Proteomic Analysis of Rap1A GTPase Signaling-Deficient C57BL/6 Mouse Pancreas and Functional Studies Identify an Essential Role of Rap1A in Pancreas Physiology
Abstract
1. Introduction
2. Results
2.1. Proteomic Analysis of Rap1A-Deficient Pancreas
2.1.1. Genotyping
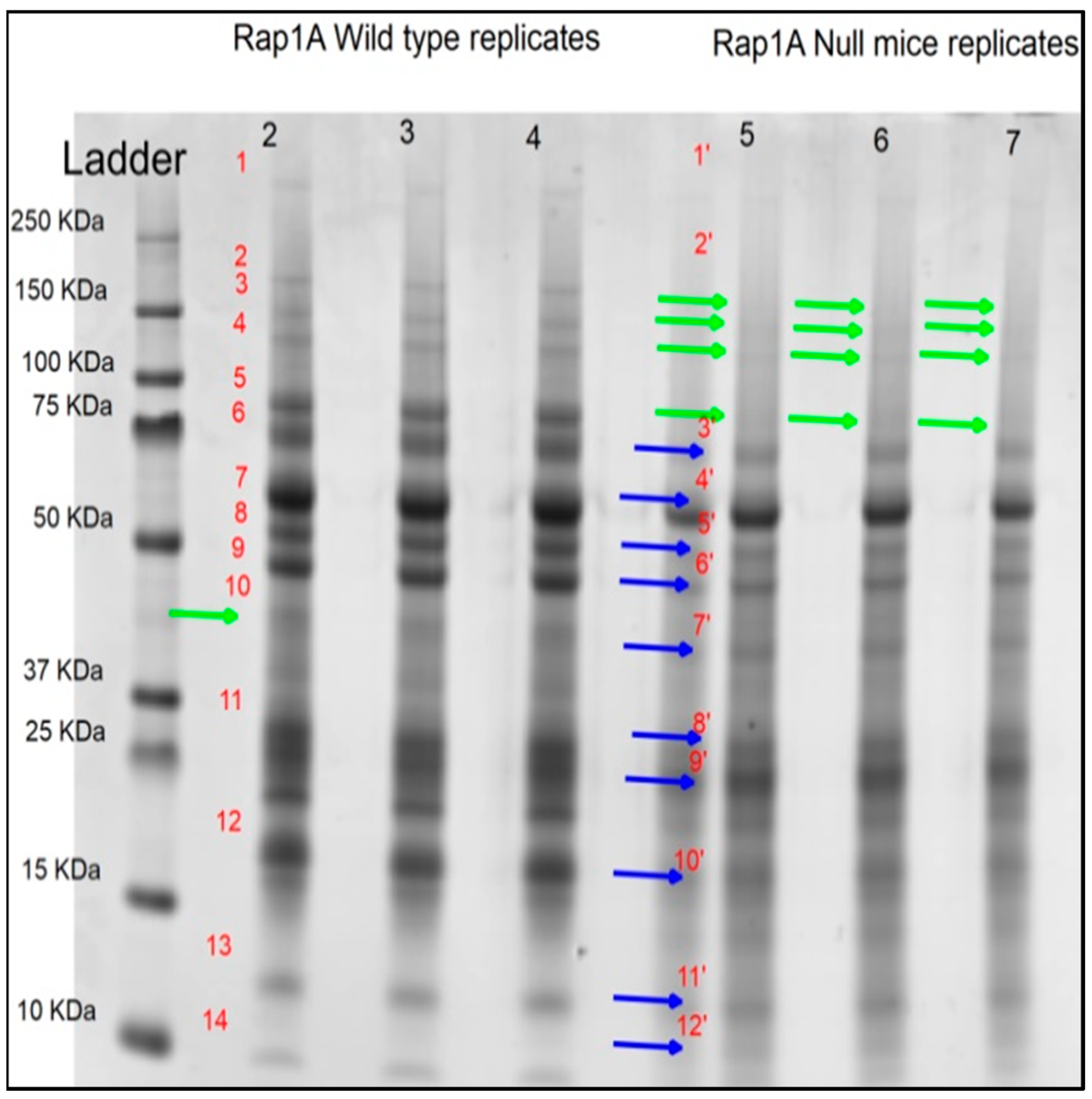
2.1.2. Pooled Sample SDS-PAGE for Detection of Differential Bands
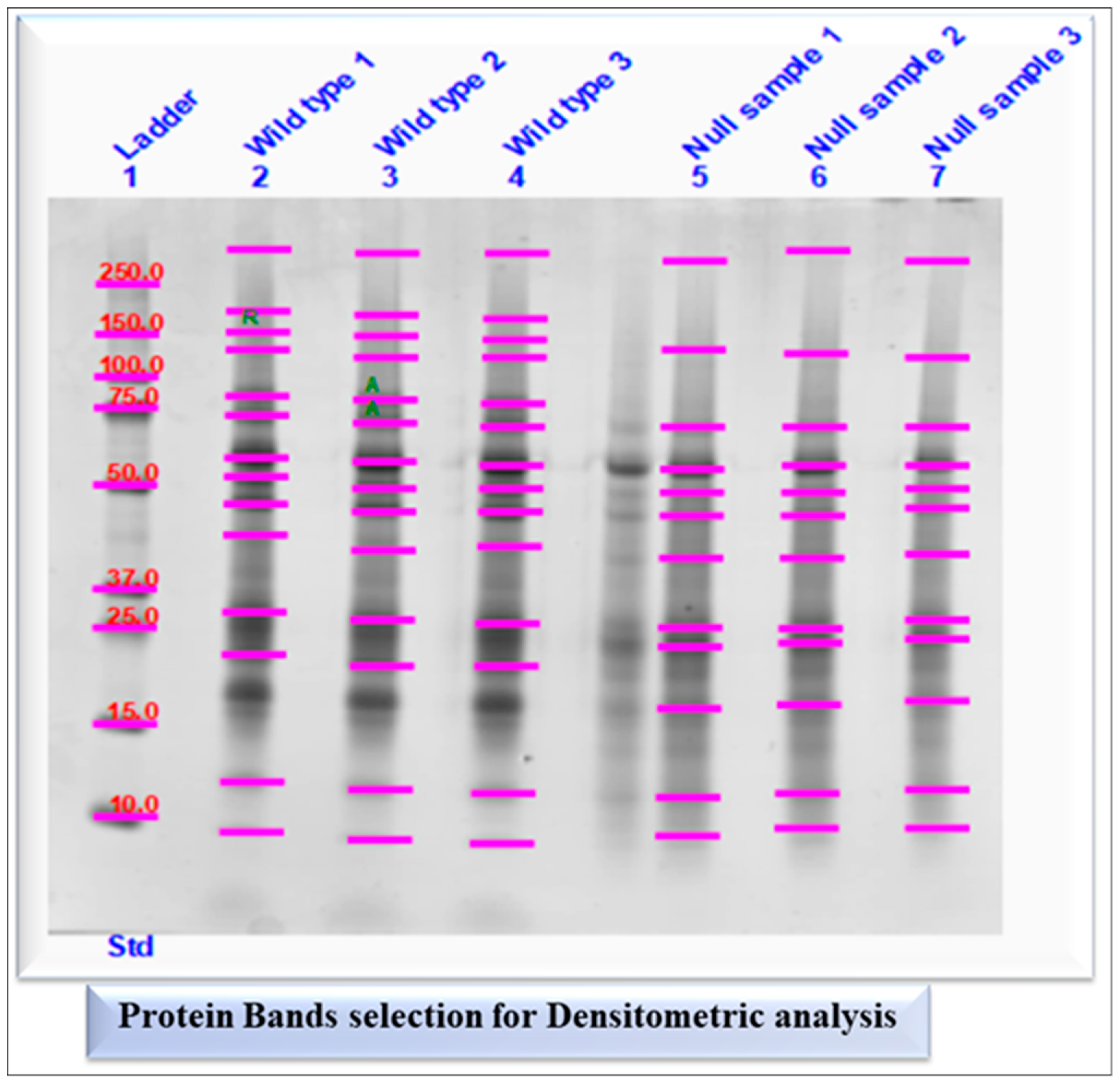
2.1.3. Densitometry Analysis of SDS-PAGE (1D Gel)
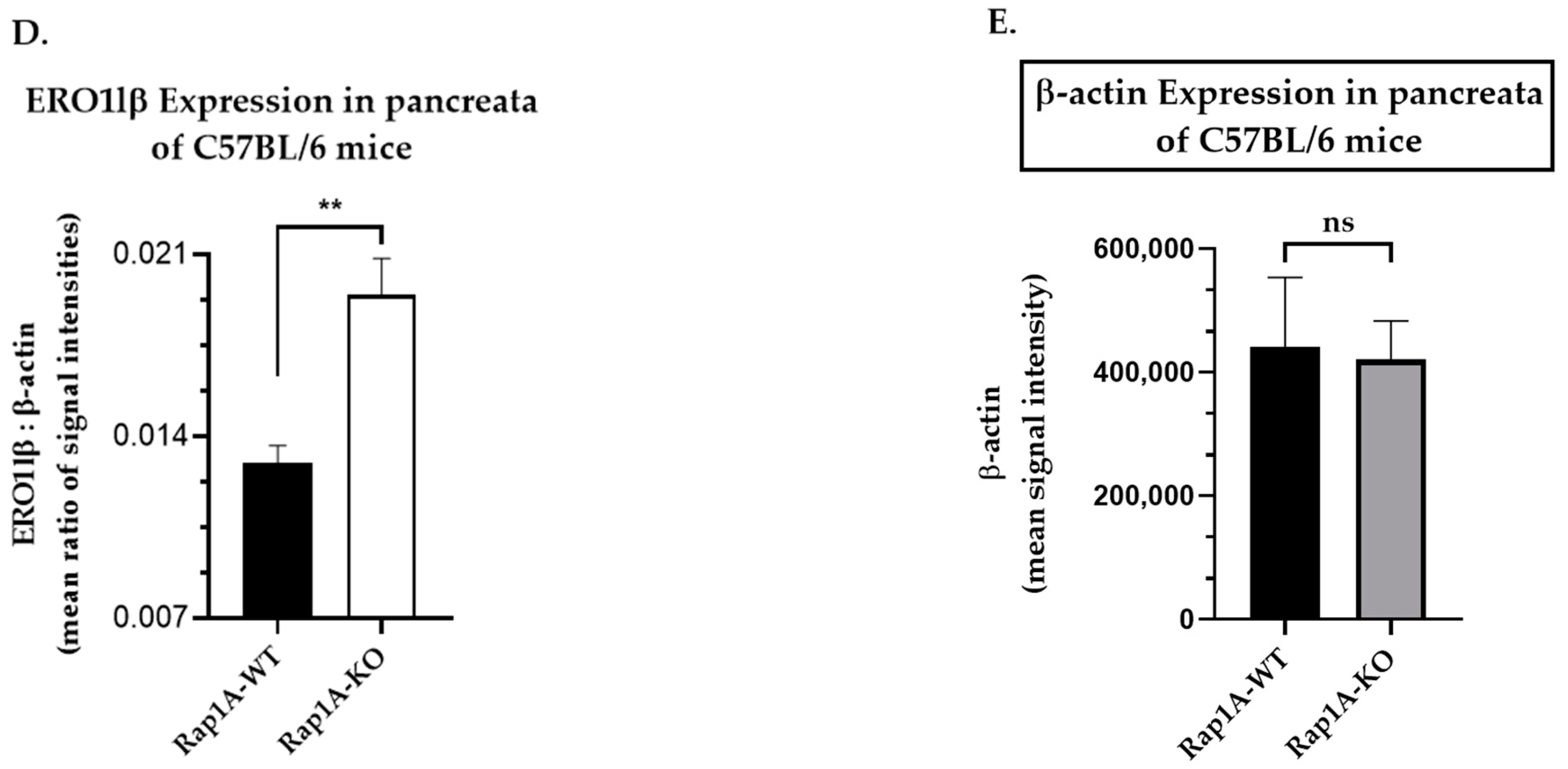
2.1.4. Semi-Quantification and Expression Analysis of Selected Protein Bands
2.1.5. Identification of Differentially Expressed Proteins by NanoLC-ESI-MS/MS
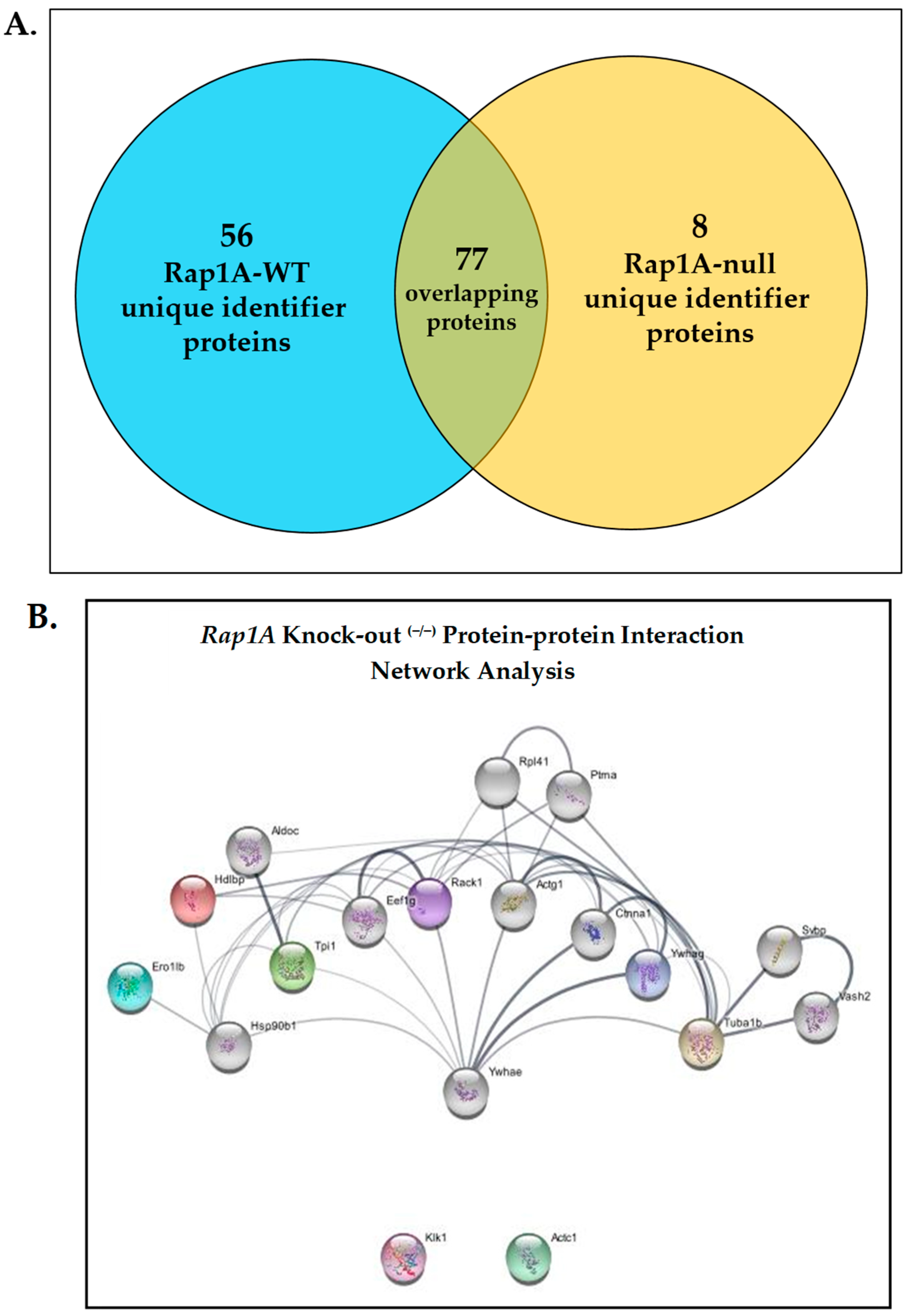
2.1.6. Protein–Protein Network Interaction Analysis
2.1.7. Gene Ontology Rap1A Knock-Out (−/−) and Wild-Type Groups in Terms of Biological Process
2.1.8. KEGG Pathway Analysis of Differentially Expressed Rap1A Knock-Out (−/−) and Wild-Type (+/+) Samples
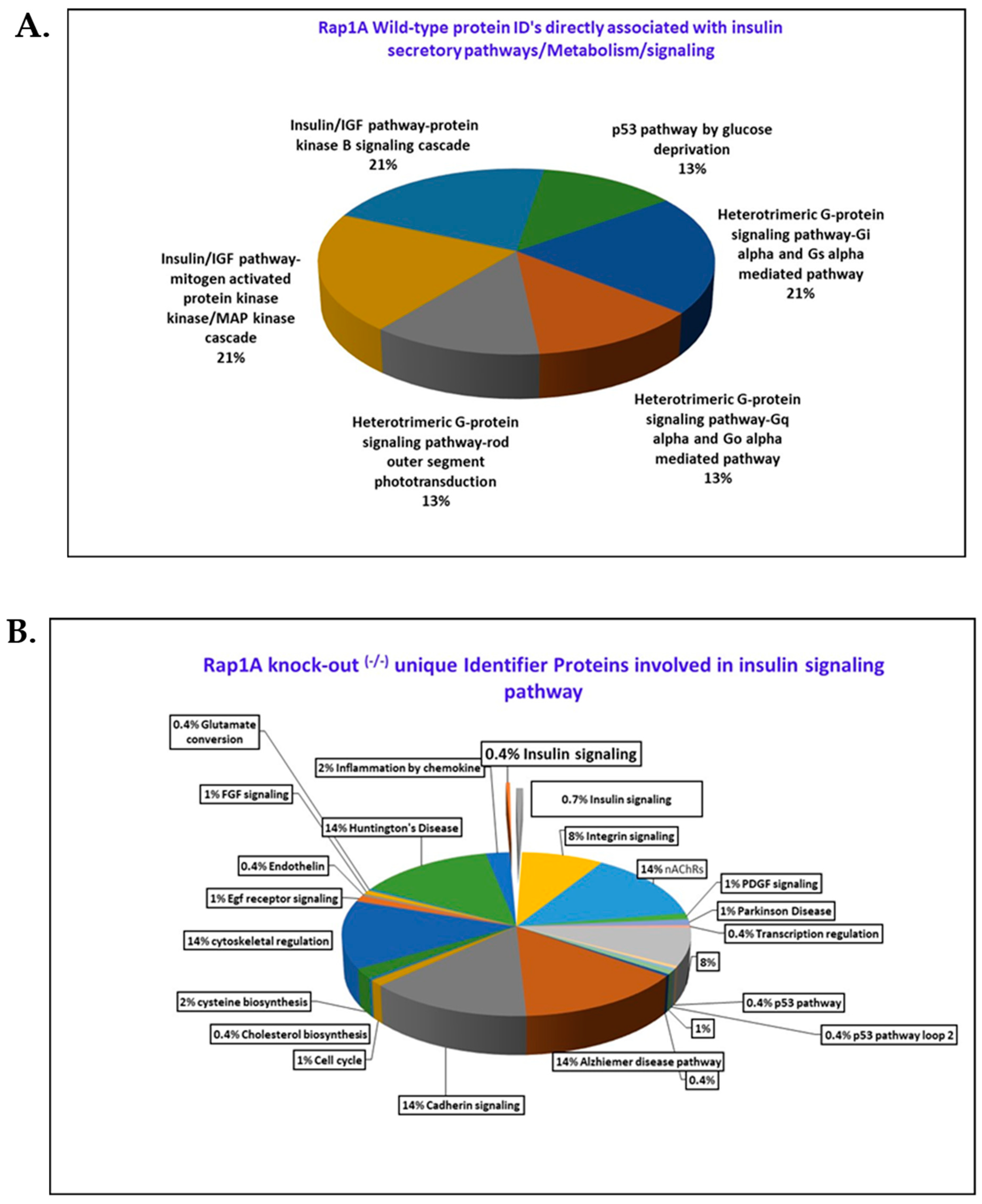
2.1.9. Wild-Type (+/+) and Rap1A Knock-Out (−/−) Unique Protein IDs’ Distinct Roles in Regulation of Insulin
2.2. Physiological Assessment of Rap1A-Deficient Mice
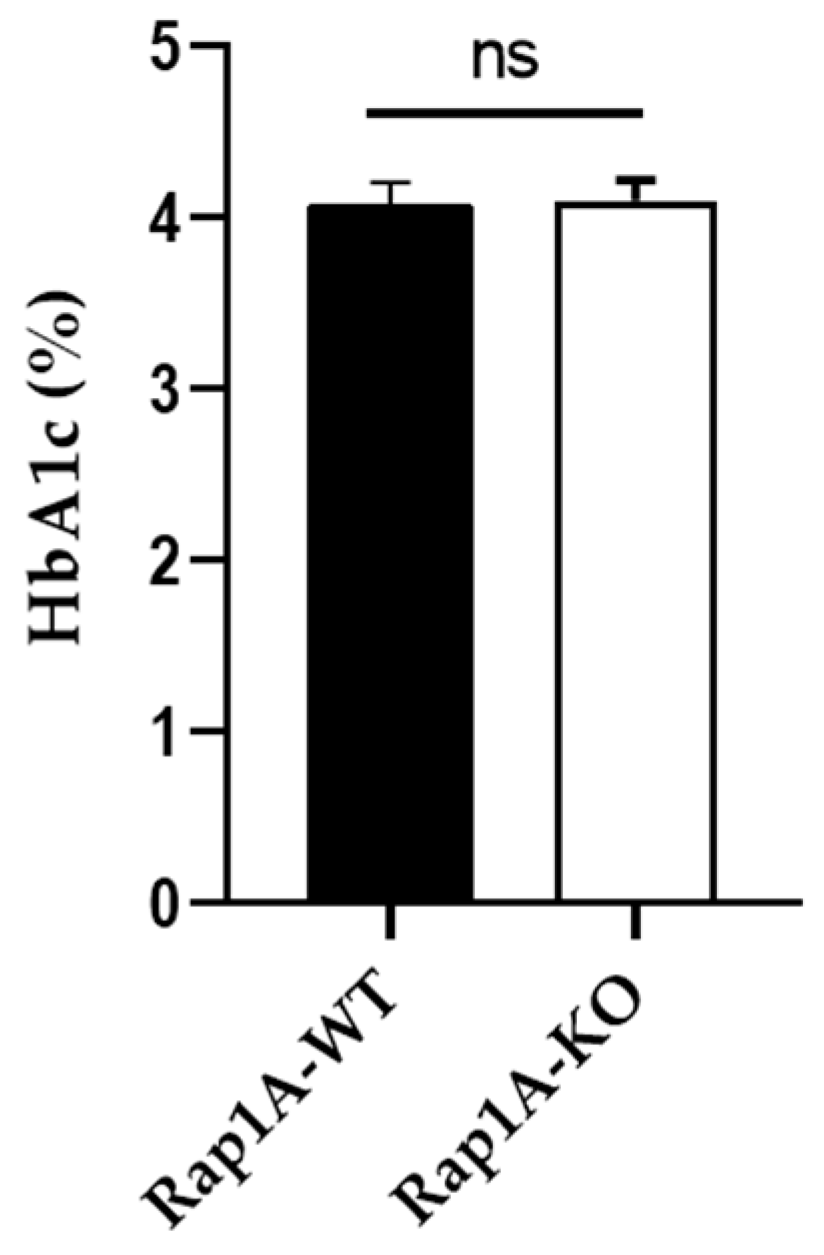
2.2.1. CBC and HbA1c
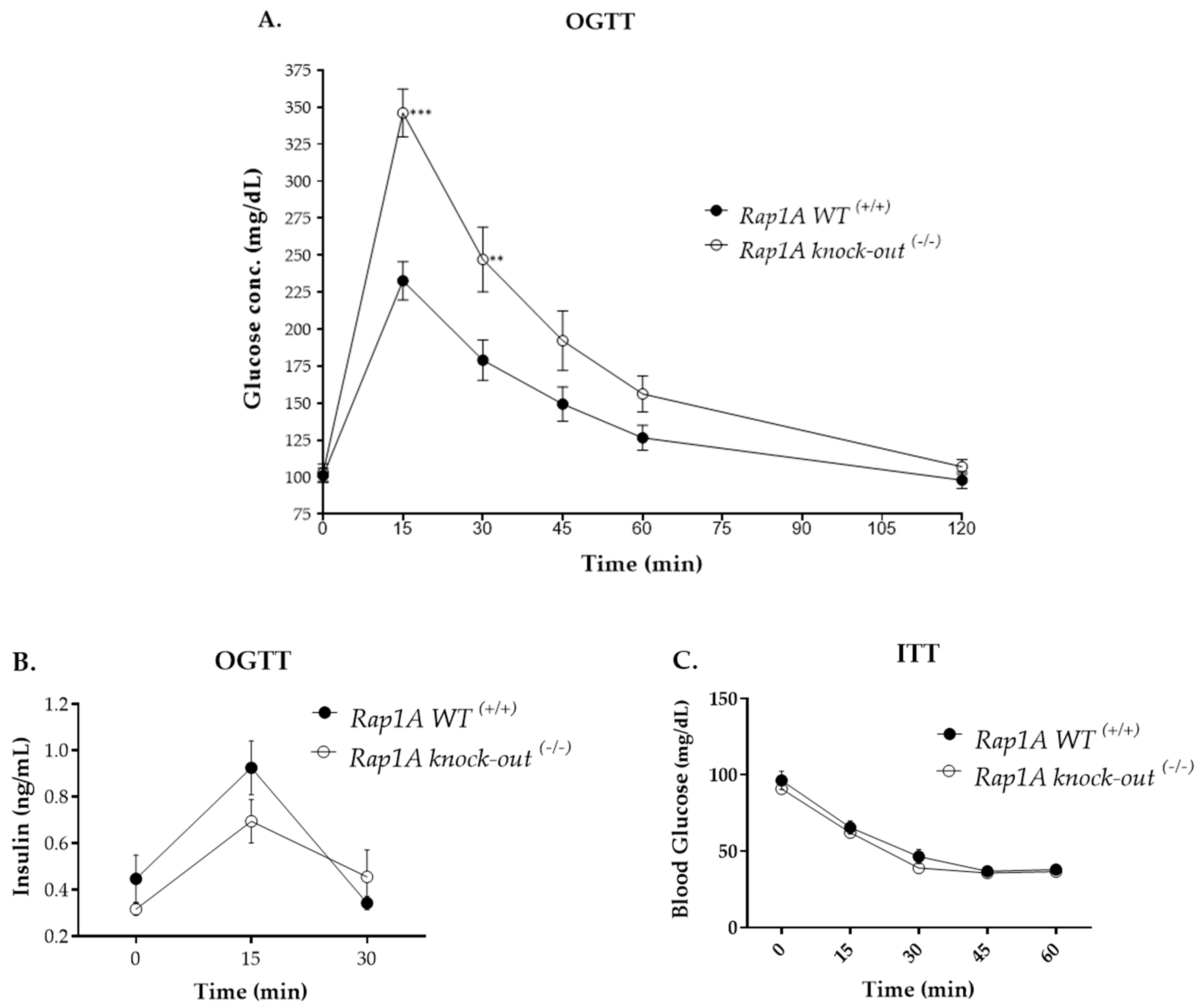
2.2.2. Oral Glucose Tolerance and Insulin Tolerance Tests
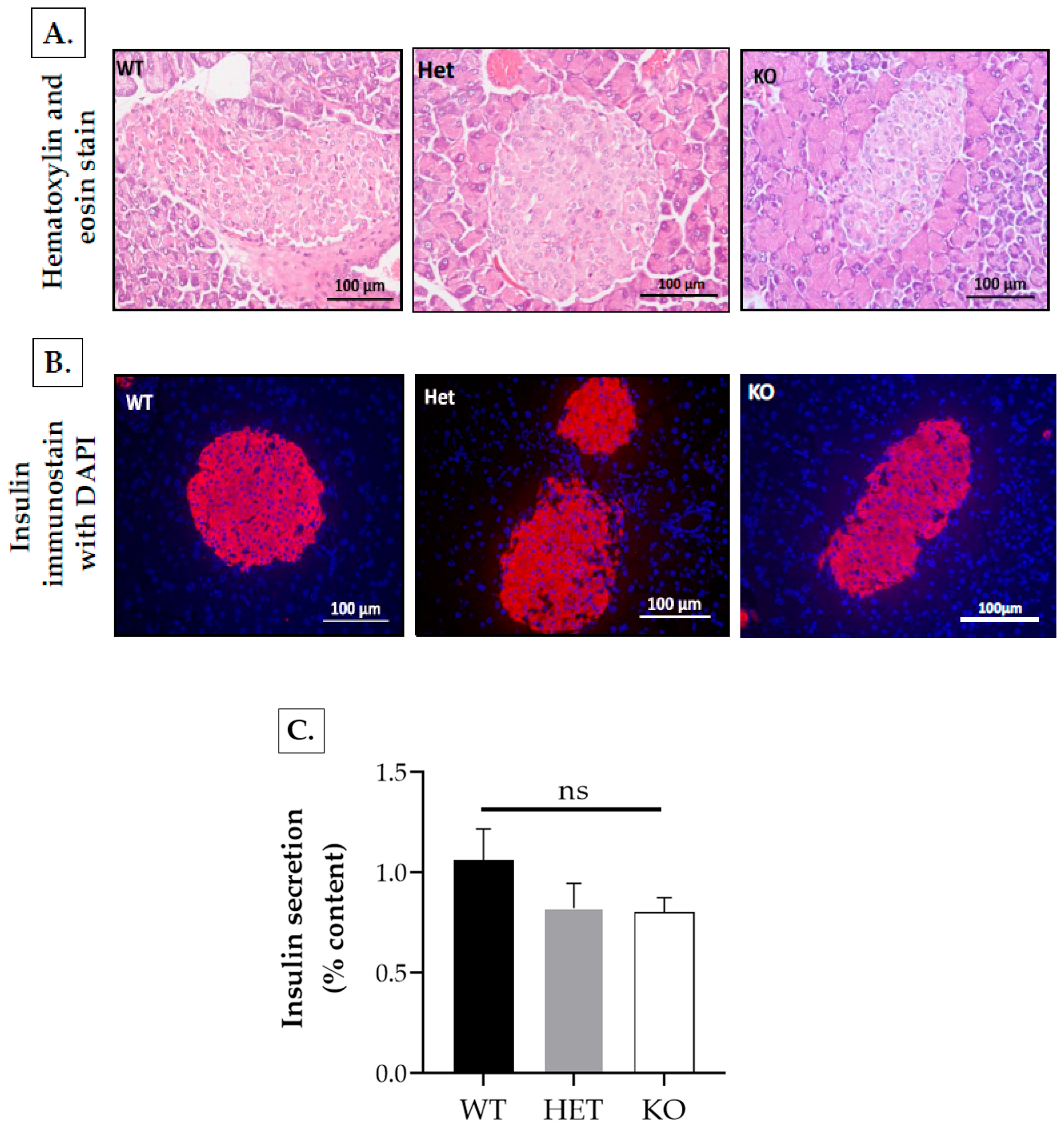
2.2.3. Insulin Secretion from Isolated Islets
3. Discussion
4. Materials and Methods
4.1. Animals
- (a)
- For proteomic studies: total of 10 male mice (5 wild type (+/+) and 5 knock-out (−/−)). Pancreatic tissue from the experimental group was harvested and stored at −80 °C.
- (b)
- For RNA analysis studies: see Section 4.6.1.
- (c)
- For Western blot analysis: see Section 4.7.
- (d)
- For physiological studies: see Section 4.8.
4.2. Proteomic Strategy
4.3. Nanoscale Liquid Chromatography Mass Spectrometry (nanoLC-ESI-MS/MS)
4.4. MS/MS Data Analysis
4.5. Bioinformatics: Functional Enrichment Analysis of DEPs
4.6. Gene Expression Analysis
4.6.1. RNA Isolation from Mouse Pancreatic Tissue
4.6.2. DNase Treatment
4.6.3. cDNA Synthesis
4.6.4. Primer Designing
4.6.5. Quantitative Real-Time PCR Analysis
4.7. Western Blot Analysis
4.8. Physiological Studies
4.8.1. CBC and HbA1c
4.8.2. Oral Glucose Tolerance Test (OGTT)
4.8.3. Insulin Tolerance Test (ITT)
4.8.4. Histology of Pancreata
4.8.5. Islet Isolation and Insulin Secretory Activity
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caron, E. Cellular functions of the Rap1 GTP-binding protein: A pattern emerges. J. Cell Sci. 2003, 116, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.L.; de Rooij, J.; Reedquist, K.A. Rap1 signalling: Adhering to new models. Nat. Rev. Mol. Cell Biol. 2001, 2, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, S.C.; Unger, N.T.; Chotani, M.A. Rap1 GTPases: An emerging role in the cardiovasculature. Life Sci. 2011, 88, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Ali, A.; Akhter, M.A.; Naeem, N.; Chotani, M.A.; Iqbal, H.; Kabir, N.; Atiq, M.; Salim, A. Epac-Rap1-activated mesenchymal stem cells improve cardiac function in rat model of myocardial infarction. Cardiovasc. Ther. 2017, 35, e12248. [Google Scholar] [CrossRef] [PubMed]
- Chotani, M.A.; Mitra, S.; Eid, A.H.; Han, S.A.; Flavahan, N.A. Distinct cAMP signaling pathways differentially regulate alpha2C-adrenoceptor expression: Role in serum induction in human arteriolar smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H69–H76. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eid, A.H.; Chotani, M.A.; Mitra, S.; Miller, T.J.; Flavahan, N.A. Cyclic AMP acts through Rap1 and JNK signaling to increase expression of cutaneous smooth muscle alpha2C-adrenoceptors. Am. J. Physiol. Heart Circ. Physiol. 2008, 295, H266–H272. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, S.C.; Unger, N.T.; Eid, A.H.; Mitra, S.; Paul El-Dahdah, N.; Quilliam, L.A.; Flavahan, N.A.; Chotani, M.A. Cyclic AMP-Rap1A signaling activates RhoA to induce α2c-adrenoceptor translocation to the cell surface of microvascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 2012, 303, C499–C511. [Google Scholar] [CrossRef] [PubMed]
- Motawea, H.K.; Jeyaraj, S.C.; Eid, A.H.; Mitra, S.; Unger, N.T.; Ahmed, A.A.; Flavahan, N.A.; Chotani, M.A. Cyclic AMP-Rap1A signaling mediates cell surface translocation of microvascular smooth muscle alpha2C-adrenoceptors through the actin-binding protein filamin-2. Am. J. Physiol. Cell Physiol. 2013, 305, C829–C845. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.; Bailey, C.L.; Fueger, P.T.; Newgard, C.B.; Casey, P.J.; Kimple, M.E. Rap1 promotes multiple pancreatic islet cell functions and signals through mammalian target of rapamycin complex 1 to enhance proliferation. J. Biol. Chem. 2010, 285, 15777–15785. [Google Scholar] [CrossRef]
- Shibasaki, T.; Takahashi, H.; Miki, T.; Sunaga, Y.; Matsumura, K.; Yamanaka, M.; Zhang, C.; Tamamoto, A.; Satoh, T.; Miyazaki, J.-i. Essential role of Epac2/Rap1 signaling in regulation of insulin granule dynamics by cAMP. Proc. Natl. Acad. Sci. USA 2007, 104, 19333–19338. [Google Scholar] [CrossRef]
- Veluthakal, R.; Thurmond, D.C. Emerging Roles of Small GTPases in Islet β-Cell Function. Cells 2021, 10, 1503. [Google Scholar] [CrossRef]
- Xiong, Q.Y.; Yu, C.; Zhang, Y.; Ling, L.; Wang, L.; Gao, J.L. Key proteins involved in insulin vesicle exocytosis and secretion. Biomed. Rep. 2017, 6, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, J.; De, P.; Chang, H.-C.; Yamauchi, A.; Christopherson, K.W.; Paranavitana, N.C.; Peng, X.; Kim, C.; Munugulavadla, V. Rap1a null mice have altered myeloid cell functions suggesting distinct roles for the closely related Rap1a and 1b proteins. J. Immunol. 2007, 179, 8322–8331. [Google Scholar] [CrossRef]
- Zito, E.; Chin, K.-T.; Blais, J.; Harding, H.P.; Ron, D. ERO1-β, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J. Cell Biol. 2010, 188, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.; Ahn, C. Low hemoglobin concentration is associated with several diabetic profiles. Korean J. Intern. Med. 2012, 27, 273–274. [Google Scholar] [CrossRef]
- Selvin, E.; Steffes, M.W.; Zhu, H.; Matsushita, K.; Wagenknecht, L.; Pankow, J.; Coresh, J.; Brancati, F.L. Glycated hemoglobin, diabetes, and cardiovascular risk in nondiabetic adults. N. Engl. J. Med. 2010, 362, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Daniels Gatward, L.F.; Kennard, M.R.; Smith, L.I.F.; King, A.J.F. The use of mice in diabetes research: The impact of physiological characteristics, choice of model and husbandry practices. Diabet. Med. 2021, 38, e14711. [Google Scholar] [CrossRef]
- Han, B.G.; Hao, C.M.; Tchekneva, E.E.; Wang, Y.Y.; Lee, C.A.; Ebrahim, B.; Harris, R.C.; Kern, T.S.; Wasserman, D.H.; Breyer, M.D.; et al. Markers of glycemic control in the mouse: Comparisons of 6-h- and overnight-fasted blood glucoses to Hb A1c. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E981–E986. [Google Scholar] [CrossRef]
- Siegel, E.G.; Wollheim, C.B.; Kikuchi, M.; Renold, A.E.; Sharp, G.W. Dependency of cyclic AMP-induced insulin release on intra- and extracellular calcium in rat islets of Langerhans. J. Clin. Investig. 1980, 65, 233–241. [Google Scholar] [CrossRef]
- Hameed, A.; Hafizur, R.M.; Khan, M.I.; Jawed, A.; Wang, H.; Zhao, M.; Matsunaga, K.; Izumi, T.; Siddiqui, S.; Khan, F.; et al. Coixol amplifies glucose-stimulated insulin secretion via cAMP mediated signaling pathway. Eur. J. Pharmacol. 2019, 858, 172514. [Google Scholar] [CrossRef]
- Hameed, A.; Raza, S.A.; Israr Khan, M.; Baral, J.; Adhikari, A.; Nur, E.A.M.; Ahmed, S.; Al-Rehaily, A.J.; Ashraf, S.; Ul-Haq, Z.; et al. Tambulin from Zanthoxylum armatum acutely potentiates the glucose-induced insulin secretion via K(ATP)-independent Ca(2+)-dependent amplifying pathway. Biomed. Pharmacother. 2019, 120, 109348. [Google Scholar] [CrossRef] [PubMed]
- Cerasi, E. Mechanisms of glucose stimulated insulin secretion in health and in diabetes: Some re-evaluations and proposals. Diabetologia 1975, 11, 1–13. [Google Scholar] [CrossRef]
- Chadt, A.; Al-Hasani, H. Glucose transporters in adipose tissue, liver, and skeletal muscle in metabolic health and disease. Pflügers Arch. Eur. J. Physiol. 2020, 472, 1273–1298. [Google Scholar] [CrossRef]
- Ashcroft, F.M.; Proks, P.; Smith, P.A.; Ämmälä, C.; Bokvist, K.; Rorsman, P. Stimulus–secretion coupling in pancreatic β cells. J. Cell. Biochem. 1994, 55, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Doliba, N.M.; Qin, W.; Najafi, H.; Liu, C.; Buettger, C.W.; Sotiris, J.; Collins, H.W.; Li, C.; Stanley, C.A.; Wilson, D.F. Glucokinase activation repairs defective bioenergetics of islets of Langerhans isolated from type 2 diabetics. Am. J. Physiol. -Endocrinol. Metab. 2012, 302, E87–E102. [Google Scholar] [CrossRef]
- Prentki, M.; Corkey, B.E.; Madiraju, S. Lipid-associated metabolic signalling networks in pancreatic beta cell function. Diabetologia 2020, 63, 10–20. [Google Scholar] [CrossRef]
- Kowluru, A. Small G proteins in islet β-cell function. Endocr. Rev. 2010, 31, 52–78. [Google Scholar] [CrossRef] [PubMed]
- Rorsman, P.; Braun, M. Regulation of insulin secretion in human pancreatic islets. Annu. Rev. Physiol. 2013, 75, 155–179. [Google Scholar] [CrossRef] [PubMed]
- Leech, C.A.; Chepurny, O.G.; Holz, G.G. Epac2-dependent rap1 activation and the control of islet insulin secretion by glucagon-like peptide-1. Vitam. Horm. 2010, 84, 279–302. [Google Scholar]
- Seino, S.; Shibasaki, T.; Minami, K. Dynamics of insulin secretion and the clinical implications for obesity and diabetes. J. Clin. Invest. 2011, 121, 2118–2125. [Google Scholar] [CrossRef]
- Holz, G.G. Epac: A new cAMP-binding protein in support of glucagon-like peptide-1 receptor-mediated signal transduction in the pancreatic β-cell. Diabetes 2004, 53, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Janoueix-Lerosey, I.; Pasheva, E.; de Tand, M.F.; Tavitian, A.; de Gunzburg, J. Identification of a specific effector of the small GTP-binding protein Rap2. Eur. J. Biochem. 1998, 252, 290–298. [Google Scholar] [CrossRef] [PubMed]
- McAvoy, T.; Zhou, M.-M.; Greengard, P.; Nairn, A.C. Phosphorylation of Rap1GAP, a striatally enriched protein, by protein kinase A controls Rap1 activity and dendritic spine morphology. Proc. Natl. Acad. Sci. USA 2009, 106, 3531–3536. [Google Scholar] [CrossRef]
- Shergalis, A.G.; Hu, S.; Bankhead III, A.; Neamati, N. Role of the ERO1-PDI interaction in oxidative protein folding and disease. Pharmacol. Ther. 2020, 210, 107525. [Google Scholar] [CrossRef] [PubMed]
- Khoo, C.; Yang, J.; Rajpal, G.; Wang, Y.; Liu, J.; Arvan, P.; Stoffers, D.A. Endoplasmic reticulum oxidoreductin-1-like β (ERO1lβ) regulates susceptibility to endoplasmic reticulum stress and is induced by insulin flux in β-cells. Endocrinology 2011, 152, 2599–2608. [Google Scholar] [CrossRef] [PubMed]
- Pagani, M.; Fabbri, M.; Benedetti, C.; Fassio, A.; Pilati, S.; Bulleid, N.J.; Cabibbo, A.; Sitia, R. Endoplasmic Reticulum Oxidoreductin 1-Lβ (ERO1-Lβ), a Human Gene Induced in the Course of the Unfolded Protein Response* 210. J. Biol. Chem. 2000, 275, 23685–23692. [Google Scholar] [CrossRef]
- Furman, B.; Ong, W.K.; Pyne, N.J. Cyclic AMP signaling in pancreatic islets. Islets Langerhans 2010, 654, 281–304. [Google Scholar]
- Tengholm, A. Cyclic AMP dynamics in the pancreatic β-cell. Upsala J. Med. Sci. 2012, 117, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Seino, S.; Sugawara, K.; Yokoi, N.; Takahashi, H. β-Cell signalling and insulin secretagogues: A path for improved diabetes therapy. Diabetes, Obes. Metab. 2017, 19, 22–29. [Google Scholar] [CrossRef]
- Nichols, C.G.; York, N.W.; Remedi, M.S. Preferential Gq signaling in diabetes: An electrical switch in incretin action and in diabetes progression? J. Clin. Investig. 2020, 130, 6235–6237. [Google Scholar] [CrossRef]
- Gucek, A.; Gandasi, N.R.; Omar-Hmeadi, M.; Bakke, M.; Doskeland, S.; Tengholm, A.; Barg, S. Fusion Pore Regulation by EPAC2/cAMP Controls Cargo Release during Insulin Exocytosis. Biophys. J. 2019, 116, 314a. [Google Scholar] [CrossRef]
- Takahashi, H.; Shibasaki, T.; Park, J.-H.; Hidaka, S.; Takahashi, T.; Ono, A.; Song, D.-K.; Seino, S. Role of Epac2A/Rap1 signaling in interplay between incretin and sulfonylurea in insulin secretion. Diabetes 2015, 64, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Lenstra, J.A.; Beintema, J.J. The amino acid sequence of mouse pancreatic ribonuclease. Extremely rapid evolutionary rates of the myomorph rodent ribonucleases. Eur. J. Biochem. 1979, 98, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Chougoni, K.K.; Grossman, S.R. Extraction of high-quality RNA from mouse pancreatic tumors. MethodsX 2020, 7, 101163. [Google Scholar] [CrossRef] [PubMed]
- Andrikopoulos, S.; Blair, A.R.; Deluca, N.; Fam, B.C.; Proietto, J. Evaluating the glucose tolerance test in mice. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E1323–E1332. [Google Scholar] [CrossRef] [PubMed]
- Vinué, Á.; González-Navarro, H. Glucose and insulin tolerance tests in the mouse. In Methods in Mouse Atherosclerosis; Springer: Berlin/Heidelberg, Germany, 2015; pp. 247–254. [Google Scholar]
- Hameed, A.; Hafizur, R.M.; Hussain, N.; Raza, S.A.; Rehman, M.; Ashraf, S.; Ul-Haq, Z.; Khan, F.; Abbas, G.; Choudhary, M.I. Eriodictyol stimulates insulin secretion through cAMP/PKA signaling pathway in mice islets. Eur. J. Pharmacol. 2018, 820, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Garcia-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Perez-Riverol, Y.; Sharma, V.; Carver, J.J.; Mendoza, L.; Kundu, D.J.; Wang, S.; Bandla, C.; Kamatchinathan, S.; et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2023, 51, D1539–D1548. [Google Scholar] [CrossRef]





| Genotype | Rap1A Knock-Out (−/−) | Rap1A WT (+/+) | |
|---|---|---|---|
| N= | 6 | 5 | |
| Hemoglobin (g/dL) | 11.25 ± 0.3981 | 10.4 ± 0.5908 | |
| Red Blood Cell Profile | Red Blood Cell Count (million/mL) | 8.043 ± 0.2714 | 7.374 ± 0.4701 |
| Mean Cell Volume (MCV), (fL) | 47.03 ± 0.65 | 48.12 ± 2.123 | |
| Mean Cell Hemoglobin Concentration (MCHC), (G/dL) | 29.73 ± 0.4240 | 29.50 ± 0.906 | |
| Total Leucocyte Count (10³/µL) | 2.050 ± 0.7361 | 2.380 ± 1.122 | |
| Differential White Blood Cell Count | Neutrophils | 16.833 ± 9.816 | 17.8 ± 12.82 |
| Lymphocytes | 79.333 ± 10.26 | 79 ± 12.759 | |
| Monocytes | 2.16 ± 0.47 | 2 ± 0.31 | |
| Eosinophils | 1.667 ± 0.33 | 1.2 ± 0.2 | |
| Platelets (10³/µL) | 608.7 ± 161 | 751.4 ± 276.8 | |
| Gene | NCBI Accession ID | Primer sequence (5’→3’) Forward (F) and Reverse (R) Primer | PCR Product Length |
|---|---|---|---|
| Actb | NM_007393.5 | F-AAGTGTGACGTTGACATCCGTAAAG | 307 bp |
| R-TGTAAAACGCAGCTCAGTAACAGTC | |||
| Ero1b | NM_026184.2 | F-CAGGGTTTAGGAACTGCCTTG | 258 bp |
| R-CCAGTGTCCAAGGCTAAAAGG | |||
| Tpi1 | NM_009415.3 | F-AGCACCCGGATCATTTATGG | 364 bp |
| R-CCACCTTGGTGACAGTTGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahwar, D.; Baqai, S.; Khan, F.; Khan, M.I.; Javaid, S.; Hameed, A.; Raza, A.; Saleem Uddin, S.; Hazrat, H.; Rahman, M.H.; et al. Proteomic Analysis of Rap1A GTPase Signaling-Deficient C57BL/6 Mouse Pancreas and Functional Studies Identify an Essential Role of Rap1A in Pancreas Physiology. Int. J. Mol. Sci. 2024, 25, 8013. https://doi.org/10.3390/ijms25158013
Shahwar D, Baqai S, Khan F, Khan MI, Javaid S, Hameed A, Raza A, Saleem Uddin S, Hazrat H, Rahman MH, et al. Proteomic Analysis of Rap1A GTPase Signaling-Deficient C57BL/6 Mouse Pancreas and Functional Studies Identify an Essential Role of Rap1A in Pancreas Physiology. International Journal of Molecular Sciences. 2024; 25(15):8013. https://doi.org/10.3390/ijms25158013
Chicago/Turabian StyleShahwar, Durrey, Sadaf Baqai, Faisal Khan, M. Israr Khan, Shafaq Javaid, Abdul Hameed, Aisha Raza, Sadaf Saleem Uddin, Hina Hazrat, M. Hafizur Rahman, and et al. 2024. "Proteomic Analysis of Rap1A GTPase Signaling-Deficient C57BL/6 Mouse Pancreas and Functional Studies Identify an Essential Role of Rap1A in Pancreas Physiology" International Journal of Molecular Sciences 25, no. 15: 8013. https://doi.org/10.3390/ijms25158013
APA StyleShahwar, D., Baqai, S., Khan, F., Khan, M. I., Javaid, S., Hameed, A., Raza, A., Saleem Uddin, S., Hazrat, H., Rahman, M. H., Musharraf, S. G., & Chotani, M. A. (2024). Proteomic Analysis of Rap1A GTPase Signaling-Deficient C57BL/6 Mouse Pancreas and Functional Studies Identify an Essential Role of Rap1A in Pancreas Physiology. International Journal of Molecular Sciences, 25(15), 8013. https://doi.org/10.3390/ijms25158013







