Proteomic Characterization of Changes in Mouse Brain Cortex Protein Expression at Different Post-Mortem Intervals: A Preliminary Study for Forensic Biomarker Identification
Abstract
1. Introduction
2. Results
2.1. Post-Mortem Mouse Brain Proteome Changes
2.2. Differences in the Mouse Cortex Protein Levels
2.2.1. Top Proteins Differentially Expressed in Mouse Cortex from 0 to 32 Post-Mortem Hours
2.2.2. Top Proteins Differentially Expressed in Mouse Cortex at Early PMI (from 0 to 12 Post-Mortem Hours)
2.2.3. Top Proteins Differentially Expressed in Mouse Cortex at Late PMI (from 24 to 32 Post-Mortem Hours)
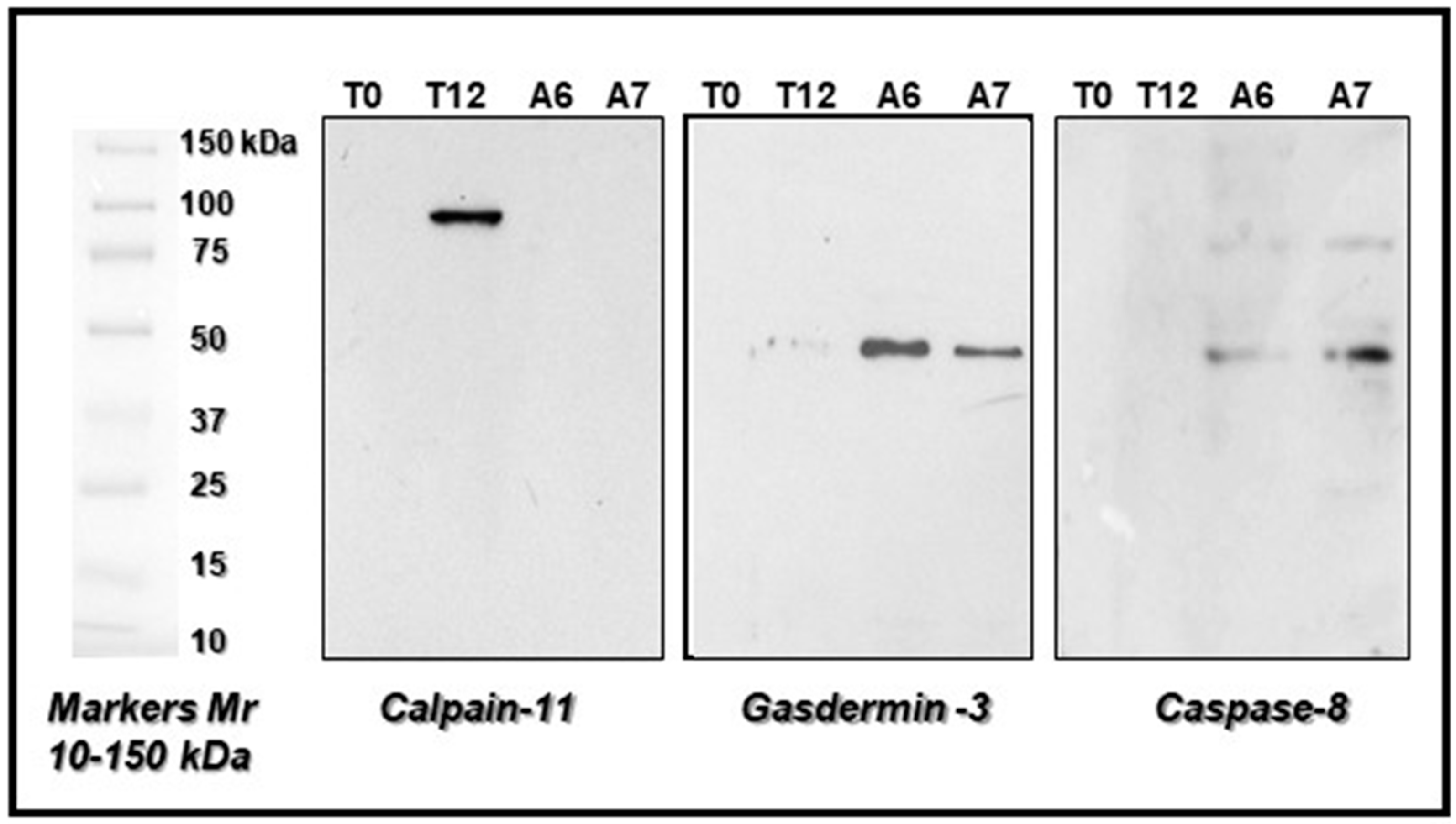
2.3. Protein Sequence Validation by LIFT Technology and Western Blot Analysis of Some Peculiar Proteins Selected among the Others in the e-PMI and l-PMI
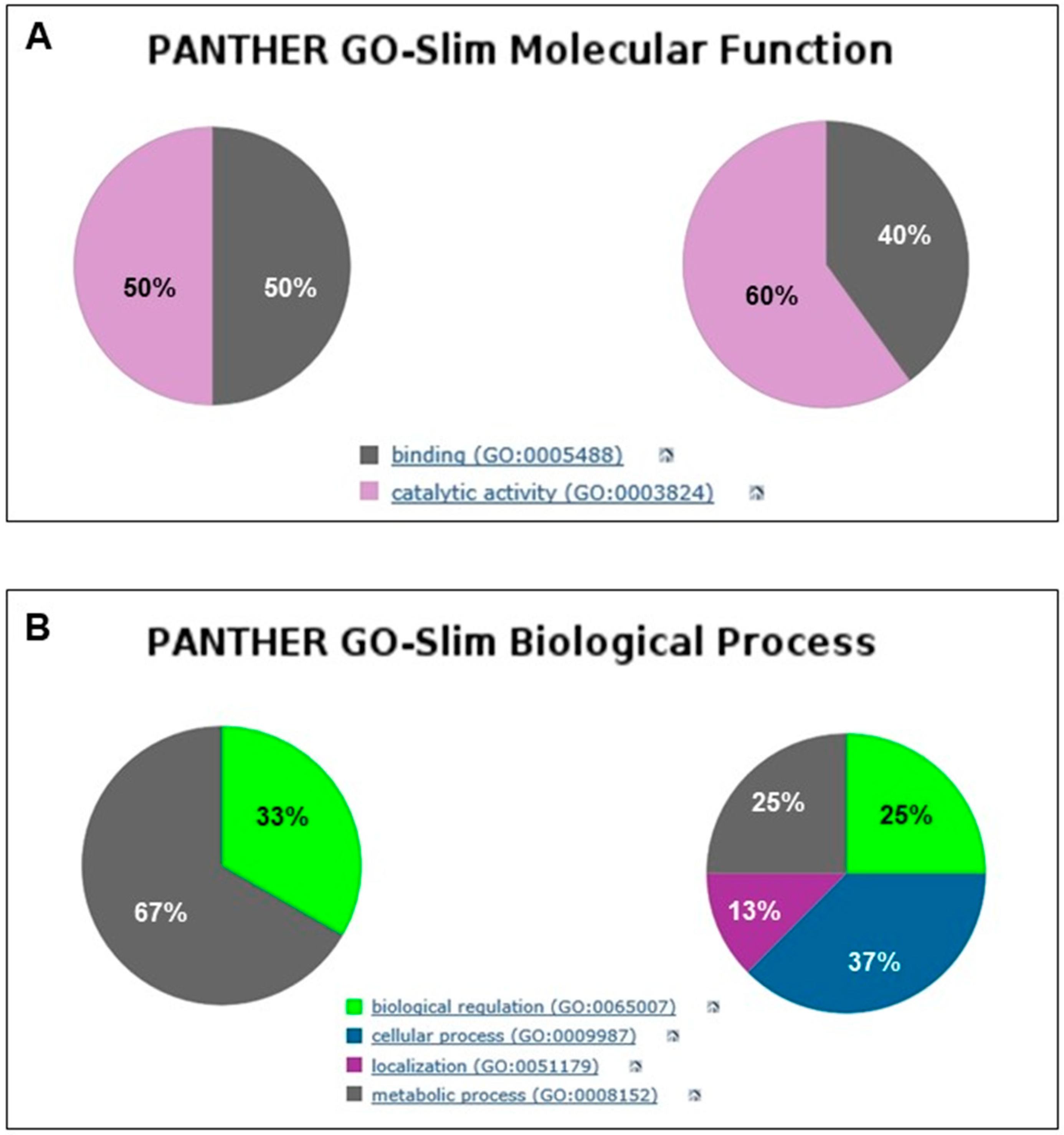
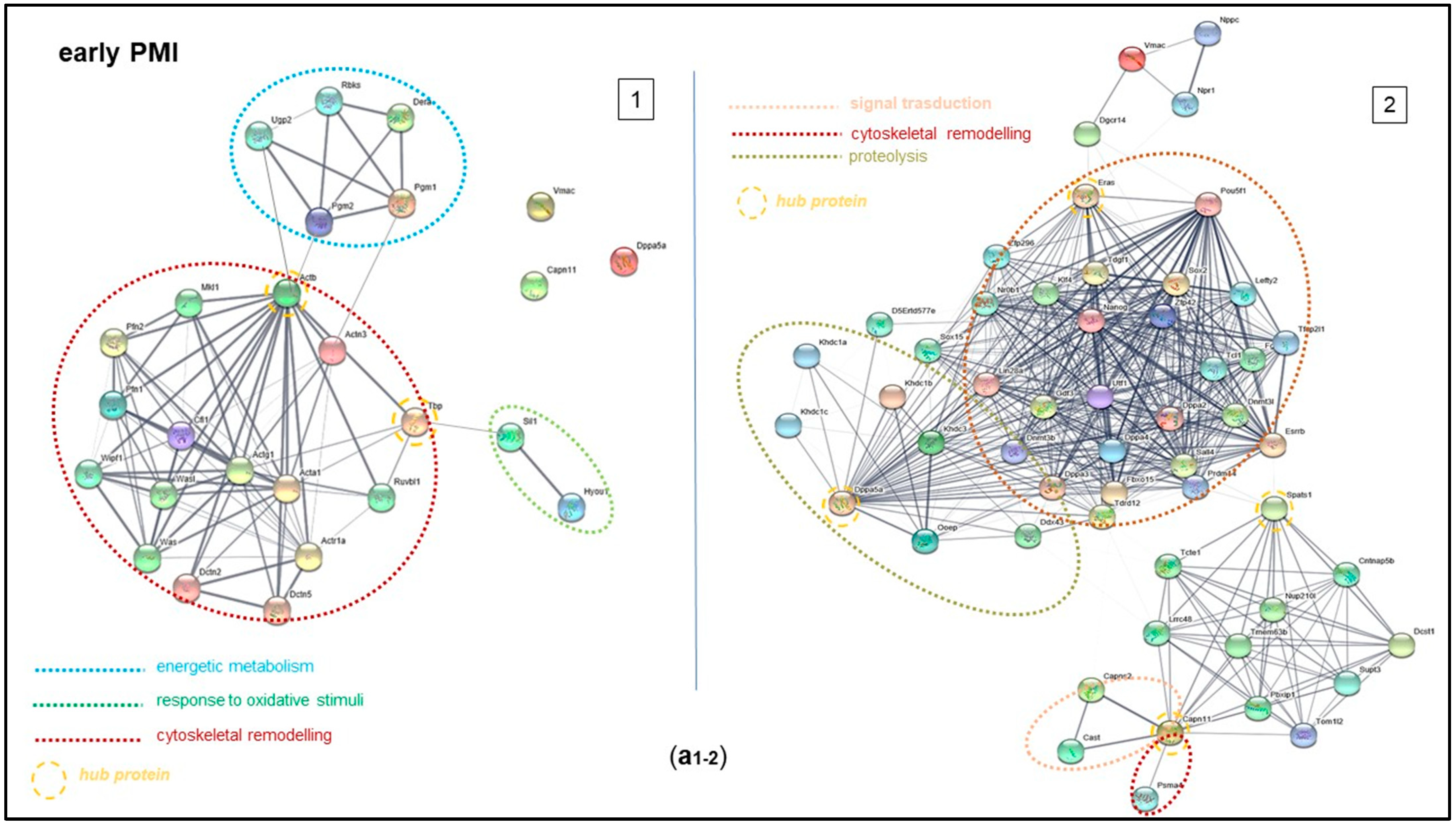
2.4. Bioinformatic Analysis of Mouse Brain Proteins from Early to Late PMI
2.5. Functional Analysis of Proteins Expressed in the Early and Late PMIs
3. Discussion
4. Materials and Methods
4.1. Animals and Tissue Samples
4.2. Bidimensional Electrophoretic (2DE) Analysis
4.3. Protein Digestion and MALDI TOF-TOF MS Analysis
4.4. Protein Validation by LIFT Technology and Western Blot Analysis
4.5. Bioinformatic Analysis
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Henßge, C.; Madea, B. Estimation of the time since death in the early post-mortem period. Forensic Sci. Int. 2004, 144, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Díaz Martín, R.D.; Camacho-Martínez, Z.; Ambrosio Hernández, J.R.; Valencia-Caballero, L. Proteomics as a new tool in forensic sciences. Span. J. Leg. Med. 2019, 45, 114–122. [Google Scholar] [CrossRef]
- Jia, X.; Hollung, K.; Therkildsen, M.; Hildrum, K.I.; Bendixen, E. Proteome analysis of early post-mortem changes in two bovine muscle types: M. longissimus dorsi and M. semitendinosis. Proteomics 2006, 6, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Lametsch, R.; Karlsson, A.; Rosenvold, K.; Andersen, H.J.; Roepstorff, P.; Bendixen, E. Postmortem proteome changes of porcine muscle related to tenderness. J. Agric. Food Chem. 2003, 51, 6992–6997. [Google Scholar] [CrossRef] [PubMed]
- Kjaersgård, I.V.H.; Jessen, F. Proteome analysis elucidating post-mortem changes in cod (Gadus morhua) muscle proteins. J. Agric. Food Chem. 2003, 51, 3985–3991. [Google Scholar] [CrossRef] [PubMed]
- Di Luca, A.; Hamill, R.M.; Mullen, A.M.; Slavov, N.; Elia, G. Comparative Proteomic Profiling of Divergent Phenotypes for Water Holding Capacity across the Post Mortem Ageing Period in Porcine Muscle Exudate. PLoS ONE 2016, 11, e0150605. [Google Scholar] [CrossRef] [PubMed]
- Gos, T.; Raszeja, S. Postmortem activity of lactate and malate dehydrogenase in human liver in relation to time after death. Int. J. Leg. Med. 1993, 106, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Kawahara, K.-I.; Biswas, K.K.; Ito, T.; Tancharoen, S.; Shiomi, N.; Koda, Y.; Matsuda, F.; Morimoto, Y.; Oyama, Y.; et al. HMGB1: A new marker for estimation of the postmortem interval. Exp. Ther. Med. 2010, 1, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kassam, N.; Gauthier, M.L.; O’Day, D.H. Post-mortem changes in calmodulin binding proteins in muscle and lung. Forensic Sci. Int. 2003, 131, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Poloz, Y.O.; O’Day, D.H. Determining time of death: Temperature-dependent postmortem changes in calcineurin A, MARCKS, CaMKII, and protein phosphatase 2A in mouse. Int. J. Leg. Med. 2009, 123, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Tavichakorntrakool, R.; Prasongwattana, V.; Sriboonlue, P.; Puapairoj, A.; Pongskul, J.; Khuntikeo, N.; Hanpanich, W.; Yenchitsomanus, P.-T.; Wongkham, C.; Thongboonkerd, V. Serial analyses of postmortem changes in human skeletal muscle: A case study of alterations in proteome profile, histology, electrolyte contents, water composition, and enzyme activity. Proteom. Clin. Appl. 2008, 2, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Kandigian, S.E.; Ethier, E.C.; Kitchen, R.R.; Lam, T.T.; Arnold, S.E.; Carlyle, B.C. Proteomic characterization of post-mortem human brain tissue following ultracentrifugation-based subcellular fractionation. Brain Commun. 2022, 4, fcac103. [Google Scholar] [CrossRef] [PubMed]
- Pla, A.; Lemus, L.; Valenzuela, A.; Villanueva, E. Postmortem activity of the key enzymes of glycolysis. In rat brain regions in relation to time after death. Z. Rechtsmed. 1986, 97, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Fountoulakis, M.; Hardmeier, R.; Höger, H.; Lubec, G. Postmortem changes in the level of brain proteins. Exp. Neurol. 2001, 167, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Lescuyer, P.; Allard, L.; Zimmermann-Ivol, C.G.; Burgess, J.A.; Hughes-Frutiger, S.; Burkhard, P.R.; Sanchez, J.-C.; Hochstrasser, D.F. Identification of post-mortem cerebrospinal fluid proteins as potential biomarkers of ischemia and neurodegeneration. Proteomics 2004, 4, 2234–2341. [Google Scholar] [CrossRef] [PubMed]
- Finehout, E.J.; Franck, Z.; Relkin, N.; Lee, K.H. Proteomic analysis of cerebrospinal fluid changes related to postmortem interval. Clin. Chem. 2006, 52, 1906–1913. [Google Scholar] [CrossRef] [PubMed]
- Sköld, K.; Svensson, M.; Norrman, N.; Sjögren, B.; Svenningsson, P.; Andrén, P.E. The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: Stathmin 2-20 and peptides as sample quality indicators. Proteomics 2007, 7, 4445–4456. [Google Scholar] [CrossRef] [PubMed]
- Hunsucker, S.W.; Solomon, B.; Gawryluk, J.; Geiger, J.D.; Vacano, G.N.; Duncan, M.W.; Patterson, D. Assessment of post-mortem-induced changes to the mouse brain proteome. J. Neurochem. 2008, 105, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Franzén, B.; Yang, Y.; Sunnemark, D.; Wickman, M.; Ottervald, J.; Oppermann, M.; Sandberg, K. Dihydropyrimidinase related protein-2 as a biomarker for temperature and time dependent post mortem changes in the mouse brain proteome. Proteomics 2003, 3, 1920–1929. [Google Scholar] [CrossRef] [PubMed]
- Samodelov, S.L.; Kullak-Ublick, G.A.; Gai, Z.; Visentin, M. Organic Cation Transporters in Human Physiology; Pharmacology, and Toxicology. Int. J. Mol. Sci. 2020, 21, 7890. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, O.; Koizumi, H.; Aoyagi, T. A novel synthetic vitamin-A-like compound (a polyprenoic acid derivative: E-5166) inhibits UVB-stimulated epidermal ornithine decarboxylase activity. Arch. Dermatol. Res. 1986, 278, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Garzón, J.; Rodríguez-Díaz, M.; López-Fando, A.; García-España, A.; Sánchez-Blázquez, P. Glycosylated phosducin-like protein long regulates opioid receptor function in mouse brain. Neuropharmacology 2002, 42, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Ikedife, J.; He, J.; Wei, Y. PEA-15 engages in allosteric interactions using a common scaffold in a phosphorylation-dependent manner. Sci. Rep. 2022, 12, 116. [Google Scholar] [CrossRef] [PubMed]
- Thomason, L.A.M.; Smithson, L.J.; Hazrati, L.-N.; McLaurin, J.; Kawaja, M.D. Reactive astrocytes associated with plaques in TgCRND8 mouse brain and in human Alzheimer brain express phosphoprotein enriched in astrocytes (PEA-15). FEBS Lett. 2013, 587, 2448–2454. [Google Scholar] [CrossRef] [PubMed]
- Kovács, Z.; Kékesi, K.A.; Bobest, M.; Török, T.; Szilágyi, N.; Szikra, T.; Szepesi, Z.; Nyilas, R.; Dobolyi, A.; Palkovits, M.; et al. Post mortem degradation of nucleosides in the brain: Comparison of human and rat brains for estimation of in vivo concentration of nucleosides. J. Neurosci. Methods 2005, 148, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, J.; Li, L.; Sun, L.; Yi, X.; Han, X.; Si, W.; Yan, R.; Chen, Z.; Xie, G.; et al. PAAT; a novel ATPase and trans-regulator of mitochondrial ABC transporters is critically involved in the maintenance of mitochondrial homeostasis. FASEB J. 2014, 28, 4821–4834. [Google Scholar] [CrossRef] [PubMed]
- Broek, J.A.; Guest, P.C.; Rahmoune, H.; Bahn, S. Proteomic analysis of post mortem brain tissue from autism patients: Evidence for opposite changes in prefrontal cortex and cerebellum in synaptic connectivity-related proteins. Mol. Autism 2014, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhu, J.; Mi, K.; Shen, Y.; Zhang, Z. Functional Role of SIL1 in Neurodevelopment and Learning. Neural Plast. 2019, 2019, 9653024. [Google Scholar] [CrossRef]
- Bröker, L.E.; Kruyt, F.A.E.; Giaccone, G. Cell death independent of caspases: A review. Clin. Cancer Res. 2005, 11, 3155–3162. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Fu, S.; Zhao, M.; Lu, L.; Lu, Q. Dysregulation of Cell Death and Its Epigenetic Mechanisms in Systemic Lupus Erythematosus. Molecules 2016, 22, 30. [Google Scholar] [CrossRef] [PubMed]
- Pfister, J.A.; D’Mello, S.R. Insights into the regulation of neuronal viability by nucleophosmin/B23. Exp. Biol. Med. 2015, 240, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, W.; Shao, F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem. Sci. 2017, 42, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, Y.; Kanneganti, T.D. Innate Immune Cell Death in Neuroinflammation and Alzheimer’s Disease. Cells 2022, 11, 1885. [Google Scholar] [CrossRef] [PubMed]
- Lian, T.; Wang, L.; Liu, Y. A New Insight into the Role of Calpains in Post-mortem Meat Tenderization in Domestic Animals: A review. Asian-Australas. J. Anim. Sci. 2013, 26, 443–454. [Google Scholar] [CrossRef]
- Beyfuss, K.; Hood, D.A. A systematic review of p53 regulation of oxidative stress in skeletal muscle. Redox Rep. 2018, 23, 100–117. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-M.; Zissler, A.; Kim, E.; Ehrenfellner, B.; Cho, E.; Lee, S.-I.; Steinbacher, P.; Yun, K.N.; Shin, J.H.; Kim, J.Y.; et al. Postmortem proteomics to discover biomarkers for forensic PMI estimation. Int. J. Leg. Med. 2019, 133, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Procopio, N.; Williams, A.; Chamberlain, A.T.; Buckley, M. Forensic proteomics for the evaluation of the post-mortem decay in bones. J. Proteom. 2018, 177, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Nagy, C.; Maheu, M.; Lopez, J.P.; Vaillancourt, K.; Cruceanu, C.; Gross, J.A.; Arnovitz, M.; Mechawar, N.; Turecki, G. Effects of postmortem interval on biomolecule integrity in the brain. J. Neuropathol. Exp. Neurol. 2015, 74, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Wuenscher, W.; Moebius, G. On brain changes in late death after strangulation. Dtsch. Z. Gesamte Gerichtl. Med. 1960, 50, 235–243. Available online: http://www.ncbi.nlm.nih.gov/pubmed/13786872 (accessed on 22 June 2024). [PubMed]
- Joyce, D. Changes in the 5-hydroxytryptamine content of rat; rabbit and human brain after death. Br. J. Pharmacol. Chemother. 1962, 18, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Santpere, G.; Arzberger, T.; Bell, J.; Blanco, R.; Boluda, S.; Budka, H.; Carmona, M.; Giaccone, G.; Krebs, B.; et al. Brain protein preservation largely depends on the postmortem storage temperature: Implications for study of proteins in human neurologic diseases and management of brain banks: A BrainNet Europe Study. J. Neuropathol. Exp. Neurol. 2007, 66, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Di Luca, A.; Elia, G.; Mullen, A.M.; Hamill, R.M. Monitoring post mortem changes in porcine muscle through 2-D DIGE proteome analysis of Longissimus muscle exudate. Proteome Sci. 2013, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, T.; Katsurano, M.; Ibaragi, S.; Shima, K.; Sasaki, A.; Hu, G.F. Ornithine decarboxylase antizyme upregulates DNA-dependent protein kinase and enhances the nonhomologous end-joining repair of DNA double-strand breaks in human oral cancer cells. Biochemistry 2007, 46, 8920–8932. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.; Kwon, E.; Paez, P.; Yan, W.; Czerwieniec, G.; Loo, J.A.; Sofroniew, M.V.; Wanner, I.B. Traumatically injured astrocytes release a proteomic signature modulated by STAT3-dependent cell survival. Glia 2016, 64, 668–694. [Google Scholar] [CrossRef] [PubMed]
- Sanoudou, D.; Kang, P.B.; Haslett, J.N.; Han, M.; Kunkel, L.M.; Beggs, A.H. Transcriptional Profile of Postmortem Skeletal Muscle. Physiol. Genom. 2004, 16, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Bonicelli, A.; Mickleburgh, H.L.; Chighine, A.; Locci, E.; Wescott, D.J.; Procopio, N. The ‘ForensOMICS’ approach for postmortem interval estimation from human bone by integrating metabolomics, lipidomics, and proteomics. Elife 2022, 11, e83658. [Google Scholar] [CrossRef] [PubMed]
- Marrone, A.; La Russa, D.; Barberio, L.; Murfuni, M.S.; Gaspari, M.; Pellegrino, D. Forensic Proteomics for the Discovery of New post mortem Interval Biomarkers: A Preliminary Study. Int. J. Mol. Sci. 2023, 24, 14627. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Schnellmann, R.G. Calpains, mitochondria, and apoptosis. Cardiovasc. Res. 2012, 96, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Sanvicens, N.; Gómez-Vicente, V.; Masip, I.; Messeguer, A.; Cotter, T.G. Oxidative Stress-induced Apoptosis in Retinal Photoreceptor Cells Is Mediated by Calpains and Caspases and Blocked by the Oxygen Radical Scavenger CR-6. J. Biol. Chem. 2004, 279, 39268–39278. [Google Scholar] [CrossRef] [PubMed]
- Broz, P.; Pelegrín, P.; Shao, F. The gasdermins; a protein family executing cell death and inflammation. Nat. Rev. Immunol. 2020, 20, 143–157. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Sobrido-Cameán, D.; Barreiro-Iglesias, A. Role of Caspase-8 and Fas in Cell Death After Spinal Cord Injury. Front. Mol. Neurosci. 2018, 11, 101. [Google Scholar] [CrossRef] [PubMed]
- Kemp, C.M.; Bardsley, R.G.; Parr, T. Changes in caspase activity during the postmortem conditioning period and its relationship to shear force in porcine longissimus muscle. J. Anim. Sci. 2006, 84, 2841–2846. [Google Scholar] [CrossRef] [PubMed]
- Angelucci, S.; Marchisio, M.; Di Giuseppe, F.; Pierdomenico, L.; Sulpizio, M.; Eleuterio, E.; Lanuti, P.; Sabatino, G.; Miscia, S.; Di Ilio, C. Proteome analysis of human Wharton’s jelly cells during in vitro expansion. Proteome Sci. 2010, 8, 18. [Google Scholar] [CrossRef]
- Gharahdaghi, F.; Weinberg, C.R.; Meagher, D.A.; Imai, B.S.; Mische, S.M. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis 1999, 20, 601–605. [Google Scholar] [CrossRef]
- Di Giuseppe, F.; Carluccio, M.; Zuccarini, M.; Giuliani, P.; Ricci-Vitiani, L.; Pallini, R.; De Sanctis, P.; Di Pietro, R.; Ciccarelli, R.; Angelucci, S. Proteomic Characterization of Two Extracellular Vesicle Subtypes Isolated from Human Glioblastoma Stem Cell Secretome by Sequential Centrifugal Ultrafiltration. Biomedicines 2021, 9, 146. [Google Scholar] [CrossRef] [PubMed]

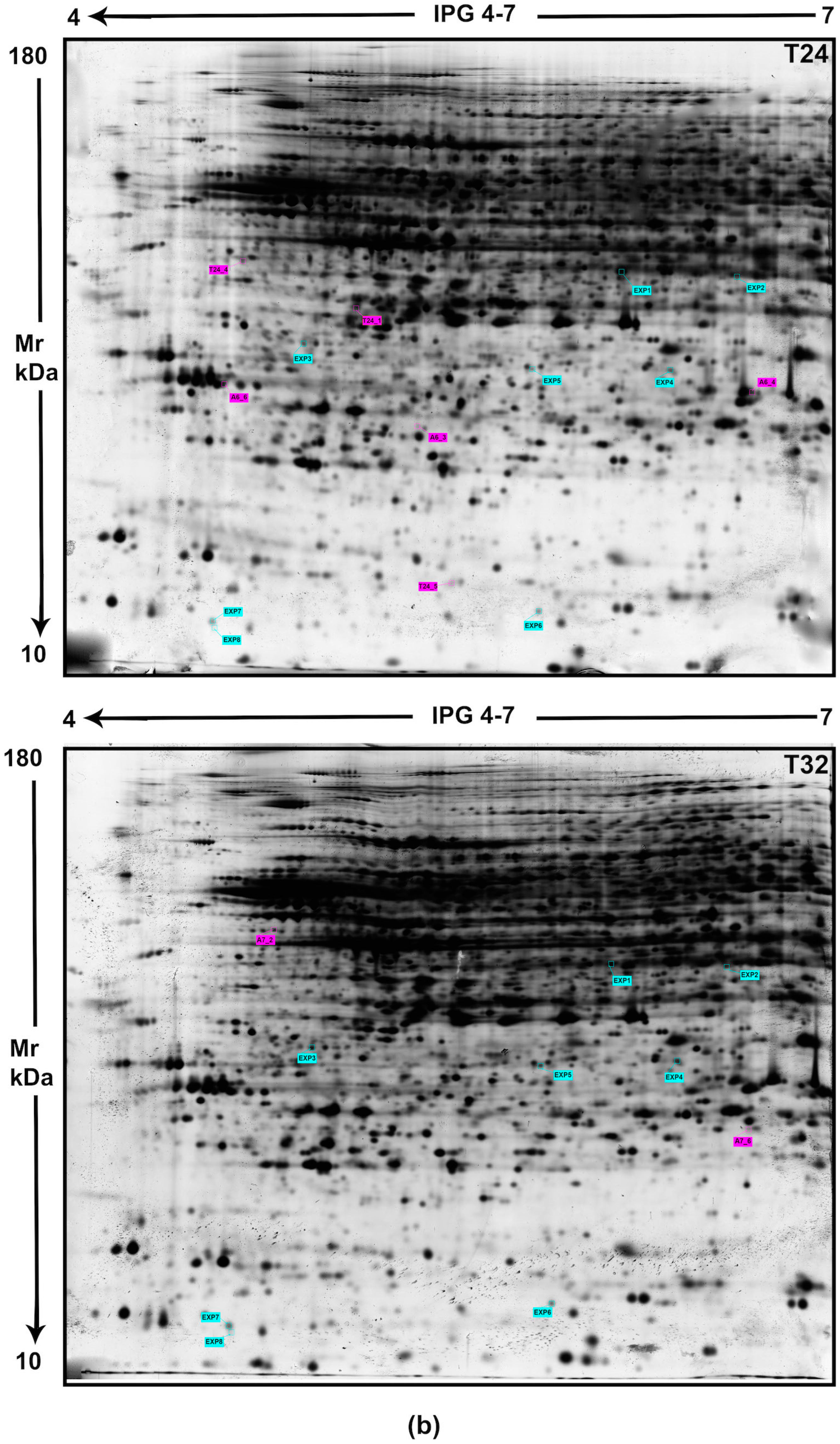



| (a) Top proteins downregulated in mouse cortex from 0 to 32 h | |||||||||
| SPOT ID | Abbreviated Name_Gene Name | AC a Swiss | Protein Description | Score b | Peptide Matched | SC c % | Theoretical (pI/Mr) | p-Value | Variation |
| EXP1 | PRS4_Psmc1 | P62192 | 26S proteasome regulatory subunit 4 | 41 | 6/38 | 49 | 5.87/59184 | 0.0076 | DOWN |
| EXP2 | CTL3_Slc44a3 | Q921V7 | Choline transporter-like protein 3 | 44 | 5/18 | 57 | 8.04/73414 | 0.0112 | DOWN |
| EXP4 | PRS10_Psmc6 | P62334 | 26S proteasome regulatory subunit 10B | 67 | 21/33 | 78 | 7.09/44172 | 0.0008 | DOWN |
| EXP5 | AZI2_Azi2 | Q9QYP6 | 5-azacytidine-induced protein 2 | 44 | 11/59 | 65 | 6.23/46062 | 0.0113 | DOWN |
| EXP6 | PDCL2_Pdcl2 | Q78Y63 | Phosducin-like protein 2 | 51 | 6/21 | 39 | 5.00/22134 | 0.0021 | DOWN |
| EXP7 | PEA15_Pea15 | Q62048 | Astrocytic phosphoprotein PEA-15 | 60 | 5/56 | 46 | 4.94/15054 | 0.0300 | DOWN |
| EXP8 | GABPB-1_Gabpb1 | Q00420 | GA-binding protein subunit beta-1 | 64 | 6/25 | 39 | 4.76/41331 | 0.0011 | DOWN |
| (b) Top proteins induced in mouse cortex at early PMI (from 0 to 24 h) | |||||||||
| SPOT ID | Abbreviated Name_Gene Name | AC a Swiss | Protein Description | Score b | Peptide Matched | SC c % | Theoretical (pI/Mr) | p-Value | Variation |
| ER1 | PGM2_Pgm2 | Q7TSV4 | Phosphoglucomutase-2 | 57 | 19/46 | 36 | 5.78/69274 | 0.0018 | UP |
| ER2 | PAAT_Paat | Q9D2Q3 | ATPase PAAT Protein associated with ABC transporters | 60 | 22/95 | 55 | 6.03/48743 | 0.0400 | UP |
| T12_1 | VMAC_Vmac | Q8BP01 | Vimentin-type intermediate filament-associated coiled-coil protein | 30 | 5/23 | 30 | 5.21/9063 | 0.0021 | UP |
| T12_2 | DPA5A_Dppa5a | Q9CQS7 | Developmental pluripotency-associated protein 5A | 30 | 8/85 | 46 | 6.15/13858 | 0.0052 | UP |
| T12_7 | SIL1_Sil1 | Q9EPK6 | Nucleotide exchange factor SIL1 precursor | 57 | 15/58 | 47 | 5.20/52429 | 0.0030 | UP |
| T12_8 | CAN11_Capn11 | Q6J756 | Calpain-11 | 50 | 11/24 | 52 | 6.03/82969 | 0.0213 | UP |
| T12_9 | ACTB_Actb | P60710 | Actin, cytoplasmic 1 | 51 | 20/99 | 48 | 5.29/42052 | 0.0051 | UP |
| (c) Top proteins induced in mouse cortex at late PMI (from 24 to 32 h) | |||||||||
| SPOT ID | Abbreviated Name_Gene Name | AC a Swiss | Protein Description | Score b | Peptide Matched | SC c % | Theoretical (pI/Mr) | p-Value | Variation |
| A6_3 | NDUFAF7_Ndufaf7 | Q9CWG8 | Protein arginine methyltransferase, mitochondrial precursor | 70 | 21/53 | 48 | 6.47/48384 | 0.0090 | UP |
| A6_4 | NPM_Npm1 | Q61937 | Nucleophosmin | 45 | 8/25 | 53 | 4.62/32560 | 0.0118 | UP |
| A6_6 | GSDA3_Gsdma3 | Q5Y4Y6 | Gasdermin-A3 | 62 | 12/46 | 59 | 5.53/51987 | 0.0015 | UP |
| A7_2 | CASP8_Casp8 | O89110 | Caspase-8 | 52 | 28/122 | 51 | 5.12/56291 | 0.0077 | UP |
| A7_6 | HS3S6_Hs3st6 | Q5GFD5 | Heparan sulfate glucosamine 3-O-sulfotransferase 6 | 58 | 8/28 | 42 | 10.55/37415 | 0.0104 | UP |
| T24_1 | CASP7_Casp7 | P97864 | Caspase-7 | 35 | 15/106 | 55 | 5.93/34666 | 0.0023 | UP |
| T24_4 | GSDA3_Gsdma3 | Q5Y4Y6 | Gasdermin-A3 | 59 | 10/57 | 44 | 5.53/51987 | 0.0103 | UP |
| T24_5 | TCAL8_Tceal8 | Q9CZY2 | Transcription elongation factor A protein-like 8 | 36 | 6/62 | 50 | 5.41/13578 | 0.0270 | UP |
| Label | Abbr. Name | Mw/pI Theor. | PMF Score a | Peptide Matched/ Peptide Searched | SC b % | Lift (MS2) Ion Parent Masses (m/z) | Score c Tof-Tof | Peptide Sequence |
|---|---|---|---|---|---|---|---|---|
| T12_8 | CAN11 | 6.03/82.96 | 50 | 11/24 | 52 | 1740.845 1453.768 1139.587 | 150 | YRDHGFSEIFINSR DADFLLRVFTEK TKGFSLEVCR |
| A6_6 | GASDA3 | 5.53/51.98 | 62 | 12/46 | 59 | 2122.195 2455.380 1390.787 | 230 | HNLCALYAGLSLLHLLSRK ILPVQLKLVESTLEQNFLQD AVTIPKGCVLAYR |
| A7_6 | CASP8 | 5.12/5629 | 52 | 28/122 | 51 | 2019.022 1101.601 909.427 | 187 | NKPRGYCLIINNHDFSK AREDITQLR SESRTSDK |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonelli, M.; Di Giuseppe, F.; Tupone, N.; Di Virgilio, V.; Catena, A.M.; Locatelli, M.; Ascani, G.; Giammaria, G.; Ciccarelli, R.; D’Ovidio, C.; et al. Proteomic Characterization of Changes in Mouse Brain Cortex Protein Expression at Different Post-Mortem Intervals: A Preliminary Study for Forensic Biomarker Identification. Int. J. Mol. Sci. 2024, 25, 8736. https://doi.org/10.3390/ijms25168736
Bonelli M, Di Giuseppe F, Tupone N, Di Virgilio V, Catena AM, Locatelli M, Ascani G, Giammaria G, Ciccarelli R, D’Ovidio C, et al. Proteomic Characterization of Changes in Mouse Brain Cortex Protein Expression at Different Post-Mortem Intervals: A Preliminary Study for Forensic Biomarker Identification. International Journal of Molecular Sciences. 2024; 25(16):8736. https://doi.org/10.3390/ijms25168736
Chicago/Turabian StyleBonelli, Martina, Fabrizio Di Giuseppe, Nicola Tupone, Vimal Di Virgilio, Antonio Maria Catena, Marcello Locatelli, Giuliano Ascani, Gianluigi Giammaria, Renata Ciccarelli, Cristian D’Ovidio, and et al. 2024. "Proteomic Characterization of Changes in Mouse Brain Cortex Protein Expression at Different Post-Mortem Intervals: A Preliminary Study for Forensic Biomarker Identification" International Journal of Molecular Sciences 25, no. 16: 8736. https://doi.org/10.3390/ijms25168736
APA StyleBonelli, M., Di Giuseppe, F., Tupone, N., Di Virgilio, V., Catena, A. M., Locatelli, M., Ascani, G., Giammaria, G., Ciccarelli, R., D’Ovidio, C., & Angelucci, S. (2024). Proteomic Characterization of Changes in Mouse Brain Cortex Protein Expression at Different Post-Mortem Intervals: A Preliminary Study for Forensic Biomarker Identification. International Journal of Molecular Sciences, 25(16), 8736. https://doi.org/10.3390/ijms25168736










