Autonomous Oscillatory Mitochondrial Respiratory Activity: Results of a Systematic Analysis Show Heterogeneity in Different In Vitro-Synchronized Cancer Cells
Abstract
1. Introduction
2. Results
2.1. Mitochondrial Respiration Undergoes Autonomous Oscillation in In Vitro Serum-Shocked Synchronized Cells
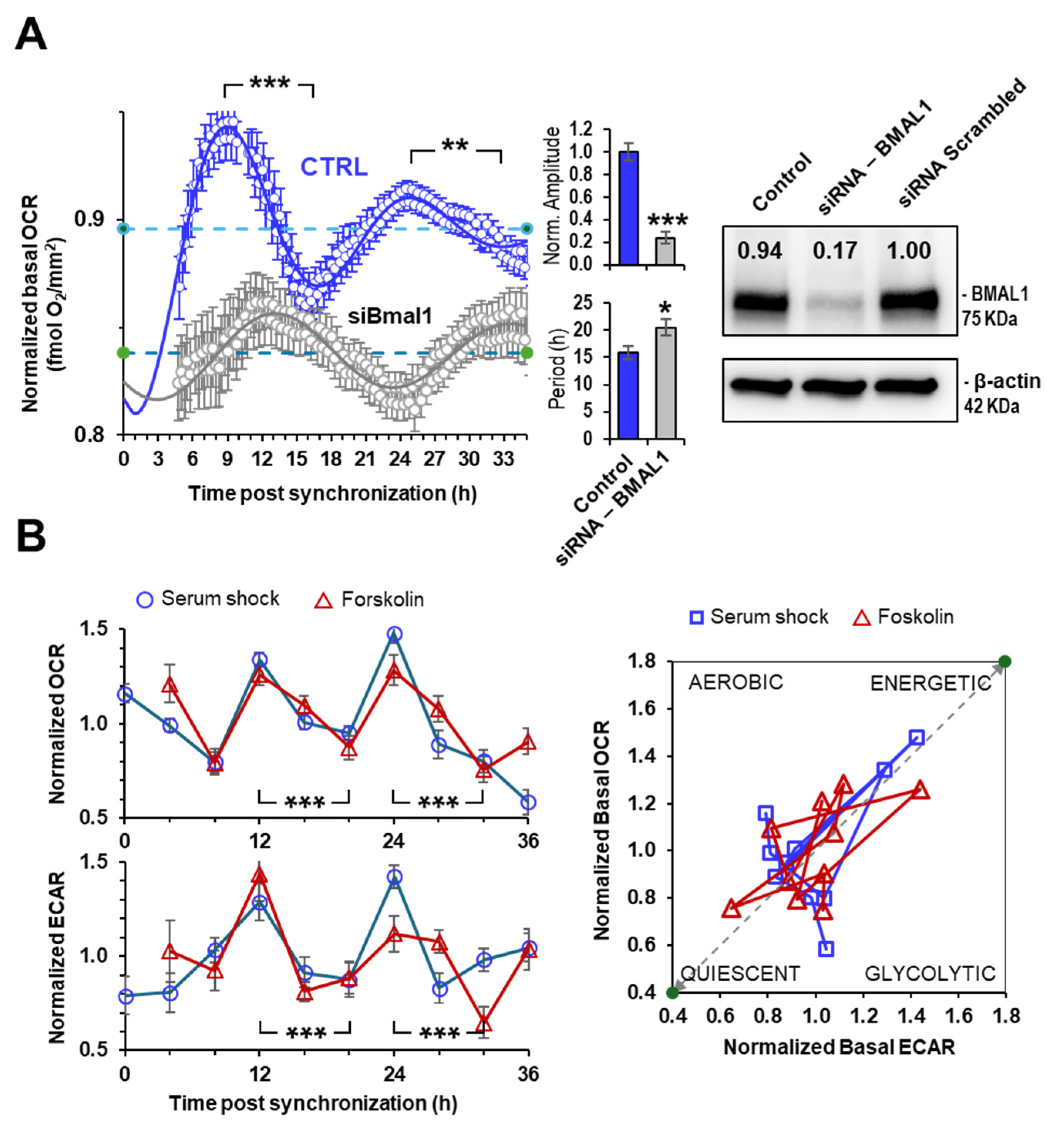
2.2. Rhythmic Mitochondrial Respiration Depends on Bmal1 Expression
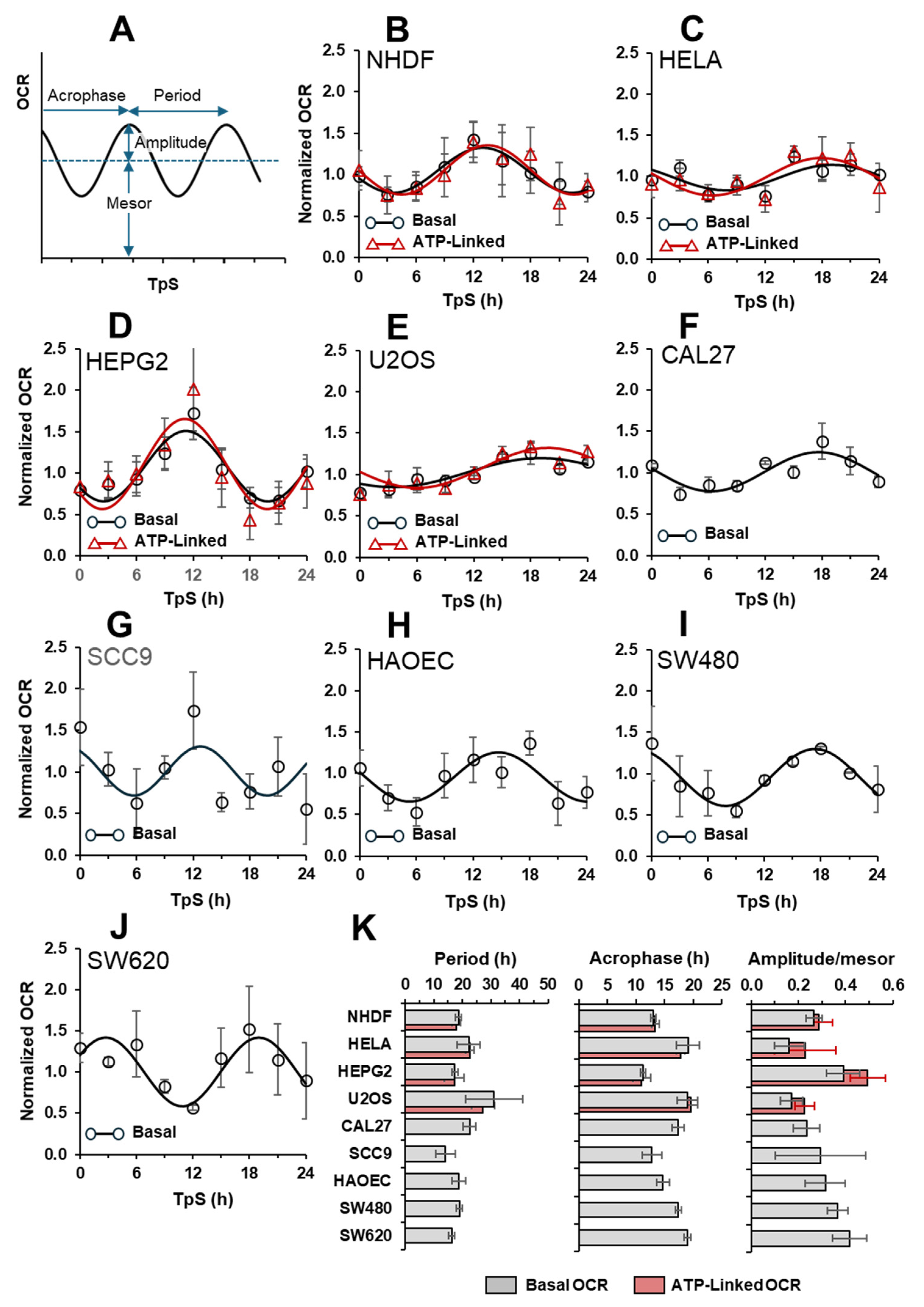
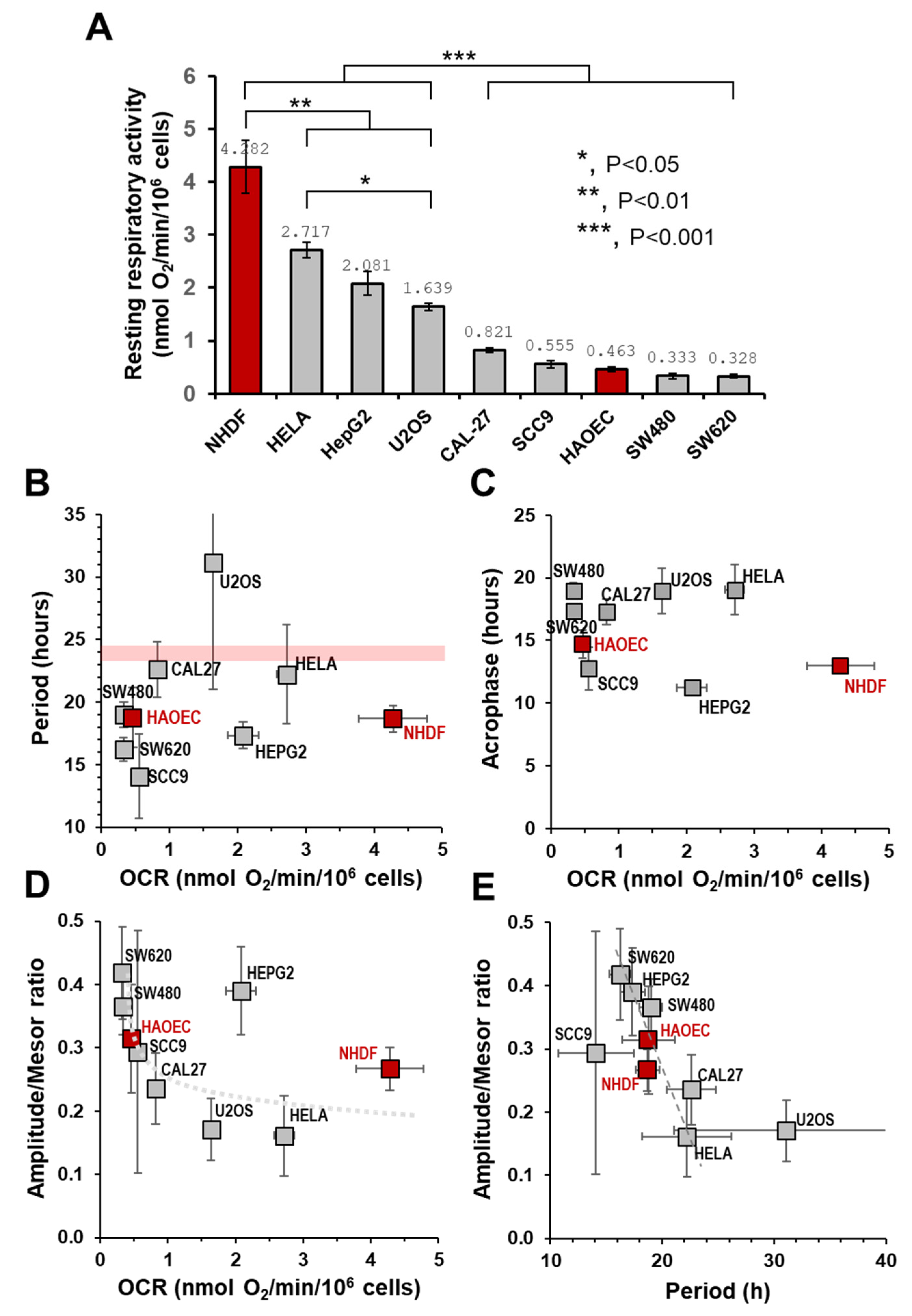
2.3. The Autonomous Oscillatory Profile of Mitochondrial Respiration Differs among Different Cell Types
3. Discussion
4. Materials and Methods
4.1. Cells and Culture Conditions
4.2. In Vitro Circadian Rhythms Synchronization
4.3. Respirometric Measurements
4.4. BMAL1 Silencing in HepG2 Cells
4.5. Western Blotting Analysis
4.6. Cosinor Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhadra, U.; Thakkar, N.; Das, P.; Bhadra, M.P. Evolution of circadian rhythms: From bacteria to human. Sleep Med. 2017, 35, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Jabbur, M.L.; Dani, C.; Spoelstra, K.; Dodd, A.N.; Johnson, C.H. Evaluating the Adaptive Fitness of Circadian Clocks and their Evolution. J. Biol. Rhythm. 2024, 39, 115–134. [Google Scholar] [CrossRef] [PubMed]
- Hastings, M.H.; Maywood, E.S.; Brancaccio, M. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 2018, 19, 453–469. [Google Scholar] [CrossRef] [PubMed]
- Menaker, M.; Murphy, Z.C.; Sellix, M.T. Central control of peripheral circadian oscillators. Curr. Opin. Neurobiol. 2013, 23, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional Architecture of the Mammalian Circadian Clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A Circadian Gene Expression Atlas in Mammals: Implications for Biology and Medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [PubMed]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 16, 359. [Google Scholar] [CrossRef] [PubMed]
- Dyar, K.A.; Lutter, D.; Artati, A.; Ceglia, N.J.; Liu, Y.; Armenta, D.; Jastroch, M.; Schneider, S.; de Mateo, S.; Cervantes, M.; et al. Atlas of Circadian Metabolism Reveals System-wide Coordination and Communication between Clocks. Cell 2018, 174, 1571–1585. [Google Scholar] [CrossRef]
- Sumová, A.; Sládek, M. Circadian Disruption as a Risk Factor for Development of Cardiovascular and Metabolic Disorders—From Animal Models to Human Population. Physiol. Res. 2024, 73 (Suppl. S1), S1–S14. [Google Scholar]
- Rutter, J.; Reick, M.; McKnight, S.L. Metabolism and the control of circadian rhythms. Annu. Rev. Biochem. 2002, 71, 307–331. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Mahan, K.; Sassone-Corsi, P. Metabolism and the Circadian Clock Converge. Physiol. Rev. 2013, 93, 107–135. [Google Scholar] [CrossRef] [PubMed]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef]
- Bass, J.; Takahashi, J.S. Circadian integration of metabolism and energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Damiola, F.; Schibler, U. A Serum Shock Induces Circadian Gene Expression in Mammalian Tissue Culture Cells. Cell 1998, 93, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Marcacci, L.; Schibler, U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 2000, 10, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Yagita, K.; Tamanini, F.; van Der Horst, G.T.; Okamura, H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 2001, 292, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Cela, O.; Scrima, R.; Pazienza, V.; Merla, G.; Benegiamo, G.; Augello, B.; Fugetto, S.; Menga, M.; Rubino, R.; Fuhr, L.; et al. Clock genes-dependent acetylation of complex I sets rhythmic activity of mitochondrial OxPhos. Biochim. Biophys. Acta 2016, 1863, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Scrima, R.; Cela, O.; Merla, G.; Augello, B.; Rubino, R.; Quarato, G.; Fugetto, S.; Menga, M.; Fuhr, L.; Relógio, A.; et al. Clock-genes and mitochondrial respiratory activity: Evidence of a reciprocal interplay. Biochim. Biophys. Acta 2016, 1857, 1344–1351. [Google Scholar] [CrossRef]
- Scrima, R.; Cela, O.; Agriesti, F.; Piccoli, C.; Tataranni, T.; Pacelli, C.; Mazzoccoli, G.; Capitanio, N. Mitochondrial calcium drives clock gene-dependent activation of pyruvate dehydrogenase and of oxidative phosphorylation. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118815. [Google Scholar] [CrossRef]
- Zhu, B.; Zhang, Q.; Pan, Y.; Mace, E.M.; York, B.; Antoulas, A.C.; Dacso, C.C.; O’Malley, B.W. A Cell-Autonomous Mammalian 12 Hr Clock Coordinates Metabolic and Stress Rhythms. Cell Metab. 2017, 25, 1305–1319. [Google Scholar] [CrossRef]
- Balance, H.; Zhu, B. Revealing the hidden reality of the mammalian 12-h ultradian rhythms. Cell Mol. Life Sci. 2021, 78, 3127–3140. [Google Scholar] [CrossRef]
- Pacelli, C.; Rotundo, G.; Lecce, L.; Menga, M.; Bidollari, E.; Scrima, R.; Cela, O.; Piccoli, C.; Cocco, T.; Vescovi, A.L.; et al. Parkin Mutation Affects Clock Gene-Dependent Energy Metabolism. Int. J. Mol. Sci. 2019, 20, 2772. [Google Scholar] [CrossRef]
- Schmitt, K.; Grimm, A.; Dallmann, R.; Oettinghaus, B.; Restelli, L.M.; Witzig, M.; Ishihara, N.; Mihara, K.; Ripperger, J.A.; Albrecht, U.; et al. Circadian Control of DRP1 Activity Regulates Mitochondrial Dynamics and Bioenergetics. Cell Metab. 2018, 27, 657–666. [Google Scholar] [CrossRef]
- Rodríguez-Santana, C.; López-Rodríguez, A.; Martinez-Ruiz, L.; Florido, J.; Cela, O.; Capitanio, N.; Ramírez-Casas, Y.; Acuña-Castroviejo, D.; Escames, G. The Relationship between Clock Genes, Sirtuin 1, and Mitochondrial Activity in Head and Neck Squamous Cell Cancer: Effects of Melatonin Treatment. Int. J. Mol. Sci. 2023, 24, 15030. [Google Scholar] [CrossRef]
- de Goede, P.; Wefers, J.; Brombacher, E.C.; Schrauwen, P.; Kalsbeek, A. Circadian rhythms in mitochondrial respiration. J. Mol. Endocrinol. 2018, 60, R115–R130. [Google Scholar] [CrossRef]
- Papa, S.; Martino, P.L.; Capitanio, G.; Gaballo, A.; De Rasmo, D.; Signorile, A.; Petruzzella, V. The oxidative phosphorylation system in mammalian mitochondria. Adv. Exp. Med. Biol. 2012, 942, 3–37. [Google Scholar]
- Vercellino, I.; Sazanov, L.A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell Biol. 2022, 23, 141–161. [Google Scholar] [CrossRef]
- Wikström, M.; Pecorilla, C.; Sharma, V. Chapter two: The mitochondrial respiratory chain. Enzymes 2023, 54, 15–36. [Google Scholar]
- Hoppe, U.C. Mitochondrial calcium channels. FEBS Lett. 2010, 584, 1975–1981. [Google Scholar] [CrossRef]
- Palmieri, F.; Pierri, C.L. Mitochondrial metabolite transport. Essays Biochem. 2010, 47, 37–52. [Google Scholar] [PubMed]
- Divakaruni, A.S.; Brand, M.D. The regulation and physiology of mitochondrial proton leak. Physiology 2011, 26, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, D.G. Mitochondrial proton leaks and uncoupling proteins. Biochim. Biophys. Acta Bioenerg. 2021, 1862, 148428. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.A.; Fisher-Wellman, K.H.; Neufer, P.D. From OCR and ECAR to energy: Perspectives on the design and interpretation of bioenergetics studies. J. Biol. Chem. 2021, 297, 101140. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D.; Nicholls, D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011, 435, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Ma, Y.; Liu, Y.; Wan, Q. Measurement of mitochondrial respiration in adherent cells by Seahorse XF96 Cell Mito Stress Test. STAR Protoc. 2020, 2, 100245. [Google Scholar] [CrossRef] [PubMed]
- Smolina, N.; Khudiakov, A.; Kostareva, A. Assaying Mitochondrial Respiration as an Indicator of Cellular Metabolism and Fitness. Methods Mol. Biol. 2023, 2644, 3–14. [Google Scholar] [PubMed]
- Zdrazilova, L.; Hansikova, H.; Gnaiger, E. Comparable respiratory activity in attached and suspended human fibroblasts. PLoS ONE 2022, 17, e0264496. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, G. Cosinor-based rhythmometry. Theor. Biol. Med. Model. 2014, 11, 16. [Google Scholar] [CrossRef]
- Westermark, P.O.; Herzel, H. Mechanism for 12 hr rhythm generation by the circadian clock. Cell Rep. 2013, 3, 1228–1238. [Google Scholar] [CrossRef]
- Zhu, B.; Dacso, C.C.; O’Malley, B.W. Unveiling “Musica Universalis” of the Cell: A Brief History of Biological 12-Hour Rhythms. J. Endocr. Soc. 2018, 2, 727–752. [Google Scholar] [CrossRef] [PubMed]
- Sardon Puig, L.; Valera-Alberni, M.; Cantó, C.; Pillon, N.J. Circadian Rhythms and Mitochondria: Connecting the Dots. Front. Genet. 2018, 9, 452. [Google Scholar] [CrossRef] [PubMed]
- Oliva-Ramírez, J.; Moreno-Altamirano, M.M.B.; Pineda-Olvera, B.; Cauich-Sánchez, P.; Sánchez-García, F.J. Crosstalk between circadian rhythmicity, mitochondrial dynamics and macrophage bactericidal activity. Immunology 2014, 143, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Ezagouri, S.; Asher, G. Circadian control of mitochondrial dynamics and functions. Curr. Opin. Physiol. 2018, 5, 25–29. [Google Scholar] [CrossRef]
- Nagoshi, E.; Brown, S.A.; Dibner, C.; Kornmann, B.; Schibler, U. Circadian gene expression in cultured cells. Methods Enzymol. 2005, 393, 543–557. [Google Scholar] [PubMed]
- Ramsey, K.M.; Yoshino, J.; Brace, C.S.; Abrassart, D.; Kobayashi, Y.; Marcheva, B.; Hong, H.K.; Chong, J.L.; Buhr, E.D.; Lee, C.; et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009, 324, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Masri, S.; Patel, V.R.; Eckel-Mahan, K.L.; Peleg, S.; Forne, I.; Ladurner, A.G.; Baldi, P.; Imhof, A.; Sassone-Corsi, P. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc. Natl. Acad. Sci. USA 2013, 110, 3339–3344. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Li, C.; Qi, X.; Song, Z.; Wu, J.; Hughes, M.E.; Li, X. The daily rhythms of mitochondrial gene expression and oxidative stress regulation are altered by aging in the mouse liver. Chronobiol. Int. 2015, 32, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Ji, Y.; Su, W.; Zhou, L.; Wu, X.; Gao, L.; Guo, J.; Liu, Y.; Zhang, Y.; Wen, X.; et al. The role of circadian clock-controlled mitochondrial dynamics in diabetic cardiomyopathy. Front. Immunol. 2023, 14, 1142512. [Google Scholar] [CrossRef]
- Latha Laxmi, I.P.; Job, A.T.; Manickam, V.; Tamizhselvi, R. Intertwined relationship of dynamin-related protein 1, mitochondrial metabolism and circadian rhythm. Mol. Biol. Rep. 2024, 51, 488. [Google Scholar] [CrossRef]
- Davis, K.; Roden, L.C.; Leaner, V.D.; van der Watt, P.J. The tumour suppressing role of the circadian clock. IUBMB Life 2019, 71, 771–780. [Google Scholar] [CrossRef]
- Hernández-Rosas, F.; López-Rosas, C.A.; Saavedra-Vélez, M.V. Disruption of the molecular circadian clock and cancer: An epigenetic link. Biochem. Genet. 2020, 58, 189–209. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Z.; Nice, E.; Huang, C.; Zhang, W.; Tang, Y. Circadian rhythms and cancers: The intrinsic links and therapeutic potentials. J. Hematol. Oncol. 2022, 15, 21. [Google Scholar] [CrossRef]
- Fuhr, L.; El-Athman, R.; Scrima, R.; Cela, O.; Carbone, A.; Knoop, H.; Li, Y.; Hoffmann, K.; Laukkanen, M.O.; Corcione, F.; et al. The Circadian Clock Regulates Metabolic Phenotype Rewiring Via HKDC1 and Modulates Tumor Progression and Drug Response in Colorectal Cancer. eBioMedicine 2018, 33, 105–121. [Google Scholar] [CrossRef]
- Altman, B.J. Cancer Clocks Out for Lunch: Disruption of Circadian Rhythm and Metabolic Oscillation in Cancer. Front. Cell Dev. Biol. 2016, 4, 62. [Google Scholar] [CrossRef]
- Straif, K.; Baan, R.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Bouvard, V.; Altieri, A.; Benbrahim-Tallaa, L.; Cogliano, V. WHO International Agency for Research on Cancer Monograph Working Group. Carcinogenicity of shiftwork, painting, and firefighting. Lancet Oncol. 2007, 8, 1065–1066. [Google Scholar] [CrossRef]
- Savvidis, C.; Koutsilieris, M. Circadian rhythm disruption in cancer biology. Mol. Med. 2012, 18, 1249–1260. [Google Scholar] [CrossRef]
- Uth, K.; Sleigh, R. Deregulation of the circadian clock constitutes a significant factor in tumorigenesis: A clockwork cancer. Part I: Clocks and clocking machinery. Biotechnol. Biotechnol. Equip. 2014, 28, 176–183. [Google Scholar] [CrossRef][Green Version]
- Uth, K.; Sleigh, R. Deregulation of the circadian clock constitutes a significant factor in tumorigenesis: A clockwork cancer. Part II. In vivo studies. Biotechnol. Biotechnol. Equip. 2014, 28, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Sahar, S.; Sassone-Corsi, P. Metabolism and cancer: The circadian clock connection. Nat. Rev. Cancer 2009, 9, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Basse, A.L.; Nielsen, K.N.; Karavaeva, I.; Ingerslev, L.R.; Ma, T.; Havelund, J.F.; Nielsen, T.S.; Frost, M.; Peics, J.; Dalbram, E.; et al. NAMPT-dependent NAD+ biosynthesis controls circadian metabolism in a tissue-specific manner. Proc. Natl. Acad. Sci. USA 2023, 120, e2220102120. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, E.K. AMP-activated protein kinase as a key molecular link between metabolism and clockwork. Exp. Mol. Med. 2013, 45, e33. [Google Scholar] [CrossRef]
- Lloyd, D.; Nanjundiah, V.; Engelmann, W.; Johnsson, A. Ultradian rhythms: Life’s dance to the music of time. J. Biosci. 2023, 48, 34. [Google Scholar] [CrossRef]
- Zhu, B.; Liu, S. Preservation of ~12-h ultradian rhythms of gene expression of mRNA and protein metabolism in the absence of canonical circadian clock. Front. Physiol. 2023, 14, 1195001. [Google Scholar] [CrossRef]
- Allada, R.; Bass, J. Circadian Mechanisms in Medicine. N. Engl. J. Med. 2021, 384, 550–561. [Google Scholar] [CrossRef]
- Ruan, W.; Yuan, X.; Eltzschig, H.K. Circadian rhythm as a therapeutic target. Nat. Rev. Drug Discov. 2021, 20, 287–307. [Google Scholar] [CrossRef]
- Wallace, D. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef]
- Wang, S.F.; Tseng, L.M.; Lee, H.C. Role of mitochondrial alterations in human cancer progression and cancer immunity. J. Biomed. Sci. 2023, 30, 61. [Google Scholar] [CrossRef]
- Fulda, S.; Galluzzi, L.; Kroemer, G. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 2010, 9, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Sainero-Alcolado, L.; Liaño-Pons, J.; Ruiz-Pérez, M.V.; Arsenian-Henriksson, M. Targeting mitochondrial metabolism for precision medicine in cancer. Cell Death Differ. 2022, 29, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Nencioni, A.; Caffa, I.; Cortellino, S.; Longo, V.D. Fasting and Cancer: Molecular Mechanisms and Clinical Application. Nat. Rev. Cancer 2018, 18, 707–719. [Google Scholar] [CrossRef]
- Clifton, K.K.; Ma, C.X.; Fontana, L.; Peterson, L.L. Intermittent Fasting in the Prevention and Treatment of Cancer. CA. Cancer J. Clin. 2021, 71, 527–546. [Google Scholar] [CrossRef]
- Mindikoglu, A.L.; Abdulsada, M.M.; Jain, A.; Choi, J.M.; Jalal, P.K.; Devaraj, S.; Mezzari, M.P.; Petrosino, J.F.; Opekun, A.R.; Jung, S.Y. Intermittent fasting from dawn to sunset for 30 consecutive days is associated with anticancer proteomic signature and upregulates key regulatory proteins of glucose and lipid metabolism, circadian clock, DNA repair, cytoskeleton remodeling, immune system and cognitive function in healthy subjects. J. Proteom. 2020, 217, 103645. [Google Scholar]
- Shi, D.; Fang, G.; Chen, Q.; Li, J.; Ruan, X.; Lian, X. Six-hour time-restricted feeding inhibits lung cancer progression and reshapes circadian metabolism. BMC Med. 2023, 21, 417. [Google Scholar] [CrossRef]


| Name | Organ | Type | Mutated Genes |
|---|---|---|---|
| HAEC | Aorta | Endothelial | N.A. |
| NHDF | Skin | Fibroblast | N.A. |
| Cal27 | Tongue | Squamous cell carcinoma | TP53, TERT, CDKN2A, KMT2A, LRBP1 |
| Hela | Cervix | Adenocarcinoma | HPV E6 E7, MED1, ERBB3 CASP8, HLA-A, TGFBR2 |
| HepG2 | Liver | Epithelial carcinoma | TP53, TERT promoter, CTNNB1 |
| SCC9 | Tongue | Squamous cell carcinoma | TP53, TERT, CDKN2A, KMT2A, LRBP1, HIF1A |
| SW480 | Colon | Adenocarcinoma | APC, TP53, ERBB2, KRAS |
| SW620 | Colon | Adenocarcinoma (met) | APC, TP53, KRAS |
| U2OS | Bone | Epithelial osteosarcoma | RB, TP53, RECQLC2, RECQLC3, RECQLC4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cela, O.; Scrima, R.; Pacelli, C.; Rosiello, M.; Piccoli, C.; Capitanio, N. Autonomous Oscillatory Mitochondrial Respiratory Activity: Results of a Systematic Analysis Show Heterogeneity in Different In Vitro-Synchronized Cancer Cells. Int. J. Mol. Sci. 2024, 25, 7797. https://doi.org/10.3390/ijms25147797
Cela O, Scrima R, Pacelli C, Rosiello M, Piccoli C, Capitanio N. Autonomous Oscillatory Mitochondrial Respiratory Activity: Results of a Systematic Analysis Show Heterogeneity in Different In Vitro-Synchronized Cancer Cells. International Journal of Molecular Sciences. 2024; 25(14):7797. https://doi.org/10.3390/ijms25147797
Chicago/Turabian StyleCela, Olga, Rosella Scrima, Consiglia Pacelli, Michela Rosiello, Claudia Piccoli, and Nazzareno Capitanio. 2024. "Autonomous Oscillatory Mitochondrial Respiratory Activity: Results of a Systematic Analysis Show Heterogeneity in Different In Vitro-Synchronized Cancer Cells" International Journal of Molecular Sciences 25, no. 14: 7797. https://doi.org/10.3390/ijms25147797
APA StyleCela, O., Scrima, R., Pacelli, C., Rosiello, M., Piccoli, C., & Capitanio, N. (2024). Autonomous Oscillatory Mitochondrial Respiratory Activity: Results of a Systematic Analysis Show Heterogeneity in Different In Vitro-Synchronized Cancer Cells. International Journal of Molecular Sciences, 25(14), 7797. https://doi.org/10.3390/ijms25147797







