Genome-Wide Identification of Seven in Absentia E3 Ubiquitin Ligase Gene Family and Expression Profiles in Response to Different Hormones in Uncaria rhynchophylla
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of UrSINA Family Members
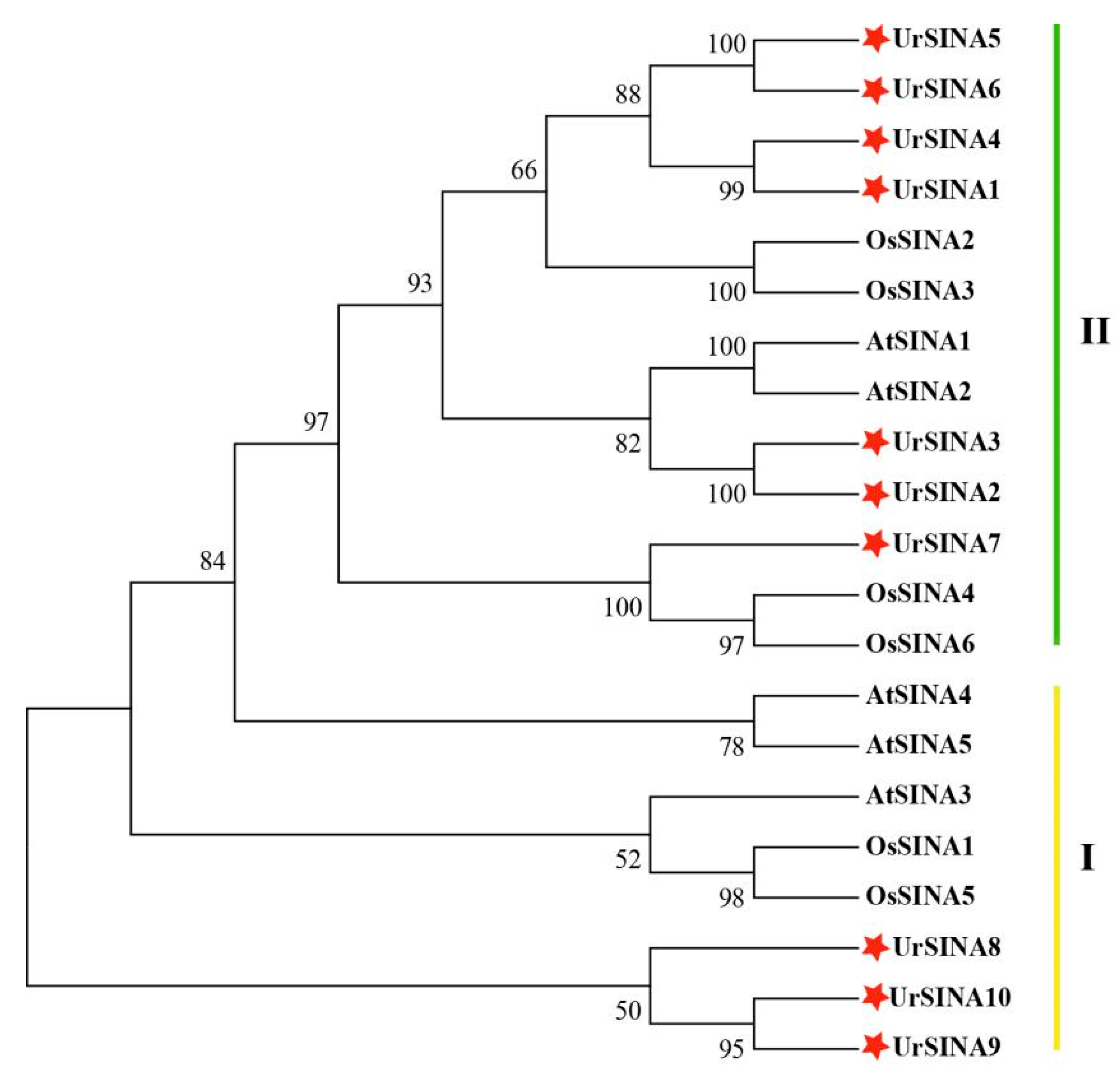
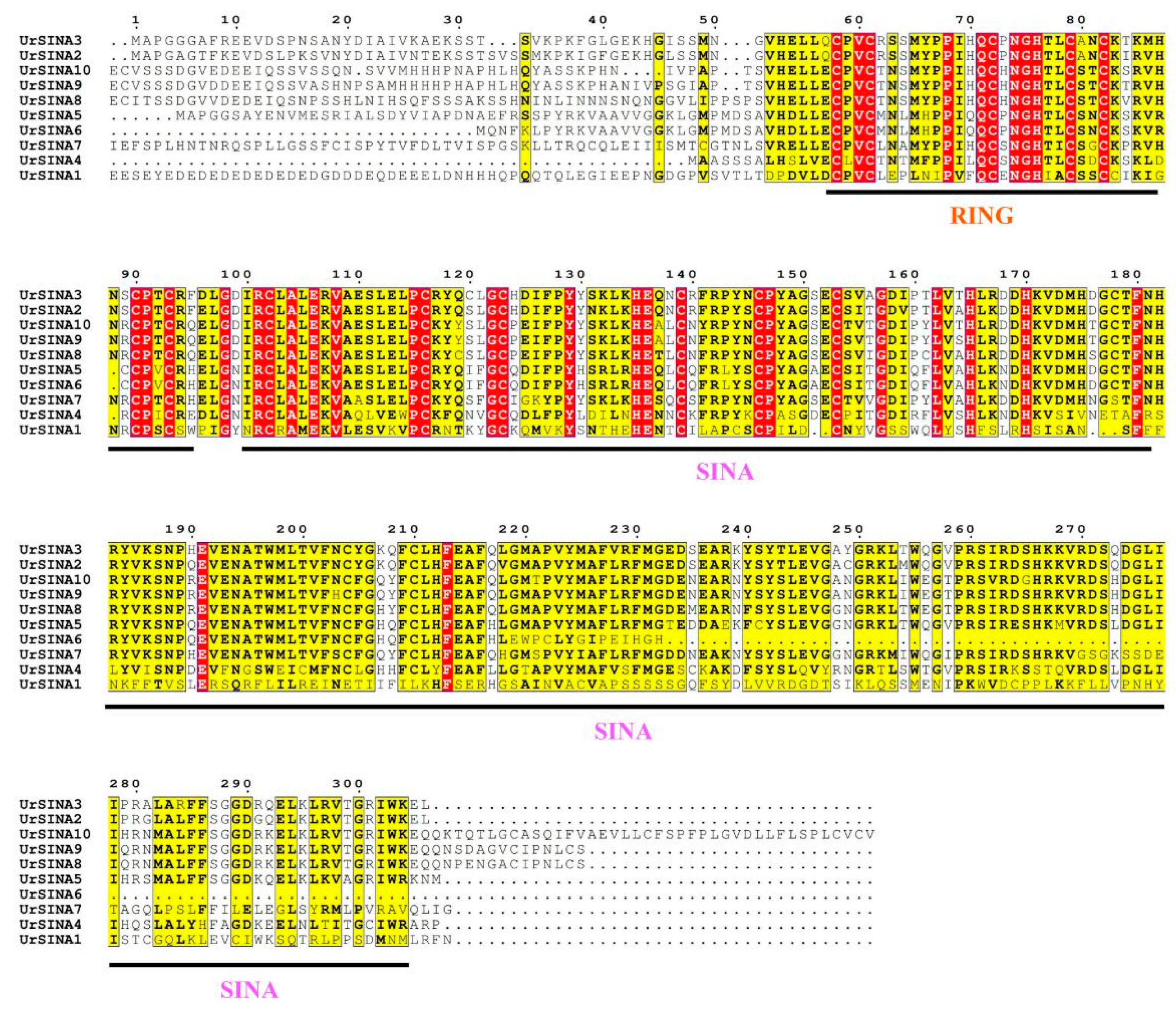
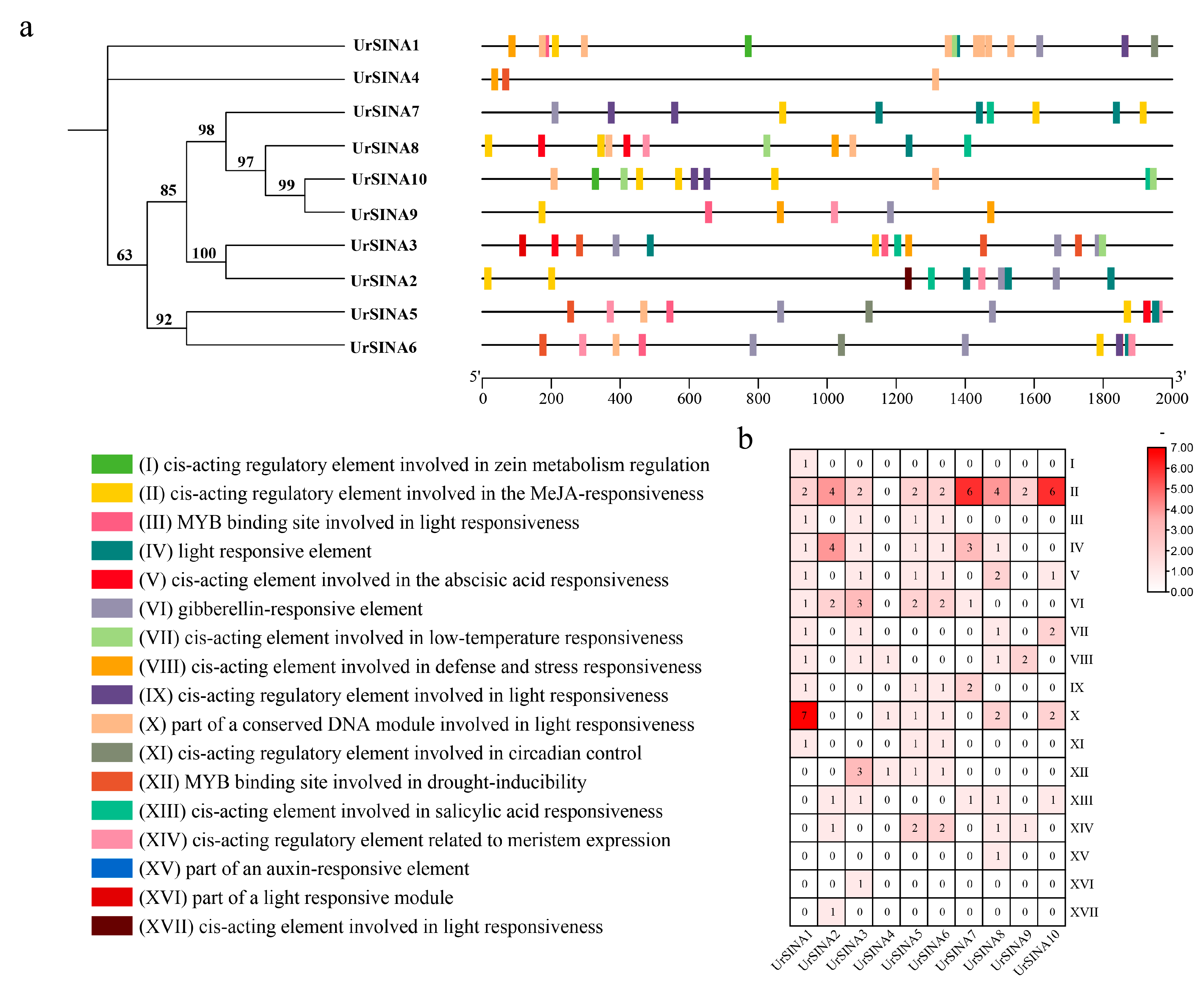
2.2. Phylogenetic Tree Construction, Classification, and Multiple Sequence Alignment of UrSINA
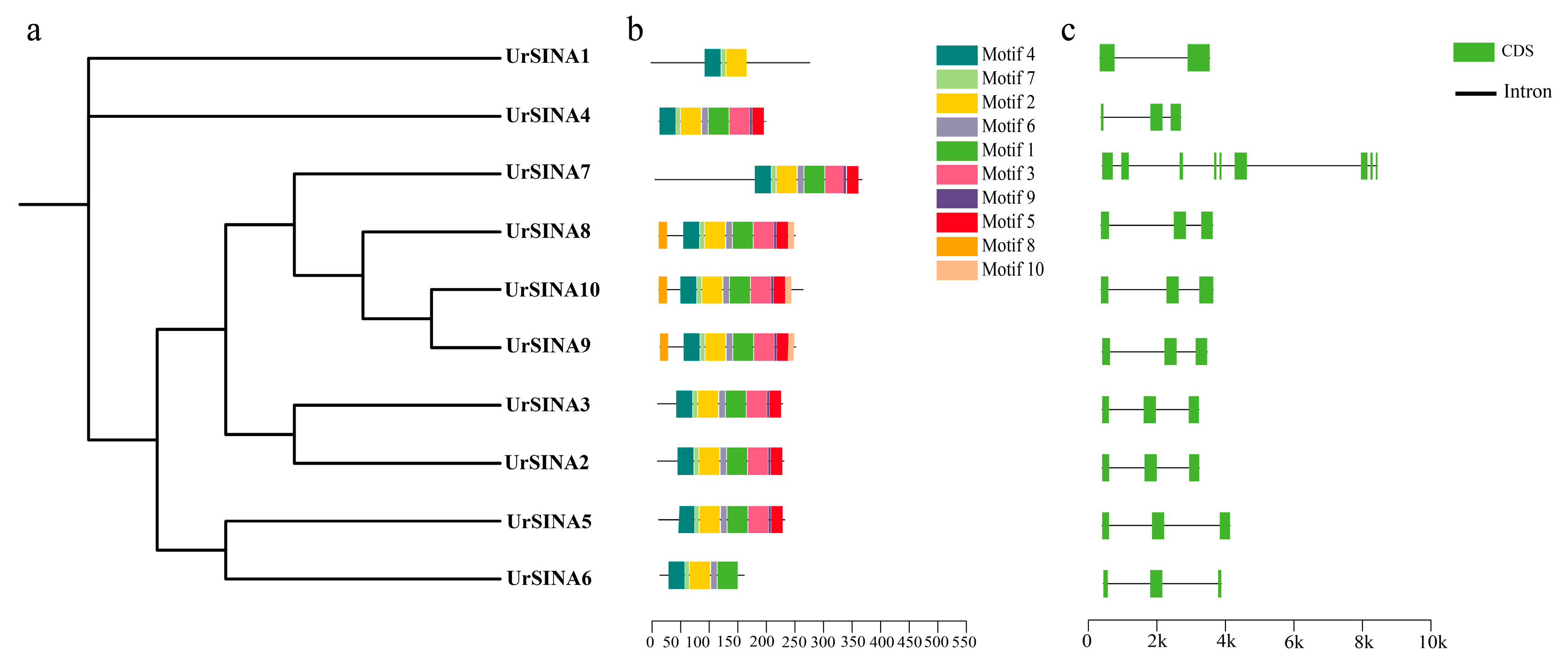
2.3. Conserved Motifs and Gene Structure of UrSINA Family Members
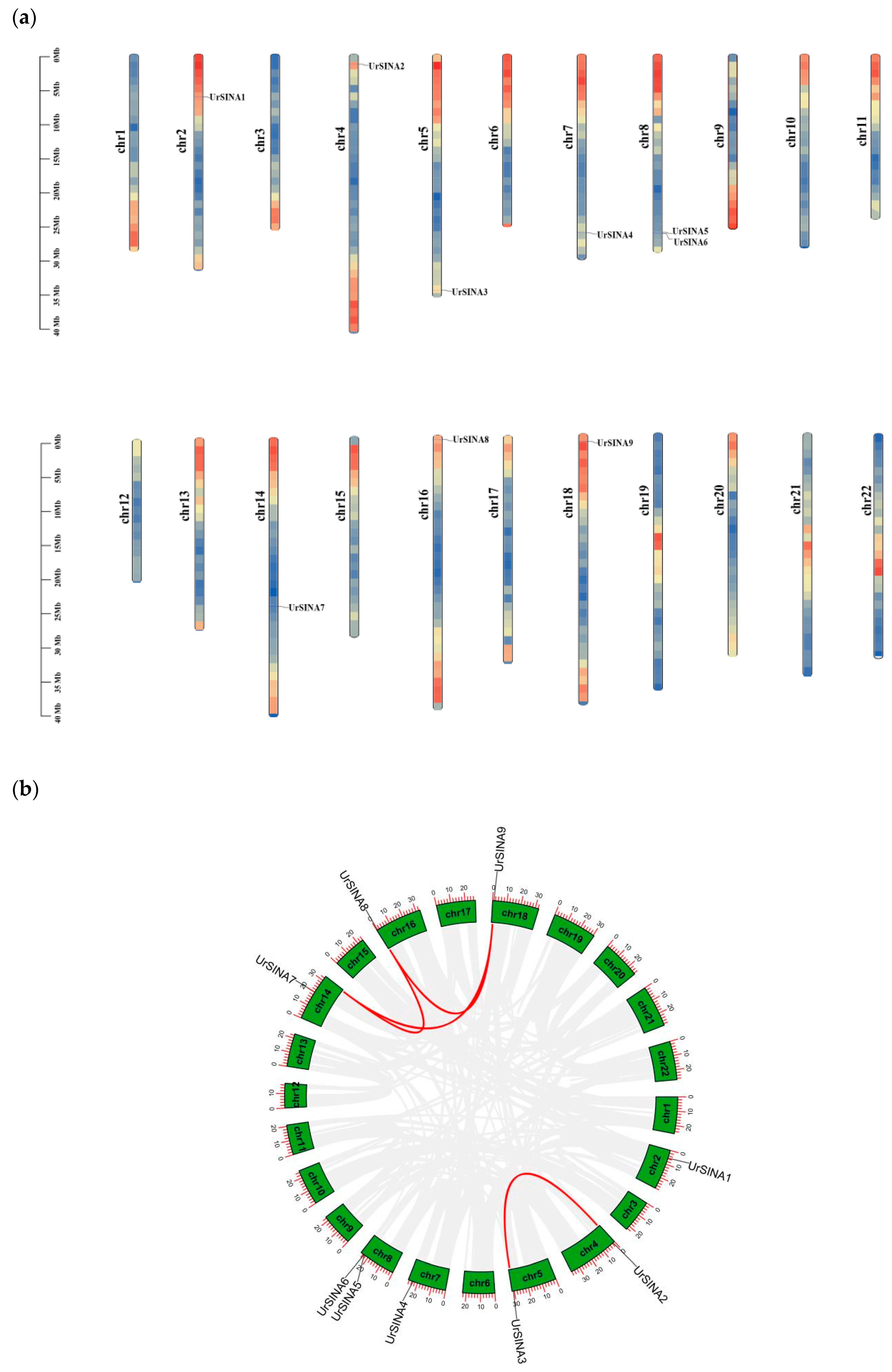
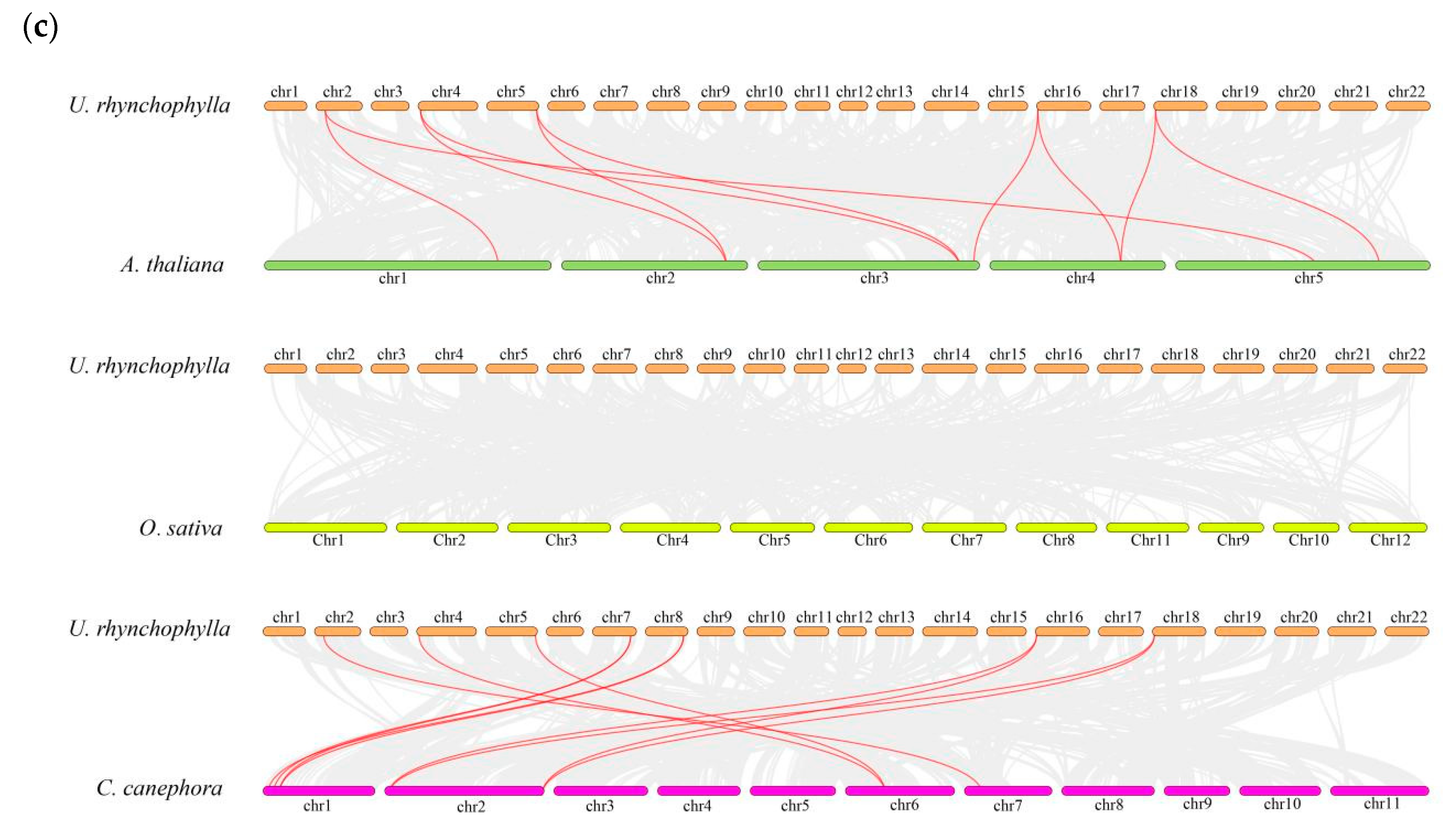
2.4. Chromosomal Localization and Intraspecific Synteny Analysis of UrSINA Gene Family
2.5. Analysis of Cis-Acting Elements in the Promoter of UrSINA Genes
2.6. Ten UrSINA Genes Are Differentially Expressed in Various Tissues
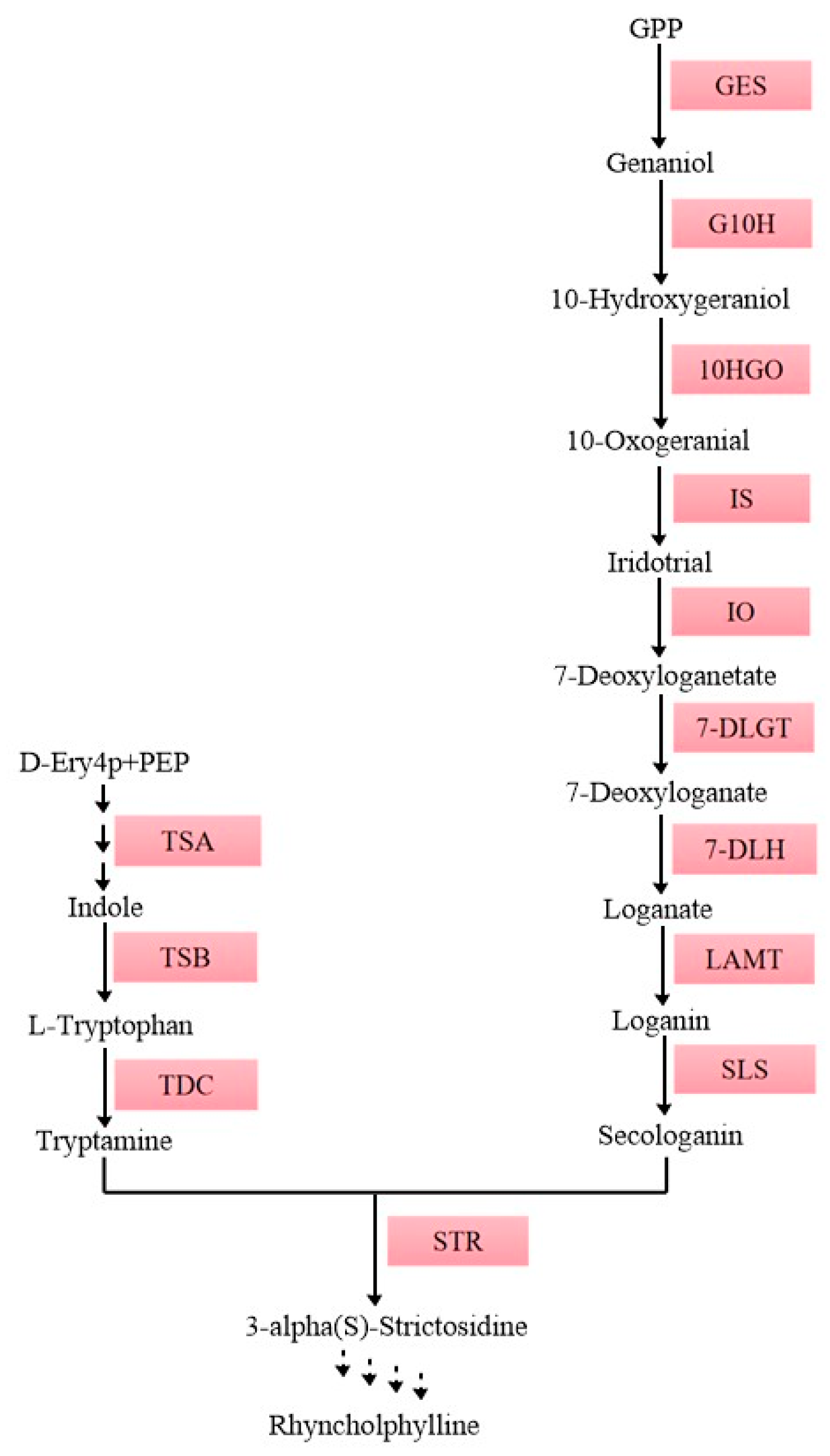
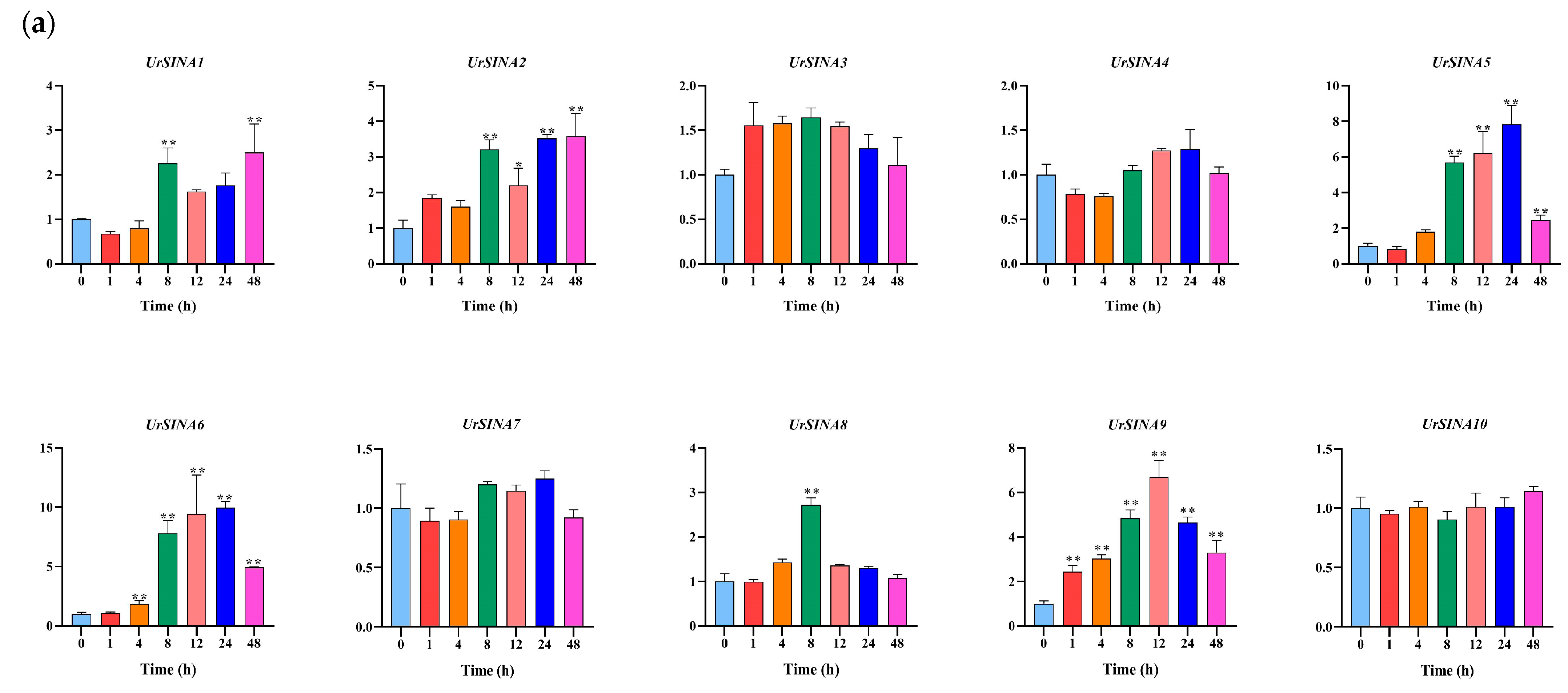
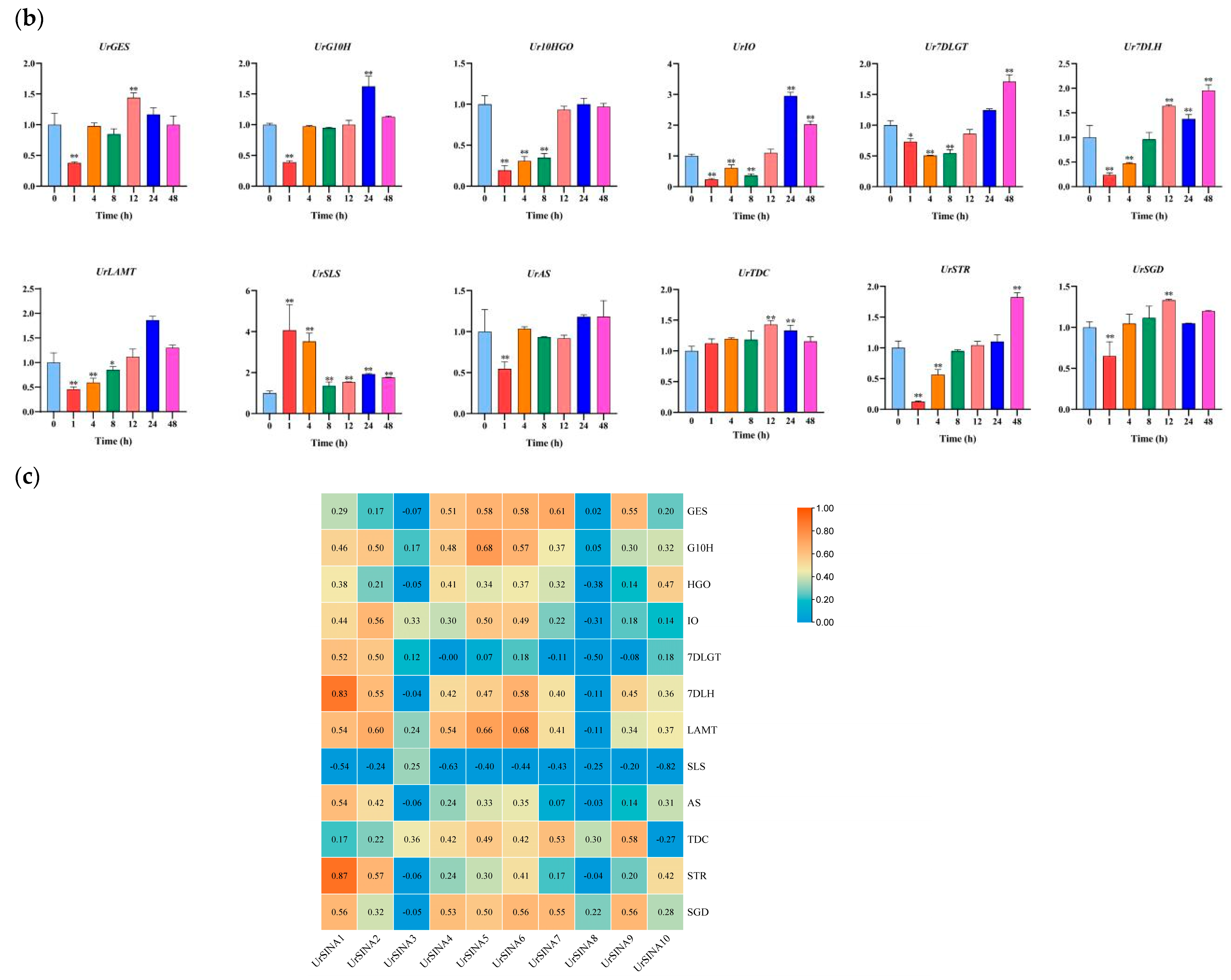
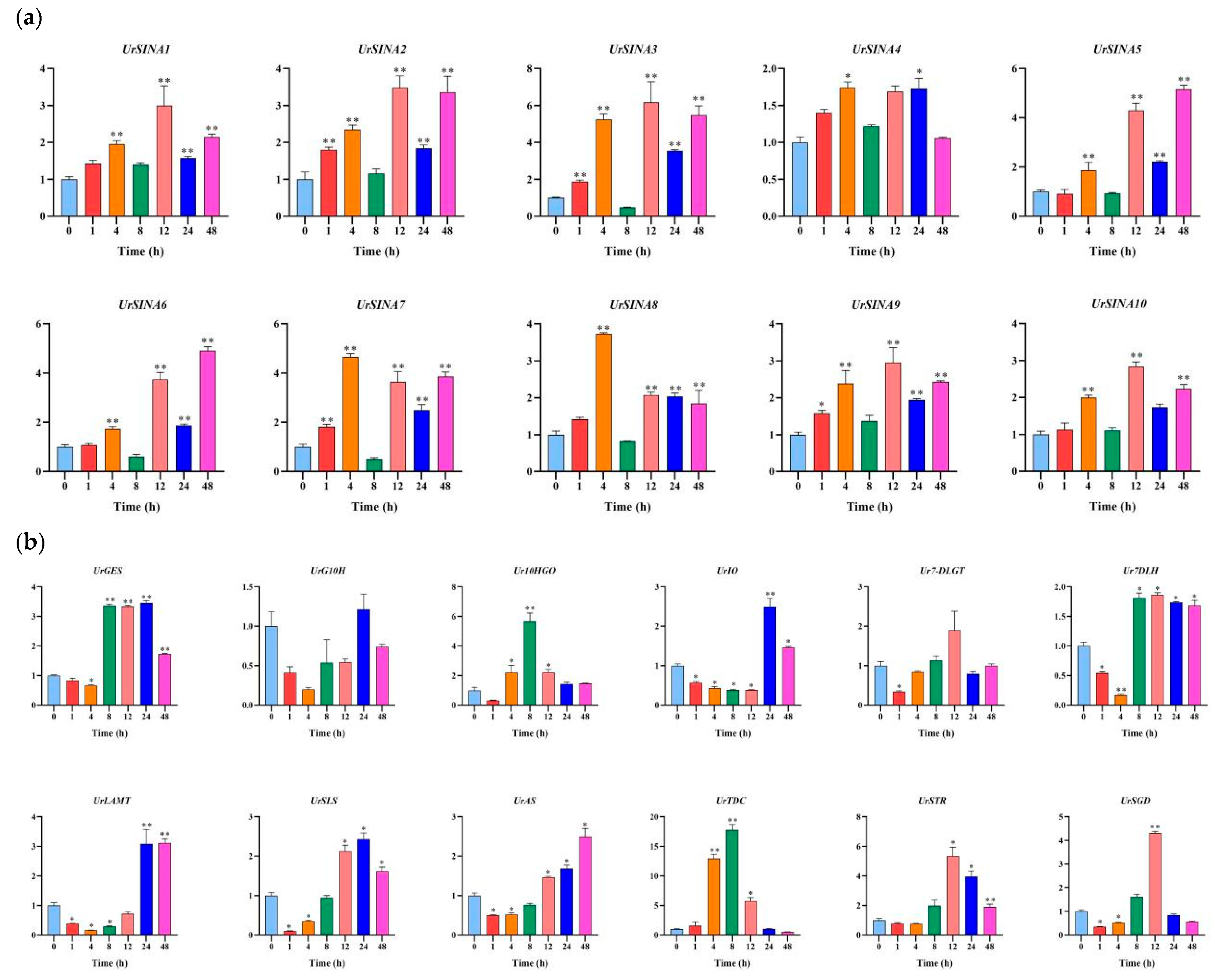
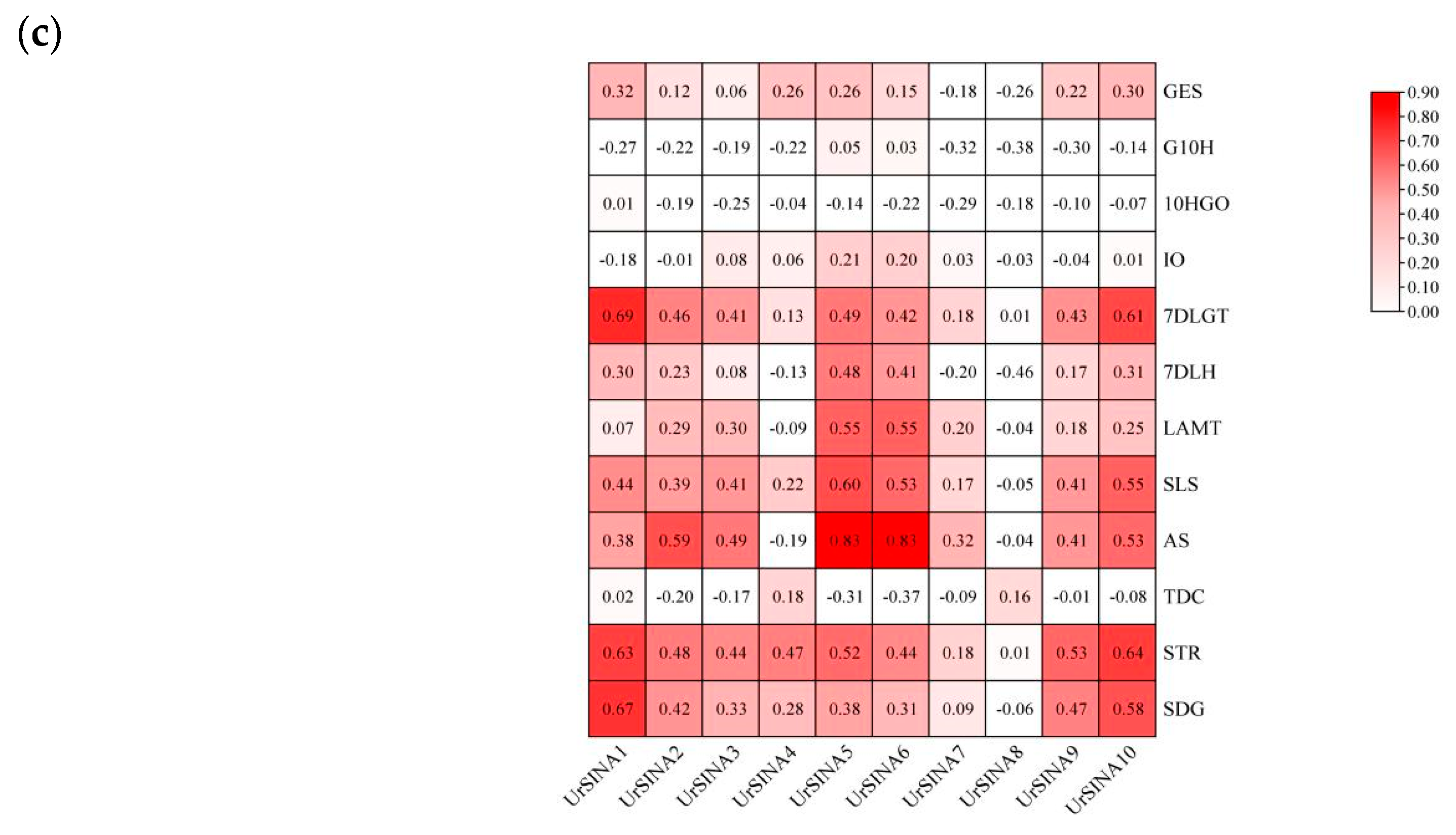
2.7. Expression Analysis of UrSINAs and Key Enzyme Genes under ABA and MeJA Hormones
3. Discussion
4. Materials and Methods
4.1. Plant Material and Hormone Treatment
4.2. Total RNA Extraction and Relative Gene Expression Analyses
4.3. Identification of SINA Genes in U. rhynchophylla and Construction of Phylogenetic Tree
4.4. Multiple Sequence Alignment, Conserved Motif, and Gene Structure Analysis
4.5. Chromosome Localization, Syntenic Analysis, and Cis-Regulatory Elements Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yin, R.; Yin, L.; Li, L.; Silva-Nash, J.; Tan, J.; Pan, Z.; Zeng, J.; Yan, L.L. Hypertension in China: Burdens, guidelines and policy responses: A state-of-the-art review. J. Hum. Hypertens. 2022, 36, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Ip, S.P.; Liu, L.; Xian, Y.F.; Lin, Z.X. Uncaria rhynchophylla and its Major Constituents on Central Nervous System: A Review on Their Pharmacological Actions. Curr. Vasc. Pharmacol. 2020, 18, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhou, Y.; Yang, L.; Mu, D.; Wilson, I.W.; Zhang, Y.; Zhu, L.; Liu, X.; Luo, L.; He, J.; et al. Genome-wide identification of GATA transcription factor family and the effect of different light quality on the accumulation of terpenoid indole alkaloids in Uncaria rhynchophylla. Plant Mol. Biol. 2024, 114, 15. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.; Shao, Y.; He, J.; Zhu, L.; Qiu, D.; Wilson, I.W.; Zhang, Y.; Pan, L.; Zhou, Y.; Lu, Y.; et al. Evaluation of Reference Genes for Normalizing RT-qPCR and Analysis of the Expression Patterns of WRKY1 Transcription Factor and Rhynchophylline Biosynthesis-Related Genes in Uncaria rhynchophylla. Int. J. Mol. Sci. 2023, 24, 16330. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhang, S.; Tao, W.; Zhang, X.; Liu, J.; Sun, J.; Zhang, H.; Pu, L.; Huang, R.; Chen, T. INDETERMINATE SPIKELET1 Recruits Histone Deacetylase and a Transcriptional Repression Complex to Regulate Rice Salt Tolerance. Plant Physiol. 2018, 178, 824–837. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef]
- Shao, Y.; Mu, D.; Zhou, Y.; Liu, X.; Huang, X.; Wilson, I.W.; Qi, Y.; Lu, Y.; Zhu, L.; Zhang, Y.; et al. Genome-Wide Mining of CULLIN E3 Ubiquitin Ligase Genes from Uncaria rhynchophylla. Plants 2024, 13, 532. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Halaly, T.; Acheampong, A.K.; Takebayashi, Y.; Jikumaru, Y.; Kamiya, Y.; Or, E. Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 2015, 66, 1527–1542. [Google Scholar] [CrossRef]
- González-Grandío, E.; Cubas, P. Identification of gene functions associated to active and dormant buds in Arabidopsis. Plant Signal. Behav. 2014, 9, e27994. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, Z.; Lu, L.; Jin, H.; Sun, L.; Yu, Q.; Xu, H.; Yang, F.; Fu, M.; Li, S.; et al. Interaction between abscisic acid and nitric oxide in PB90-induced catharanthine biosynthesis of catharanthus roseus cell suspension cultures. Biotechnol. Prog. 2013, 29, 994–1001. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Li, X.; Chen, C.; Wang, C.; Fu, Y.; Zheng, Z.; Shi, M.; Hao, X.; Zhao, L.; Qiu, M.; et al. Mining of the CULLIN E3 ubiquitin ligase genes in the whole genome of Salvia miltiorrhiza. Curr. Res. Food Sci. 2022, 5, 1760–1768. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, S.; Cui, S.; Hou, H.; Wu, H.; Hao, B.; Cai, L.; Xu, Z.; Liu, L.; Jiang, L.; et al. Transcriptional and post-transcriptional regulation of heading date in rice. New Phytol. 2021, 230, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, C.; Li, J.; Gao, X.; Huang, Q.; Gong, Y.; Hao, X.; Maoz, I.; Kai, G.; Zhou, W. Genome-Wide Analysis of U-box E3 Ubiquitin Ligase Family in Response to ABA Treatment in Salvia miltiorrhiza. Front. Plant Sci. 2022, 13, 829447. [Google Scholar] [CrossRef]
- Jmii, S.; Cappadocia, L. Plant SUMO E3 Ligases: Function, Structural Organization, and Connection with DNA. Front. Plant Sci. 2021, 12, 652170. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.Y.; Ahammed, G.J.; Jiang, C.Y.; Li, C.X.; Zhou, J. The E3 ubiquitin ligase gene SlRING1 is essential for plant tolerance to cadmium stress in Solanum lycopersicum. J. Biotechnol. 2020, 324, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Siswanto, F.M.; Jawi, I.M.; Kartiko, B.H. The role of E3 ubiquitin ligase seven in absentia homolog in the innate immune system: An overview. Vet. World 2018, 11, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cui, F.; Wu, Y.; Lou, L.; Liu, L.; Tian, M.; Ning, Y.; Shu, K.; Tang, S.; Xie, Q. The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 2015, 27, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Yang, W. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef]
- Xie, Q.; Guo, H.S.; Dallman, G.; Fang, S.; Weissman, A.M.; Chua, N.H. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 2002, 419, 167–170. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Gao, Z.; Xu, X.; Wang, Y.; Lin, Y.; Ye, P.; Huang, T. Plant U-box E3 ligases PUB25 and PUB26 control organ growth in Arabidopsis. New Phytol. 2021, 229, 403–413. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, W.T. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells 2011, 31, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jin, Y.; Fu, J.; Zhu, Y.; Zheng, J.; Hu, J.; Wang, G. Genome-wide analysis of SINA family in plants and their phylogenetic relationships. DNA Seq. J. DNA Seq. Mapp. 2008, 19, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Hou, Y.; Jiang, F.; Lang, H.; Li, J.; Cheng, J.; Wang, L.; Liu, X.; Zhang, H. Genome-wide characterization of SINA E3 ubiquitin ligase family members and their expression profiles in response to various abiotic stresses and hormones in kiwifruit. Plant Physiol. Biochem. 2023, 201, 107891. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Wang, X.; Ji, X.L.; Qiao, Z.W.; You, C.X.; Hao, Y.J. Genome-Wide Identification of Apple Ubiquitin SINA E3 Ligase and Functional Characterization of MdSINA2. Front. Plant Sci. 2020, 11, 1109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Garreton, V.; Chua, N.H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 2005, 19, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Zhang, C.L.; Li, H.L.; Wang, G.L.; You, C.X. Apple SINA E3 ligase MdSINA3 negatively mediates JA-triggered leaf senescence by ubiquitinating and degrading the MdBBX37 protein. Plant J. 2022, 111, 457–472. [Google Scholar] [CrossRef]
- Wang, W.; Fan, Y.; Niu, X.; Miao, M.; Kud, J.; Zhou, B.; Zeng, L.; Liu, Y.; Xiao, F. Functional analysis of the seven in absentia ubiquitin ligase family in tomato. Plant Cell Environ. 2018, 41, 689–703. [Google Scholar] [CrossRef]
- Thomelin, P.; Bonneau, J.; Brien, C.; Suchecki, R.; Baumann, U.; Kalambettu, P.; Langridge, P.; Tricker, P.; Fleury, D. The wheat Seven in absentia gene is associated with increases in biomass and yield in hot climates. J. Exp. Bot. 2021, 72, 3774–3791. [Google Scholar] [CrossRef]
- Yu, C.C.; Sun, P.W.; Rong, M.; Gao, Z.H.; Liu, Y.; Xiao, M.J.; Jiang, J.M.; Xu, Y.H.; Wei, J.H. E3 ubiquitin ligase RING3 mediates AsWRKY44 degradation to promote wound-induced sesquiterpene biosynthesis in Aquilaria sinensis. Ind. Crops Prod. 2023, 191, 115908. [Google Scholar] [CrossRef]
- Chen, J.; Hou, S.; Zhang, Q.; Meng, J.; Zhang, Y.; Du, J.; Wang, C.; Liang, D.; Guo, Y. Genome-Wide Identification and Analysis of the WRKY Gene Family in Asparagus officinalis. Genes 2023, 14, 1704. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.; Chen, W.; Shao, Y.; Wilson, I.W.; Zhao, H.; Luo, Z.; Lin, X.; He, J.; Zhang, Y.; Mo, C.; et al. Genome-Wide Identification and Expression Analysis of WRKY Transcription Factors in Siraitia siamensis. Plants 2023, 12, 288. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Lang, Y.; Liu, Z. Basic Helix-Loop-Helix (bHLH) transcription factor family in Yellow horn (Xanthoceras sorbifolia Bunge): Genome-wide characterization, chromosome location, phylogeny, structures and expression patterns. Int. J. Biol. Macromol. 2020, 160, 711–723. [Google Scholar] [CrossRef]
- Den Herder, G.; De Keyser, A.; De Rycke, R.; Rombauts, S.; Van de Velde, W.; Clemente, M.R.; Verplancke, C.; Mergaert, P.; Kondorosi, E.; Holsters, M.; et al. Seven in absentia proteins affect plant growth and nodulation in Medicago truncatula. Plant Physiol. 2008, 148, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Zhou, W.; Liu, Y.; Song, Z.; Zheng, S.; Wang, F.; Lu, Z.; Qi, D.; Li, B.; Sun, N.; et al. Characterization of RING-type ubiquitin SINA E3 ligases and their responsive expression to salt and osmotic stresses in Brassica napus. Plant Cell Rep. 2023, 42, 859–877. [Google Scholar] [CrossRef] [PubMed]
- Den Herder, G.; Yoshida, S.; Antolín-Llovera, M.; Ried, M.K.; Parniske, M. Lotus japonicus E3 ligase SEVEN IN ABSENTIA4 destabilizes the symbiosis receptor-like kinase SYMRK and negatively regulates rhizobial infection. Plant Cell 2012, 24, 1691–1707. [Google Scholar] [CrossRef]
- Fan, Z.Q.; Chen, J.Y.; Kuang, J.F.; Lu, W.J.; Shan, W. The Banana Fruit SINA Ubiquitin Ligase MaSINA1 Regulates the Stability of MaICE1 to be Negatively Involved in Cold Stress Response. Front. Plant Sci. 2017, 8, 995. [Google Scholar] [CrossRef]
- Mishra, S.K.; Thakran, P. Intron specificity in pre-mRNA splicing. Curr. Genet. 2018, 64, 777–784. [Google Scholar] [CrossRef]
- Lu, J.; Peatman, E.; Tang, H.; Lewis, J.; Liu, Z. Profiling of gene duplication patterns of sequenced teleost genomes: Evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications. BMC Genom. 2012, 13, 246. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Peng, T.; Dai, W. Critical cis-acting elements and interacting transcription factors: Key players associated with abiotic stress responses in plants. Plant Mol. Biol. Report. 2014, 32, 303–317. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Hu, Y.; Yang, M.; Hu, X.; Wang, X. Light-Induced Dynamic Change of Phytochrome B and Cryptochrome 1 Stabilizes SINATs in Arabidopsis. Front. Plant Sci. 2021, 12, 722733. [Google Scholar] [CrossRef]
- Lee, H.Y.; Park, H.L.; Park, C.; Chen, Y.C.; Yoon, G.M. Reciprocal antagonistic regulation of E3 ligases controls ACC synthase stability and responses to stress. Proc. Natl. Acad. Sci. USA 2021, 118, e2011900118. [Google Scholar] [CrossRef]
- Ning, Y.; Jantasuriyarat, C.; Zhao, Q.; Zhang, H.; Chen, S.; Liu, J.; Liu, L.; Tang, S.; Park, C.H.; Wang, X.; et al. The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol. 2011, 157, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xing, C.; Yao, Z.; Huang, X. PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 2017, 15, 1186–1203. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.T.; Liu, H.; Gao, X.S.; Zhang, H.X. Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production. Plant Cell Rep. 2010, 29, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Gene Name | Chr | Start | End | Amino Acids (aa) | Molecular Weight (kDa) | Theoretical pI | Instability Index | Subcelluar Localization |
|---|---|---|---|---|---|---|---|---|---|
| g6840.t1 | UrSINA1 | chr2 | 5552559 | 5556024 | 388 | 44,051.84 | 4.51 | 74.28 | Nucleus. |
| g29173.t1 | UrSINA2 | chr4 | 1326540 | 1329600 | 309 | 34,835.01 | 8.39 | 45.37 | Mitochondrion. Nucleus. |
| g21951.t1 | UrSINA3 | chr5 | 30647250 | 30650297 | 306 | 34,688.69 | 8.5 | 45.78 | Mitochondrion. Nucleus. |
| g29806.t1 | UrSINA4 | chr7 | 23211096 | 23213614 | 262 | 29,574.07 | 6.73 | 38.09 | Nucleus. |
| g26498.t1 | UrSINA5 | chr8 | 23132312 | 23136340 | 308 | 34,923.39 | 7.84 | 48.27 | Mitochondrion. Nucleus. |
| g41916.t1 | UrSINA6 | chr8 | 23289827 | 23293540 | 206 | 23,637.45 | 6.92 | 40.87 | Mitochondrion. Nucleus. |
| g27550.t1 | UrSINA7 | chr14 | 20226758 | 20235432 | 506 | 57,458.84 | 6.82 | 37.63 | Mitochondrion. Nucleus. |
| g4133.t1 | UrSINA8 | chr16 | 514255 | 517777 | 334 | 37,684.73 | 6.37 | 52.39 | Mitochondrion. Nucleus. |
| g41269.t1 | UrSINA9 | chr18 | 927300 | 930607 | 332 | 37,683.68 | 6.53 | 48.82 | Mitochondrion. Nucleus. |
| g26576.t1 | UrSINA10 | Scaffold9 | 387231 | 390770 | 353 | 40,128.82 | 6.46 | 51.41 | Mitochondrion. Nucleus. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, J.; Lian, C.; Shao, Y.; Chen, S.; Lu, Y.; Zhu, L.; Mu, D.; Tang, Q. Genome-Wide Identification of Seven in Absentia E3 Ubiquitin Ligase Gene Family and Expression Profiles in Response to Different Hormones in Uncaria rhynchophylla. Int. J. Mol. Sci. 2024, 25, 7636. https://doi.org/10.3390/ijms25147636
Lan J, Lian C, Shao Y, Chen S, Lu Y, Zhu L, Mu D, Tang Q. Genome-Wide Identification of Seven in Absentia E3 Ubiquitin Ligase Gene Family and Expression Profiles in Response to Different Hormones in Uncaria rhynchophylla. International Journal of Molecular Sciences. 2024; 25(14):7636. https://doi.org/10.3390/ijms25147636
Chicago/Turabian StyleLan, Jinxu, Conglong Lian, Yingying Shao, Suiqing Chen, Ying Lu, Lina Zhu, Detian Mu, and Qi Tang. 2024. "Genome-Wide Identification of Seven in Absentia E3 Ubiquitin Ligase Gene Family and Expression Profiles in Response to Different Hormones in Uncaria rhynchophylla" International Journal of Molecular Sciences 25, no. 14: 7636. https://doi.org/10.3390/ijms25147636
APA StyleLan, J., Lian, C., Shao, Y., Chen, S., Lu, Y., Zhu, L., Mu, D., & Tang, Q. (2024). Genome-Wide Identification of Seven in Absentia E3 Ubiquitin Ligase Gene Family and Expression Profiles in Response to Different Hormones in Uncaria rhynchophylla. International Journal of Molecular Sciences, 25(14), 7636. https://doi.org/10.3390/ijms25147636






