang, X.; Qiu, X.; Qiao, Z.; Wang, L.; Zhu, X.; Sui, H.; Ma, J. CREB1 Gene Polymorphisms Combined with Environmental Risk Factors Increase Susceptibility to Major Depressive Disorder (MDD). Int. J. Clin. Exp. Pathol. 2015, 8, 906–913. [Google Scholar]
- Rafa-Zabłocka, K.; Kreiner, G.; Bagińska, M.; Nalepa, I. Selective Depletion of CREB in Serotonergic Neurons Affects the Upregulation of Brain-Derived Neurotrophic Factor Evoked by Chronic Fluoxetine Treatment. Front. Neurosci. 2018, 12, 637. [Google Scholar] [CrossRef]
- Errico, F.; Napolitano, F.; Squillace, M.; Vitucci, D.; Blasi, G.; de Bartolomeis, A.; Bertolino, A.; D’Aniello, A.; Usiello, A. Decreased Levels of D-Aspartate and NMDA in the Prefrontal Cortex and Striatum of Patients with Schizophrenia. J. Psychiatr. Res. 2013, 47, 1432–1437. [Google Scholar] [CrossRef]
- Duan, K.; Gu, Q.; Petralia, R.S.; Wang, Y.-X.; Panja, D.; Liu, X.; Lehmann, M.L.; Zhu, H.; Zhu, J.; Li, Z. Mitophagy in the Basolateral Amygdala Mediates Increased Anxiety Induced by Aversive Social Experience. Neuron 2021, 109, 3793–3809.e8. [Google Scholar] [CrossRef]
- Gao, L.; Peng, L.; Wang, J.; Zhang, J.H.; Xia, Y. Mitochondrial Stress: A Key Role of Neuroinflammation in Stroke. J. Neuroinflamm. 2024, 21, 44. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, Z.; Yu, L.; Ding, Y.; Xu, Y.; Xu, N.; Li, R.; Tang, J.; Chen, G.; Zhang, J.H. GCN2 Reduces Inflammation by P-EIF2α/ATF4 Pathway after Intracerebral Hemorrhage in Mice. Exp. Neurol. 2019, 313, 16–25. [Google Scholar] [CrossRef]
- Lee, W.S.; Lee, W.-H.; Bae, Y.C.; Suk, K. Axon Guidance Molecules Guiding Neuroinflammation. Exp. Neurobiol. 2019, 28, 311–319. [Google Scholar] [CrossRef]
- Crupi, R.; Impellizzeri, D.; Cuzzocrea, S. Role of Metabotropic Glutamate Receptors in Neurological Disorders. Front. Mol. Neurosci. 2019, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wang, S.; Dong, Y.; Yuan, Z. The Role and Regulatory Mechanism of Hippo Signaling Components in the Neuronal System. Front. Immunol. 2020, 11, 281. Available online: https://www.frontiersin.org/articles/10.3389/fimmu.2020.00281 (accessed on 23 February 2022). [CrossRef]
- Aloi, M.S.; Su, W.; Garden, G.A. The P53 Transcriptional Network Influences Microglia Behavior and Neuroinflammation. Crit. Rev. Immunol. 2015, 35, 401–415. [Google Scholar] [CrossRef]
- Zhang, H.; Li, N.; Li, Z.; Li, Y.; Yu, Y.; Zhang, L. The Involvement of Caspases in Neuroinflammation and Neuronal Apoptosis in Chronic Pain and Potential Therapeutic Targets. Front. Pharmacol. 2022, 13, 898574. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, X.; Liu, B.; Xia, Y.; Xin, Z.; Deng, B.; He, L.; Deng, J.; Ren, W. Aspartate Metabolism Facilitates IL-1β Production in Inflammatory Macrophages. Front. Immunol. 2021, 12, 753092. [Google Scholar] [CrossRef]
- Wu, Y.-H.; Hsieh, H.-L. Roles of Heme Oxygenase-1 in Neuroinflammation and Brain Disorders. Antioxidants 2022, 11, 923. [Google Scholar] [CrossRef]
- Syapin, P.J. Regulation of Haeme Oxygenase-1 for Treatment of Neuroinflammation and Brain Disorders. Br. J. Pharmacol. 2008, 155, 623–640. [Google Scholar] [CrossRef]
- Shih, R.-H.; Yang, C.-M. Induction of Heme Oxygenase-1 Attenuates Lipopolysaccharide-Induced Cyclooxygenase-2 Expression in Mouse Brain Endothelial Cells. J. Neuroinflamm. 2010, 7, 86. [Google Scholar] [CrossRef]
- Simpson, D.S.A.; Oliver, P.L. ROS Generation in Microglia: Understanding Oxidative Stress and Inflammation in Neurodegenerative Disease. Antioxidants 2020, 9, 743. [Google Scholar] [CrossRef]
- Dehghani, A.; Karatas, H.; Can, A.; Erdemli, E.; Yemisci, M.; Eren-Kocak, E.; Dalkara, T. Nuclear Expansion and Pore Opening Are Instant Signs of Neuronal Hypoxia and Can Identify Poorly Fixed Brains. Sci. Rep. 2018, 8, 14770. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastawy, E.M.; Eraslan, I.M.; Voglsanger, L.; Suphioglu, C.; Walker, A.J.; Dean, O.M.; Read, J.L.; Ziemann, M.; Smith, C.M. Novel Insights into Changes in Gene Expression within the Hypothalamus in Two Asthma Mouse Models: A Transcriptomic Lung–Brain Axis Study. Int. J. Mol. Sci. 2024, 25, 7391. https://doi.org/10.3390/ijms25137391
Bastawy EM, Eraslan IM, Voglsanger L, Suphioglu C, Walker AJ, Dean OM, Read JL, Ziemann M, Smith CM. Novel Insights into Changes in Gene Expression within the Hypothalamus in Two Asthma Mouse Models: A Transcriptomic Lung–Brain Axis Study. International Journal of Molecular Sciences. 2024; 25(13):7391. https://doi.org/10.3390/ijms25137391
Chicago/Turabian StyleBastawy, Eslam M., Izel M. Eraslan, Lara Voglsanger, Cenk Suphioglu, Adam J. Walker, Olivia M. Dean, Justin L. Read, Mark Ziemann, and Craig M. Smith. 2024. "Novel Insights into Changes in Gene Expression within the Hypothalamus in Two Asthma Mouse Models: A Transcriptomic Lung–Brain Axis Study" International Journal of Molecular Sciences 25, no. 13: 7391. https://doi.org/10.3390/ijms25137391
APA StyleBastawy, E. M., Eraslan, I. M., Voglsanger, L., Suphioglu, C., Walker, A. J., Dean, O. M., Read, J. L., Ziemann, M., & Smith, C. M. (2024). Novel Insights into Changes in Gene Expression within the Hypothalamus in Two Asthma Mouse Models: A Transcriptomic Lung–Brain Axis Study. International Journal of Molecular Sciences, 25(13), 7391. https://doi.org/10.3390/ijms25137391









