Characterization of Chemoresistance in Pancreatic Cancer: A Look at MDR-1 Polymorphisms and Expression in Cancer Cells and Patients
Abstract
1. Introduction
2. Results
2.1. MDR-1 Is Highly Conserved across the Vertebrate Evolutionary Lineage
2.2. Pancreatic Cancer and Non-Cancer Cell Lines Exhibit Different MDR-1 Polymorphisms
2.3. The Analysis of MDR-1 Polymorphisms from Patients with Pancreatic Ductal Cancers Reveals a High Frequency of the Selected Variants
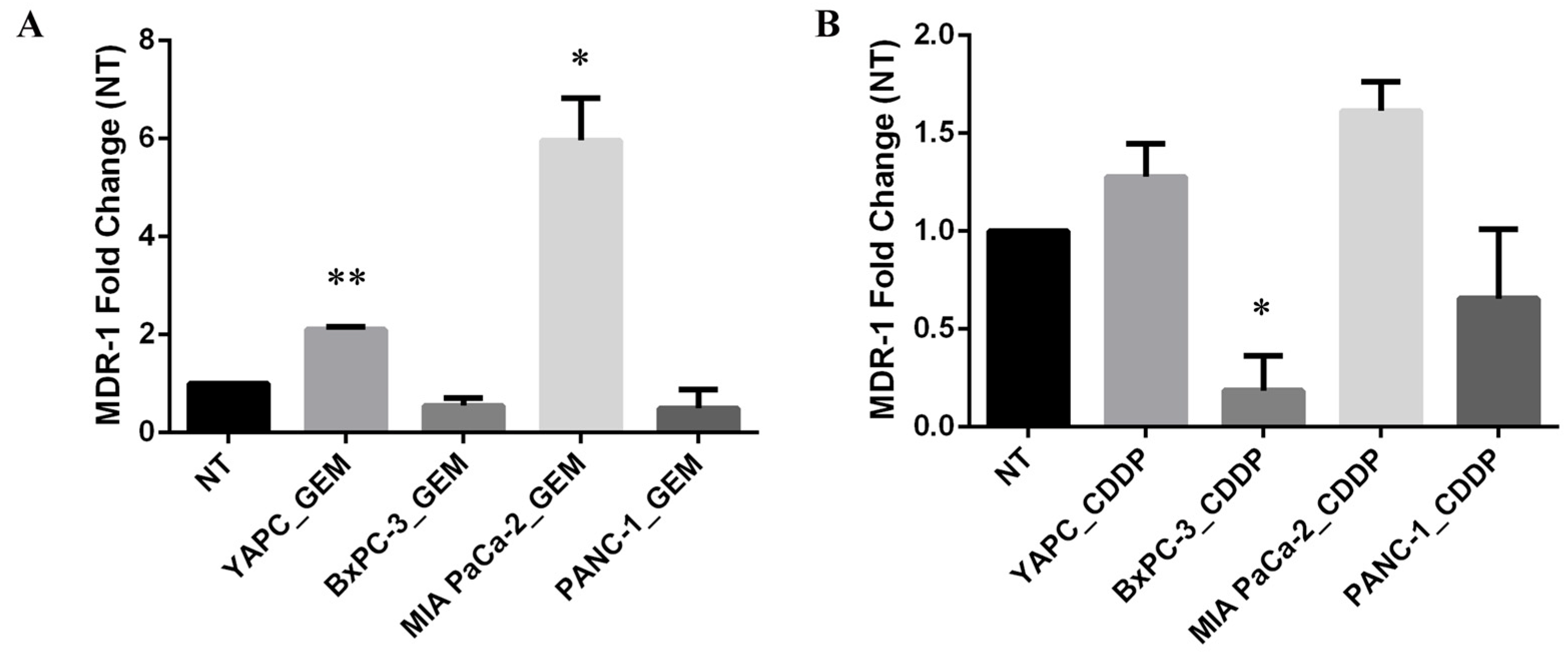
2.4. MDR-1 Expression Is Altered in Pancreatic Cancer Cell Lines by Chemotherapeutic Drug Treatments
2.5. Analysis of MDR-1 Expression in Pancreatic Cancer Patients
3. Discussion
4. Materials and Methods
4.1. Case Series and Samples
4.2. Cell Lines
4.3. DNA, RNA, and Protein Extraction from Patient Tissues
4.4. DNA, RNA, and Protein Extraction from Cell Lines
4.5. Genotyping using the Polymerase Chain Reaction (PCR)
4.6. Cell Treatment and Viability Measurement in the Presence of Cisplatin
4.7. Quantitative Real-Time PCR (qRT-PCR)
4.8. Western Blotting
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic Cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic Cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef]
- Kaur, S.; Smith, L.M.; Patel, A.; Menning, M.; Watley, D.C.; Malik, S.S.; Krishn, S.R.; Mallya, K.; Aithal, A.; Sasson, A.R.; et al. A Combination of MUC5AC and CA19-9 Improves the Diagnosis of Pancreatic Cancer: A Multicenter Study. Am. J. Gastroenterol. 2017, 112, 172–183. [Google Scholar] [CrossRef]
- Kaur, S.; Baine, M.J.; Jain, M.; Sasson, A.R.; Batra, S.K. Early Diagnosis of Pancreatic Cancer: Challenges and New Developments. Biomark. Med. 2012, 6, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Romano, R.; Picca, A.; Eusebi, L.H.U.; Marzetti, E.; Calvani, R.; Moro, L.; Bucci, C.; Guerra, F. Extracellular Vesicles and Pancreatic Cancer: Insights on the Roles of miRNA, lncRNA, and Protein Cargos in Cancer Progression. Cells 2021, 10, 1361. [Google Scholar] [CrossRef] [PubMed]
- Girolimetti, G.; Pelisenco, I.A.; Eusebi, L.H.; Ricci, C.; Cavina, B.; Kurelac, I.; Verri, T.; Calcagnile, M.; Alifano, P.; Salvi, A.; et al. Dysregulation of a Subset of Circulating and Vesicle-Associated miRNA in Pancreatic Cancer. Noncoding RNA 2024, 10, 29. [Google Scholar] [CrossRef]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic Cancer: A Review of Clinical Diagnosis, Epidemiology, Treatment and Outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef]
- Labori, K.J.; Katz, M.H.; Tzeng, C.W.; Bjørnbeth, B.A.; Cvancarova, M.; Edwin, B.; Kure, E.H.; Eide, T.J.; Dueland, S.; Buanes, T.; et al. Impact of Early Disease Progression and Surgical Complications on Adjuvant Chemotherapy Completion Rates and Survival in Patients Undergoing the Surgery First Approach for Resectable Pancreatic Ductal Adenocarcinoma—A Population-Based Cohort Study. Acta Oncol. 2016, 55, 265–277. [Google Scholar] [CrossRef]
- Zeng, S.; Pöttler, M.; Lan, B.; Grützmann, R.; Pilarsky, C.; Yang, H. Chemoresistance in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 4504. [Google Scholar] [CrossRef] [PubMed]
- Burris, H.A.; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Cripps, M.C.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P.; et al. Improvements in Survival and Clinical Benefit with Gemcitabine as First-Line Therapy for Patients with Advanced Pancreas Cancer: A Randomized Trial. J. Clin. Oncol. 1997, 15, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.-L.; Gourgou-Bourgade, S.; de la Fouchardière, C.; et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Hwang, I.; Yoo, C.; Kim, K.-P.; Jeong, J.H.; Chang, H.-M.; Lee, S.S.; Park, D.H.; Song, T.J.; Seo, D.W.; et al. Nab-Paclitaxel plus Gemcitabine versus FOLFIRINOX as the First-Line Chemotherapy for Patients with Metastatic Pancreatic Cancer: Retrospective Analysis. Investig. New Drugs 2018, 36, 732–741. [Google Scholar] [CrossRef]
- McBride, A.; Bonafede, M.; Cai, Q.; Princic, N.; Tran, O.; Pelletier, C.; Parisi, M.; Patel, M. Comparison of Treatment Patterns and Economic Outcomes among Metastatic Pancreatic Cancer Patients Initiated on Nab-Paclitaxel plus Gemcitabine versus FOLFIRINOX. Expert Rev. Clin. Pharmacol. 2017, 10, 1153–1160. [Google Scholar] [CrossRef]
- Peixoto, R.D.; Ho, M.; Renouf, D.J.; Lim, H.J.; Gill, S.; Ruan, J.Y.; Cheung, W.Y. Eligibility of Metastatic Pancreatic Cancer Patients for First-Line Palliative Intent Nab-Paclitaxel Plus Gemcitabine Versus FOLFIRINOX. Am. J. Clin. Oncol. 2017, 40, 507–511. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.-L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef]
- Quiñonero, F.; Mesas, C.; Doello, K.; Cabeza, L.; Perazzoli, G.; Jimenez-Luna, C.; Rama, A.R.; Melguizo, C.; Prados, J. The Challenge of Drug Resistance in Pancreatic Ductal Adenocarcinoma: A Current Overview. Cancer Biol. Med. 2019, 16, 688–699. [Google Scholar] [CrossRef]
- Pang, L.; Word, B.; Xu, J.; Wang, H.; Hammons, G.; Huang, S.-M.; Lyn-Cook, B. ATP-Binding Cassette Genes. Genotype and Expression: A Potential. Association with Pancreatic Cancer Development and Chemoresistance? Gastroenterol. Res. Pr. 2014, 2014, 414931. [Google Scholar] [CrossRef]
- Thiebaut, F.; Tsuruo, T.; Hamada, H.; Gottesman, M.M.; Pastan, I.; Willingham, M.C. Cellular Localization of the Multidrug-Resistance Gene Product P-Glycoprotein in Normal Human Tissues. Proc. Natl. Acad. Sci. USA 1987, 84, 7735–7738. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.G.; Monte, M.J.; Macias, R.I.R.; Romero, M.R.; Herraez, E.; Asensio, M.; Ortiz-Rivero, S.; Cives-Losada, C.; Di Giacomo, S.; Gonzalez-Gallego, J.; et al. Expression of Chemoresistance-Associated ABC Proteins in Hepatobiliary, Pancreatic and Gastrointestinal Cancers. Cancers 2022, 14, 3524. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, S.; Briz, O.; Monte, M.J.; Sanchez-Vicente, L.; Abete, L.; Lozano, E.; Mazzanti, G.; Di Sotto, A.; Marin, J.J.G. Chemosensitization of Hepatocellular Carcinoma Cells to Sorafenib by β-Caryophyllene Oxide-Induced Inhibition of ABC Export Pumps. Arch. Toxicol. 2019, 93, 623–634. [Google Scholar] [CrossRef]
- Wolf, S.J.; Bachtiar, M.; Wang, J.; Sim, T.S.; Chong, S.S.; Lee, C.G.L. An Update on ABCB1 Pharmacogenetics: Insights from a 3D Model into the Location and Evolutionary Conservation of Residues Corresponding to SNPs Associated with Drug Pharmacokinetics. Pharmacogenomics J. 2011, 11, 315–325. [Google Scholar] [CrossRef]
- Kasuya, K.; Tsuchida, A.; Nagakawa, Y.; Suzuki, Y.; Suzuki, M.; Aoki, T.; Abe, Y.; Shimazu, M.; Itoi, T.; Sofuni, A. Prediction of a Side Effect and Efficacy of Adjuvant Chemotherapy with Gemcitabine for Post Operative Patient of Pancreatic Cancer by a Genetic Polymorphism Analysis. Hepatogastroenterology 2012, 59, 1609–1613. [Google Scholar] [CrossRef]
- Chen, M.; Xue, X.; Wang, F.; An, Y.; Tang, D.; Xu, Y.; Wang, H.; Yuan, Z.; Gao, W.; Wei, J.; et al. Expression and Promoter Methylation Analysis of ATP-Binding Cassette Genes in Pancreatic Cancer. Oncol. Rep. 2012, 27, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, A.; Onishi, Y.; Hirano, H.; Seigneuret, M.; Obanayama, K.; Kim, G.; Liew, E.L.; Sakaeda, T.; Yoshiura, K.-I.; Niikawa, N.; et al. Quantitative Structure--Activity Relationship Analysis and Molecular Dynamics Simulation to Functionally Validate Nonsynonymous Polymorphisms of Human ABC Transporter ABCB1 (P-Glycoprotein/MDR1). Biochemistry 2007, 46, 7678–7693. [Google Scholar] [CrossRef]
- Kimchi-Sarfaty, C.; Oh, J.M.; Kim, I.-W.; Sauna, Z.E.; Calcagno, A.M.; Ambudkar, S.V.; Gottesman, M.M. A “Silent” Polymorphism in the MDR1 Gene Changes Substrate Specificity. Science 2007, 315, 525–528. [Google Scholar] [CrossRef]
- Wilbur, H.C.; Durham, J.N.; Lim, S.J.; Purtell, K.; Bever, K.M.; Laheru, D.A.; De Jesus-Acosta, A.; Azad, N.S.; Wilt, B.; Diaz, L.A.; et al. Gemcitabine, Docetaxel, Capecitabine, Cisplatin, Irinotecan as First-Line Treatment for Metastatic Pancreatic Cancer. Cancer Res. Commun. 2023, 3, 1672–1677. [Google Scholar] [CrossRef]
- De Troia, B.; Dalu, D.; Filipazzi, V.; Isabella, L.; Tosca, N.; Ferrario, S.; Gambaro, A.R.; Somma, L.; Fasola, C.; Cheli, S.; et al. ABCB1 c.3435C>T Polymorphism Is Associated with Platinum Toxicity: A Preliminary Study. Cancer Chemother. Pharmacol. 2019, 83, 803–808. [Google Scholar] [CrossRef]
- Ieiri, I.; Takane, H.; Otsubo, K. The MDR1 (ABCB1) Gene Polymorphism and Its Clinical Implications. Clin. Pharmacokinet. 2004, 43, 553–576. [Google Scholar] [CrossRef] [PubMed]
- Mugosa, S.; Todorovic, Z.; Cukic, J.; Sahman-Zaimovic, M.; Djordjevic, N. ABCB1 Polymorphism in Clopidogrel-Treated Montenegrin Patients. Open Life Sci. 2021, 16, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Gréen, H.; Falk, I.J.; Lotfi, K.; Paul, E.; Hermansson, M.; Rosenquist, R.; Paul, C.; Nahi, H. Association of ABCB1 Polymorphisms with Survival and in Vitro Cytotoxicty in de Novo Acute Myeloid Leukemia with Normal Karyotype. Pharmacogenomics J. 2012, 12, 111–118. [Google Scholar] [CrossRef]
- Bogacz, A.; Mrozikiewicz, P.M.; Deka-Pawlik, D.; Seremak-Mrozikiewicz, A.; Bartkowiak-Wieczorek, J.; Barlik, M.; Drews, K.; Kowalska, A.; Grześkowiak, E. Frequency of G2677T/A and C3435T Polymorphisms of MDR1 Gene in Preeclamptic Women. Ginekol. Pol. 2013, 84, 781–787. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting Protein-Chemical Interaction Networks with Tissue and Affinity Data. Nucleic Acids Res. 2016, 44, D380-4. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A Pathology Atlas of the Human Cancer Transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef]
- Leschziner, G.D.; Andrew, T.; Pirmohamed, M.; Johnson, M.R. ABCB1 Genotype and PGP Expression, Function and Therapeutic Drug Response: A Critical Review and Recommendations for Future Research. Pharmacogenomics J. 2007, 7, 154–179. [Google Scholar] [CrossRef]
- Hodges, L.M.; Markova, S.M.; Chinn, L.W.; Gow, J.M.; Kroetz, D.L.; Klein, T.E.; Altman, R.B. Very Important Pharmacogene Summary: ABCB1 (MDR1, P-Glycoprotein). Pharmacogenet Genom. 2011, 21, 152–161. [Google Scholar] [CrossRef]
- Wang, D.; Johnson, A.D.; Papp, A.C.; Kroetz, D.L.; Sadée, W. Multidrug Resistance Polypeptide 1 (MDR1, ABCB1) Variant 3435C>T Affects mRNA Stability. Pharmacogenet Genom. 2005, 15, 693–704. [Google Scholar] [CrossRef]
- Razi, B.; Anani Sarab, G.; Omidkhoda, A.; Alizadeh, S. Multidrug Resistance 1 (MDR1/ABCB1) Gene Polymorphism (Rs1045642 C > T) and Susceptibility to Multiple Myeloma: A Systematic Review and Meta-Analysis. Hematology 2018, 23, 456–462. [Google Scholar] [CrossRef]
- Vesel, M.; Rapp, J.; Feller, D.; Kiss, E.; Jaromi, L.; Meggyes, M.; Miskei, G.; Duga, B.; Smuk, G.; Laszlo, T.; et al. ABCB1 and ABCG2 Drug Transporters Are Differentially Expressed in Non-Small Cell Lung Cancers (NSCLC) and Expression Is Modified by Cisplatin Treatment via Altered Wnt Signaling. Respir. Res. 2017, 18, 52. [Google Scholar] [CrossRef] [PubMed]
- Bergman, A.M.; Pinedo, H.M.; Talianidis, I.; Veerman, G.; Loves, W.J.P.; van der Wilt, C.L.; Peters, G.J. Increased Sensitivity to Gemcitabine of P-Glycoprotein and Multidrug Resistance-Associated Protein-Overexpressing Human Cancer Cell Lines. Br. J. Cancer 2003, 88, 1963–1970. [Google Scholar] [CrossRef]
- Soranzo, N.; Cavalleri, G.L.; Weale, M.E.; Wood, N.W.; Depondt, C.; Marguerie, R.; Sisodiya, S.M.; Goldstein, D.B. Identifying Candidate Causal Variants Responsible for Altered Activity of the ABCB1 Multidrug Resistance Gene. Genome Res. 2004, 14, 1333–1344. [Google Scholar] [CrossRef]
- Sito, H.; Tan, S.C. Genetic Polymorphisms as Potential Pharmacogenetic Biomarkers for Platinum-Based Chemotherapy in Non-Small Cell Lung Cancer. Mol. Biol. Rep. 2024, 51, 102. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to Build a Bridge from a Population-Based to a More “Personalized” Approach to Cancer Staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A. The Cellosaurus, a Cell-Line Knowledge Resource. J. Biomol. Tech. 2018, 29, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Okajima, F.; Adachi, M.; Ohwada, S.; Kondo, Y. Growth Dependency of a New Human Pancreatic Cancer Cell Line, YAPC, on Autocrine Interleukin-1alpha Stimulation. Int. J. Cancer 1998, 76, 141–147. [Google Scholar] [CrossRef]
- Deer, E.L.; González-Hernández, J.; Coursen, J.D.; Shea, J.E.; Ngatia, J.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. Phenotype and Genotype of Pancreatic Cancer Cell Lines. Pancreas 2010, 39, 425–435. [Google Scholar] [CrossRef]
- Yunis, A.A.; Arimura, G.K.; Russin, D.J. Human Pancreatic Carcinoma (MIA PaCa-2) in Continuous Culture: Sensitivity to Asparaginase. Int. J. Cancer 1977, 19, 128–135. [Google Scholar] [CrossRef]
- Shichi, Y.; Gomi, F.; Sasaki, N.; Nonaka, K.; Arai, T.; Ishiwata, T. Epithelial and Mesenchymal Features of Pancreatic Ductal Adenocarcinoma Cell Lines in Two- and Three-Dimensional Cultures. J. Pers. Med. 2022, 12, 746. [Google Scholar] [CrossRef]
- Tan, M.H.; Nowak, N.J.; Loor, R.; Ochi, H.; Sandberg, A.A.; Lopez, C.; Pickren, J.W.; Berjian, R.; Douglass, H.O.; Chu, T.M. Characterization of a New Primary Human Pancreatic Tumor Line. Cancer Investig. 1986, 4, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.; Mazzetta, J.; Nelson-Rees, W.; Kaplan, M.; Todaro, G. Establishment of a Continuous Tumor-Cell Line (Panc-1) from a Human Carcinoma of the Exocrine Pancreas. Int. J. Cancer 1975, 15, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Jang, S.D.; Kim, H.; Chung, S.; Park, J.K.; Kuh, H.-J. Phenotypic Heterogeneity and Plasticity of Cancer Cell Migration in a Pancreatic Tumor Three-Dimensional Culture Model. Cancers 2020, 12, 1305. [Google Scholar] [CrossRef]
- De Santis, M.C.; Gozzelino, L.; Margaria, J.P.; Costamagna, A.; Ratto, E.; Gulluni, F.; Di Gregorio, E.; Mina, E.; Lorito, N.; Bacci, M.; et al. Lysosomal Lipid Switch Sensitises to Nutrient Deprivation and mTOR Targeting in Pancreatic Cancer. Gut 2023, 72, 360–371. [Google Scholar] [CrossRef]
- Moro, L.; Simoneschi, D.; Kurz, E.; Arbini, A.A.; Jang, S.; Guaragnella, N.; Giannattasio, S.; Wang, W.; Chen, Y.-A.; Pires, G.; et al. Epigenetic Silencing of the Ubiquitin Ligase Subunit FBXL7 Impairs C-SRC Degradation and Promotes Epithelial-to-Mesenchymal Transition and Metastasis. Nat. Cell Biol. 2020, 22, 1130–1142. [Google Scholar] [CrossRef]
- Guerra, F.; Paiano, A.; Migoni, D.; Girolimetti, G.; Perrone, A.M.; De Iaco, P.; Fanizzi, F.P.; Gasparre, G.; Bucci, C. Modulation of RAB7A Protein Expression Determines Resistance to Cisplatin through Late Endocytic Pathway Impairment and Extracellular Vesicular Secretion. Cancers 2019, 11, 52. [Google Scholar] [CrossRef]
- Romano, R.; Calcagnile, M.; Margiotta, A.; Franci, L.; Chiariello, M.; Alifano, P.; Bucci, C. RAB7A Regulates Vimentin Phosphorylation through AKT and PAK. Cancers 2021, 13, 2220. [Google Scholar] [CrossRef]
- Gagliardi, S.; Mitruccio, M.; Di Corato, R.; Romano, R.; Aloisi, A.; Rinaldi, R.; Alifano, P.; Guerra, F.; Bucci, C. Defects of Mitochondria-Lysosomes Communication Induce Secretion of Mitochondria-Derived Vesicles and Drive Chemoresistance in Ovarian Cancer Cells. Cell Commun. Signal. 2024, 22, 165. [Google Scholar] [CrossRef] [PubMed]
- Vergara, D.; Stanca, E.; Guerra, F.; Priore, P.; Gaballo, A.; Franck, J.; Simeone, P.; Trerotola, M.; De Domenico, S.; Fournier, I.; et al. β-Catenin Knockdown Affects Mitochondrial Biogenesis and Lipid Metabolism in Breast Cancer Cells. Front. Physiol. 2017, 8, 544. [Google Scholar] [CrossRef]



| Cell Lines | Exons | Analyzed Polymorphism | Genotype |
|---|---|---|---|
| H6c7 | Exon 12 | rs1128503 (1236T>C) | Heterozygote T/C |
| Exon 21 | rs2032582 (2677T>G) | Heterozygote T/G | |
| Exon 26 | rs1045642 (3435T>C) | no variation (T/T wt) | |
| BXPC3 | Exon 12 | rs1128503 (1236T>C) | Not determined |
| Exon 21 | rs2032582 (2677T>G) | no variation (T/T wt) | |
| Exon 26 | rs1045642 (3435T>C) | no variation (T/T wt) | |
| YAPC | Exon 12 | rs1128503 (1236T>C) | no variation (T/T wt) |
| Exon 21 | rs2032582 (2677T>G) | Homozygote G/G | |
| Exon 26 | rs1045642 (3435T>C) | Homozygote C/C | |
| PANC-1 | Exon 12 | rs1128503 (1236T>C) | Heterozygote T/C |
| Exon 21 | rs2032582 (2677T>G) | Heterozygote T/G | |
| Exon 26 | rs1045642 (3435T>C) | no variation (T/T wt) | |
| MIA PaCa-2 | Exon 12 | rs1128503 (1236T>C) | no variation (T/T wt) |
| Exon 21 | rs2032582 (2677T>G) | no variation (T/T wt) | |
| Exon 26 | rs1045642 (3435T>C) | no variation (T/T wt) |
| Characteristics | |
|---|---|
| Gender (F/M) | 3/5 |
| Age, mean years (range) | 69.9 (59–85) |
| Size of lesion, median mm (range) | 36.9 (15–59) |
| Tumor stage, n (%) | |
| II | 1 (12.5%) |
| III | 1 (12.5%) |
| IV | 6 (75%) |
| Lesion location, n (%) | |
| Pancreatic head | 6 (75%) |
| Pancreatic body | 1 (12.5%) |
| Pancreatic tail | 1 (12.5%) |
| Distant metastases, n (%) | |
| M0 | 2 (25%) |
| M1 | 6 (75%) |
| Patients | Exon | Analyzed Polymorphism | Genotype |
|---|---|---|---|
| PC1 | Exon 21 Exon 12 | rs2032582 (2677T>G) rs1128503 (1236T>C) | No variation (T/T wt) No variation (T/T wt) |
| PC2 | Exon 21 Exon 12 | rs2032582 (2677T>G) rs1128503 (1236T>C) | Heterozygote T/G Heterozygote T/C |
| PC3 | Exon 21 Exon 12 | rs2032582 (2677T>G) rs1128503 (1236T>C) | Heterozygote T/G Heterozygote T/C |
| PC4 | Exon 21 Exon 12 | rs2032582 (2677T>G) rs1128503 (1236T>C) | Heterozygote T/G Heterozygote T/C |
| PC5 | Exon 21 Exon 12 | rs2032582 (2677T>G) rs1128503 (1236T>C) | Homozygote G/G Homozygote C/C |
| PC6 | Exon 21 Exon 12 | rs2032582 (2677T>G) rs1128503 (1236T>C) | Heterozygote T/G Heterozygote T/C |
| PC7 | Exon 21 Exon 12 | rs2032582 (2677T>G) rs1128503 (1236T>C) | Homozygote G/G Homozygote C/C |
| PC8 | Exon 21 Exon 12 | rs2032582 (2677T>G) rs1128503 (1236T>C) | Heterozygote T/G Heterozygote T/C |
| Cell line | EC50 GEM (nM) [9] | EC50 CDDP (µM) |
|---|---|---|
| BxPC-3 | 168 ± 32.02 | 6 ± 1.5 |
| YAPC | 800 ± 125 | 6 ± 1.51 |
| PANC-1 | 187 ± 24.63 | 12 ± 1.47 |
| MIA PaCa-2 | 18 ± 1.64 | 8 ± 1.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Girolimetti, G.; Balena, B.; Cordella, P.; Verri, T.; Eusebi, L.H.; Bozzetti, M.P.; Bucci, C.; Guerra, F. Characterization of Chemoresistance in Pancreatic Cancer: A Look at MDR-1 Polymorphisms and Expression in Cancer Cells and Patients. Int. J. Mol. Sci. 2024, 25, 8515. https://doi.org/10.3390/ijms25158515
Girolimetti G, Balena B, Cordella P, Verri T, Eusebi LH, Bozzetti MP, Bucci C, Guerra F. Characterization of Chemoresistance in Pancreatic Cancer: A Look at MDR-1 Polymorphisms and Expression in Cancer Cells and Patients. International Journal of Molecular Sciences. 2024; 25(15):8515. https://doi.org/10.3390/ijms25158515
Chicago/Turabian StyleGirolimetti, Giulia, Barbara Balena, Paola Cordella, Tiziano Verri, Leonardo Henry Eusebi, Maria Pia Bozzetti, Cecilia Bucci, and Flora Guerra. 2024. "Characterization of Chemoresistance in Pancreatic Cancer: A Look at MDR-1 Polymorphisms and Expression in Cancer Cells and Patients" International Journal of Molecular Sciences 25, no. 15: 8515. https://doi.org/10.3390/ijms25158515
APA StyleGirolimetti, G., Balena, B., Cordella, P., Verri, T., Eusebi, L. H., Bozzetti, M. P., Bucci, C., & Guerra, F. (2024). Characterization of Chemoresistance in Pancreatic Cancer: A Look at MDR-1 Polymorphisms and Expression in Cancer Cells and Patients. International Journal of Molecular Sciences, 25(15), 8515. https://doi.org/10.3390/ijms25158515







