High Resolution HLA-A, HLA-B, and HLA-C Allele Frequencies in Romanian Hematopoietic Stem Cell Donors
Abstract
1. Introduction
2. Results
2.1. Allele Frequencies
2.2. HLA-A Alleles
2.3. HLA-B Alleles
2.4. HLA-C Alleles
3. Discussion
- -
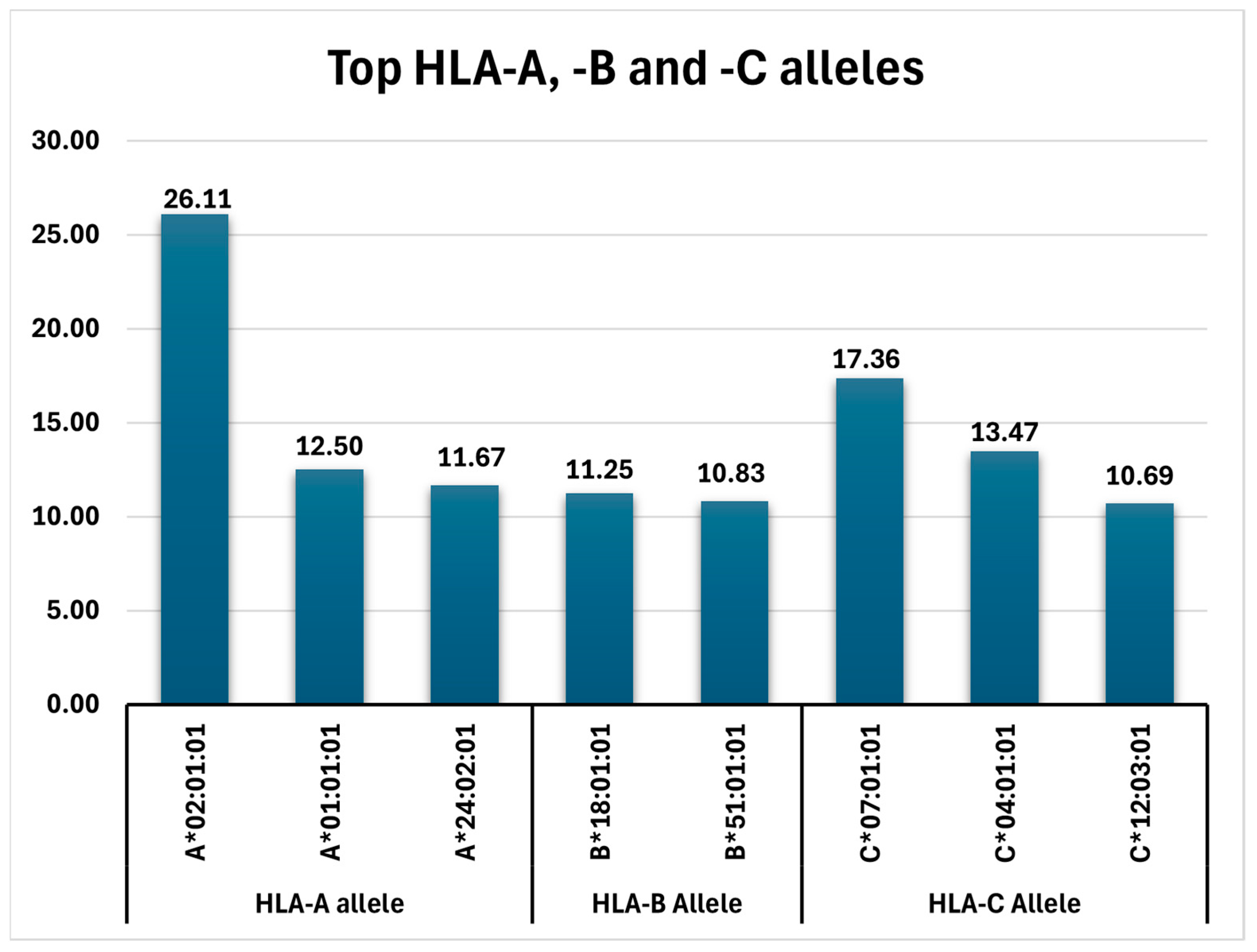
- HLA-A: A*02:01:01 (26.11%), A*01:01:01 (12.5%), A*24:02:01 (11.7%), A*03:01:01 (9.72%), A*11:01:01, and A*32:01:01 (each with 6.25%);
- -
- HLA-B: B*18:01:01 (11.25%), B*51:01:01 (10.83%), and B*08:01:01 (7.78%);
- -
- HLA-C: C*07:01:01 (17.36%), C*04:01:01 (13.47%), and C*12:03:01 (10.69%).
3.1. HLA-A Alleles
3.2. HLA-B Alleles
3.3. HLA-C Alleles
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Constantinescu, I. Imunologia Transplantului (Transplantation Immunology); Editura Universitara UMF “Carol Davila”: Bucharest, Romania, 2009; pp. 4–31. [Google Scholar]
- Mihaescu, G.; Chifiriuc, C. Cap. 4. Moleculele Complexului Major de Histocompatibilitate (CMH) (Molecules of the Major Hystocompatibility Complex, MHC) si Cap. 5. Raspunsul Imun (Immune Response). In Imunologie şi Imunopatologie (Immunology and Immunopathology), 1st ed.; Editura Medicala: Bucharest, Romania, 2015; pp. 261–305. [Google Scholar]
- Olinescu, A. Determinismul genetic al functiilor imune (Genetic determinism of immune functions). In Imunologie (Immunology); Editura Didactică şi Pedagogică: Bucharest, Romania, 1995; pp. 270–304. [Google Scholar]
- Delves, P.J.; Martin, S.J.; Burton, D.R.; Roitt, I.M. Part. 1. Fundamentals of Immunology. In Roitt’s Essential Immunology, 13th ed.; Delves, P.J., Martin, Eds.; John Wiley and Sons, Ltd.: Oxford, UK, 2017; pp. 1–291. [Google Scholar]
- Rich, R.R.; Fleisher, T.A.; Shearer, W.T.; Schroeder, H.W., Jr.; Frew, A.J.; Weyand, C.M. Part One: Principles of immune response. In Clinical Immunology. Principles and Practice, 4th ed.; Rich, R.R., Ed.; Elsevier Saunders: Exeter, UK, 2013; pp. 3–183. [Google Scholar]
- Bravo-Egana, V.; Sanders, H.; Chitnis, N. New challenges, new opportunities: Next generation sequencing and its place in the advancement of HLA typing. Hum. Immunol. 2021, 82, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Ursu, R.I.; Cucu, N.; Ursu, G.F.; Crăciunescu, I.; Severin, E.; Puiu, M.; Alexandrescu, L. Frequency study of the FTO and ADRB3 genotypes in a Romanian cohort of obese children. Rom. Biotechnol. Lett. 2016, 21, 11610–11620. [Google Scholar]
- Ursu, R.; Iordache, P.; Radoi, V.E.; Ursu, G.F.; Cucu, N.; Craciunescu, I.; Bohiltea, R.; Calota, V.; Voinoiu, A.; Staicu, C.; et al. Novel loci associated with hypertension in a Romanian cohort of elder male individuals. Eur. Heart J. 2017, 38 (Suppl. S1), 559. [Google Scholar] [CrossRef]
- Krebs, J.E.; Goldstein, E.S.; Kilpatrick, S.T. Somatic DNA Recombination and Hypermutation in the Immune System. In Lewin’s Genes XII; Casali, P., Ed.; Jones & Bartlett Learning, LLC: Burlington, MA, USA, 2018; pp. 1505–1671. [Google Scholar]
- Shiina, T.; Inoko, H.; Kulski, J.K. An update of the HLA genomic region, locus information and disease association. Tissue Antigens 2004, 64, 631–649. [Google Scholar] [CrossRef] [PubMed]
- Rodey, G.E. Part One. Chapter 2. Structure and Function of the HLA Complex. In HLA Beyond Tears: Introduction to Human Histocompatibility, 2nd ed.; Ed DeNovo: Hesperus, CO, USA, 2000; pp. 13–43. [Google Scholar]
- UniProt. Available online: https://www.uniprot.org (accessed on 7 January 2024).
- Constantinescu, I.; Boscaiu, V.; Cianga, P.; Dinu, A.A.; Gai, E.; Melinte, M.; Moise, A. The frequency of HLA alleles in the Romanian Population. Immunogenetics 2016, 68, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Vică, M.L.; Matei, H.V.; Bondor, C.I.; Nicula, G.Z.; Siserman, C.V.; Loga, L.; Dican, L. HLA Polymorphisms and Haplotype Diversity in Transylvania, Romania. J. Immunol. Res. 2019, 30, 1342762. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Allele Frequency Net Database. Available online: http://www.allelefrequencies.net/default.asp (accessed on 9 January 2024).
- DiBrino, M.; Tsuchida, T.; Turner, R.V.; Parker, K.C.; Coligan, J.E.; Biddison, W.E. HLA-A1 and HLA-A3 T cell epitopes derived from influenza virus proteins predicted from peptide binding motifs. J. Immunol. 1993, 151, 5930–5935. [Google Scholar] [CrossRef] [PubMed]
- Traversari, C.; van der Bruggen, P.; Luescher, I.F.; Lurquin, C.; Chomez, P.; Van Pel, A.; De Plaen, E.; Amar-Costesec, A.; Boon, T. A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J. Exp. Med. 1992, 176, 1453–1457. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Asemissen, A.M.; Keilholz, U.; Tenzer, S.; Müller, M.; Walter, S.; Stevanovic, S.; Schild, H.; Letsch, A.; Thiel, E.; Rammensee, H.G.; et al. Identification of a highly immunogenic HLA-A*01-binding T cell epitope of WT1. Clin. Cancer Res. 2006, 12, 7476–7482. [Google Scholar] [CrossRef] [PubMed]
- Parmentier, N.; Stroobant, V.; Colau, D.; de Diesbach, P.; Morel, S.; Chapiro, J.; van Endert, P.; Van den Eynde, B.J. Production of an antigenic peptide by insulin-degrading enzyme. Nat. Immunol. 2010, 11, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Keib, A.; Mei, Y.F.; Cicin-Sain, L.; Busch, D.H.; Dennehy, K.M. Measuring Antiviral Capacity of T Cell Responses to Adenovirus. J. Immunol. 2019, 202, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Vahedi-Faridi, A.; Saenger, W.; Ziegler, A.; Uchanska-Ziegler, B. Conformational changes within the HLA-A1:MAGE-A1 complex induced by binding of a recombinant antibody fragment with TCR-like specificity. Protein. Sci. 2009, 18, 37–49. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Quiñones-Parra, S.; Grant, E.; Loh, L.; Nguyen, T.H.; Campbell, K.A.; Tong, S.Y.; Miller, A.; Doherty, P.C.; Vijaykrishna, D.; Rossjohn, J.; et al. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc. Natl. Acad. Sci. USA 2014, 111, 1049–1054. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Raman, M.C.; Rizkallah, P.J.; Simmons, R.; Donnellan, Z.; Dukes, J.; Bossi, G.; Le Provost, G.S.; Todorov, P.; Baston, E.; Hickman, E.; et al. Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy. Sci. Rep. 2016, 6, 18851. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ikeda, H.; Lethé, B.; Lehmann, F.; van Baren, N.; Baurain, J.F.; de Smet, C.; Chambost, H.; Vitale, M.; Moretta, A.; Boon, T.; et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity 1997, 6, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Stern, M.; Ruggeri, L.; Capanni, M.; Mancusi, A.; Velardi, A. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood 2008, 112, 708–710. [Google Scholar] [CrossRef] [PubMed]
- Tălăngescu, A.; Calenic, B.; Mihăilescu, D.F.; Tizu, M.; Marunțelu, I.; Constantinescu, A.E.; Constantinescu, I. Molecular Analysis of HLA Genes in Romanian Patients with Chronic Hepatitis B Virus Infection. Curr. Issues Mol. Biol. 2024, 46, 1064–1077. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bulek, A.M.; Cole, D.K.; Skowera, A.; Dolton, G.; Gras, S.; Madura, F.; Fuller, A.; Miles, J.J.; Gostick, E.; Price, D.A.; et al. Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes. Nat. Immunol. 2012, 13, 283–289. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Skowera, A.; Ellis, R.J.; Varela-Calviño, R.; Arif, S.; Huang, G.C.; Van-Krinks, C.; Zaremba, A.; Rackham, C.; Allen, J.S.; Tree, T.I.; et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J. Clin. Investig. 2009, 118, 3390–3402, Erratum in J. Clin. Investig. 2009, 119, 2844. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nakanishi, K.; Inoko, H. Combination of HLA-A24, -DQA1*03, and -DR9 contributes to acute-onset and early complete beta-cell destruction in type 1 diabetes: Longitudinal study of residual beta-cell function. Diabetes 2006, 55, 1862–1868. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, D.; Knight, R.R.; Estorninho, M.; Ellis, R.J.; Kester, M.G.; de Ru, A.; Eichmann, M.; Huang, G.C.; Powrie, J.; Dayan, C.M.; et al. Circulating preproinsulin signal peptide-specific CD8 T cells restricted by the susceptibility molecule HLA-A24 are expanded at onset of type 1 diabetes and kill β-cells. Diabetes 2012, 61, 1752–1759. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fogdell-Hahn, A.; Ligers, A.; Grønning, M.; Hillert, J.; Olerup, O. Multiple sclerosis: A modifying influence of HLA class I genes in an HLA class II associated autoimmune disease. Tissue Antigens 2000, 55, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Friese, M.A.; Jakobsen, K.B.; Friis, L.; Etzensperger, R.; Craner, M.J.; McMahon, R.M.; Jensen, L.T.; Huygelen, V.; Jones, E.Y.; Bell, J.I.; et al. Opposing effects of HLA class I molecules in tuning autoreactive CD8+ T cells in multiple sclerosis. Nat. Med. 2008, 14, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, J.; Meguro, A.; Ishii, G.; Mihara, T.; Takeuchi, M.; Mizuki, Y.; Yuda, K.; Yamane, T.; Kawagoe, T.; Ota, M.; et al. The association analysis between HLA-A*26 and Behçet’s disease. Sci. Rep. 2019, 9, 4426. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- LeHoang, P.; Ozdemir, N.; Benhamou, A.; Tabary, T.; Edelson, C.; Betuel, H.; Semiglia, R.; Cohen, J.H. HLA-A29.2 subtype associated with birdshot retinochoroidopathy. Am. J. Ophthalmol. 1992, 113, 33–35. [Google Scholar] [CrossRef] [PubMed]
- Barber, L.D.; Gillece-Castro, B.; Percival, L.; Li, X.; Clayberger, C.; Parham, P. Overlap in the repertoires of peptides bound in vivo by a group of related class I HLA-B allotypes. Curr. Biol. 1995, 5, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Hickman, H.D.; Luis, A.D.; Buchli, R.; Few, S.R.; Sathiamurthy, M.; VanGundy, R.S.; Giberson, C.F.; Hildebrand, W.H. Toward a definition of self: Proteomic evaluation of the class I peptide repertoire. J. Immunol. 2004, 172, 2944–2952. [Google Scholar] [CrossRef] [PubMed]
- Rist, M.J.; Theodossis, A.; Croft, N.P.; Neller, M.A.; Welland, A.; Chen, Z.; Sullivan, L.C.; Burrows, J.M.; Miles, J.J.; Brennan, R.M.; et al. HLA peptide length preferences control CD8+ T cell responses. J. Immunol. 2013, 191, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Dazert, E.; Neumann-Haefelin, C.; Bressanelli, S.; Fitzmaurice, K.; Kort, J.; Timm, J.; McKiernan, S.; Kelleher, D.; Gruener, N.; Tavis, J.E.; et al. Loss of viral fitness and cross-recognition by CD8+ T cells limit HCV escape from a protective HLA-B27-restricted human immune response. J. Clin. Investig. 2009, 119, 376–386. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schneidewind, A.; Brockman, M.A.; Sidney, J.; Wang, Y.E.; Chen, H.; Suscovich, T.J.; Li, B.; Adam, R.I.; Allgaier, R.L.; Mothé, B.R.; et al. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 2008, 82, 5594–5605. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Varnavidou-Nicolaidou, A.; Karpasitou, K.; Georgiou, D.; Stylianou, G.; Kokkofitou, A.; Michalis, C.; Constantina, C.; Gregoriadou, C.; Kyriakides, G. HLA-B27 in the Greek Cypriot population: Distribution of subtypes in patients with ankylosing spondylitis and other HLA-B27-related diseases. The possible protective role of B*2707. Hum. Immunol. 2004, 65, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Hiby, S.E.; Apps, R.; Sharkey, A.M.; Farrell, L.E.; Gardner, L.; Mulder, A.; Claas, F.H.; Walker, J.J.; Redman, C.W.; Morgan, L.; et al. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J. Clin. Investig. 2011, 120, 4102–4110, Erratum in J. Clin. Investig. 2011, 121, 455. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Makadzange, A.T.; Gillespie, G.; Dong, T.; Kiama, P.; Bwayo, J.; Kimani, J.; Plummer, F.; Easterbrook, P.; Rowland-Jones, S.L. Characterization of an HLA-C-restricted CTL response in chronic HIV infection. Eur. J. Immunol. 2010, 40, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.P.; Stuart, P.E.; Nistor, I.; Hiremagalore, R.; Chia, N.V.C.; Jenisch, S.; Weichenthal, M.; Abecasis, G.R.; Lim, H.W.; Christophers, E.; et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am. J. Hum. Genet. 2006, 78, 827–851. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Arakawa, A.; Siewert, K.; Stöhr, J.; Besgen, P.; Kim, S.M.; Rühl, G.; Nickel, J.; Vollmer, S.; Thomas, P.; Krebs, S.; et al. Melanocyte antigen triggers autoimmunity in human psoriasis. J. Exp. Med. 2015, 212, 2203–2212. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Allele HLA-A | No. | AF (Allele Frequency) |
|---|---|---|
| A*02:01:01 | 188 | 26.11% |
| A*01:01:01 | 90 | 12.50% |
| A*24:02:01 | 84 | 11.67% |
| A*03:01:01 | 70 | 9.72% |
| A*11:01:01 | 45 | 6.25% |
| A*32:01:01 | 45 | 6.25% |
| A*26:01:01 | 33 | 4.58% |
| A*68:01:02 | 21 | 2.92% |
| A*23:01:01 | 18 | 2.50% |
| A*25:01:01 | 17 | 2.36% |
| A*29:02:01 | 16 | 2.22% |
| A*33:01:01 | 12 | 1.67% |
| A*33:03:01 | 12 | 1.67% |
| A*30:01:01 | 10 | 1.39% |
| A*31:01:02 | 10 | 1.39% |
| A*29:01:01 | 8 | 1.11% |
| A*02:05:01 | 6 | 0.83% |
| A*02:02:01 | 4 | 0.56% |
| A*66:01:01 | 4 | 0.56% |
| A*68:02:01 | 4 | 0.56% |
| A*30:02:01 | 4 | 0.56% |
| A*02:06:01 | 3 | 0.42% |
| A*02:11:01 | 3 | 0.42% |
| A*24:03:01 | 3 | 0.42% |
| A*02:17:02 | 2 | 0.28% |
| A*03:02:01 | 2 | 0.28% |
| A*30:04:01 | 2 | 0.28% |
| A*02:09:01 | 1 | 0.14% |
| A*24:07:01 | 1 | 0.14% |
| A*34:02:01 | 1 | 0.14% |
| A*02:18:02 | 1 | 0.14% |
| HLA-B Allele | N | AF |
|---|---|---|
| B*18:01:01 | 81 | 11.25% |
| B*51:01:01 | 78 | 10.83% |
| B*08:01:01 | 56 | 7.78% |
| B*35:01:01 | 36 | 5% |
| B*35:03:01 | 33 | 4.58% |
| B*07:02:01 | 27 | 3.75% |
| B*13:02:01 | 27 | 3.75% |
| B*38:01:01 | 22 | 3.06% |
| B*44:02:01 | 22 | 3.06% |
| B*14:02:01 | 21 | 2.92% |
| B*27:05:02 | 21 | 2.92% |
| B*44:03:01 | 21 | 2.92% |
| B*35:02:01 | 17 | 2.36% |
| B*41:01:01 | 16 | 2.22% |
| B*52:01:01 | 16 | 2.22% |
| B*40:02:01 | 15 | 2.08% |
| B*57:01:01 | 15 | 2.08% |
| B*39:01:01 | 14 | 1.94% |
| B*44:05:01 | 14 | 1.94% |
| B*49:01:01 | 14 | 1.94% |
| B*27:02:01 | 13 | 1.81% |
| B*55:01:01 | 13 | 1.81% |
| B*15:01:01 | 12 | 1.67% |
| B*58:01:01 | 12 | 1.67% |
| B*40:01:01 | 11 | 1.53% |
| B*50:01:01 | 10 | 1.39% |
| B*37:01:01 | 8 | 1.11% |
| B*47:01:01 | 8 | 1.11% |
| B*18:05:01 | 7 | 0.97% |
| B*40:06:01 | 7 | 0.97% |
| B*44:27:01 | 7 | 0.97% |
| B*56:01:01 | 7 | 0.97% |
| B*41:02:01 | 5 | 0.69% |
| B*07:05:01 | 4 | 0.56% |
| B*53:01:01 | 4 | 0.56% |
| B*35:08:01 | 3 | 0.42% |
| B*39:06:02 | 3 | 0.42% |
| B*51:07:01 | 3 | 0.42% |
| B*15:03:01 | 2 | 0.28% |
| B*51:05:01 | 2 | 0.28% |
| B*15:17:01 | 2 | 0.28% |
| B*18:03:01 | 2 | 0.28% |
| B*45:01:01 | 2 | 0.28% |
| B*54:01:01 | 2 | 0.28% |
| B*46:01:01 | 1 | 0.14% |
| B*48:01:01 | 1 | 0.14% |
| B*18:04:01 | 1 | 0.14% |
| B*39:05:01 | 1 | 0.14% |
| B*58:02:01 | 1 | 0.14% |
| HLA-C Allele | N | AF |
|---|---|---|
| C*07:01:01 | 125 | 17.36% |
| C*04:01:01 | 97 | 13.47% |
| C*12:03:01 | 77 | 10.69% |
| C*06:02:01 | 68 | 9.44% |
| C*02:02:02 | 63 | 8.75% |
| C*01:02:01 | 39 | 5.41% |
| C*07:02:01 | 37 | 5.13% |
| C*15:02:01 | 36 | 5% |
| C*03:04:01 | 20 | 2.77% |
| C*08:02:01 | 20 | 2.77% |
| C*14:02:01 | 20 | 2.77% |
| C*03:03:01 | 19 | 2.63% |
| C*12:02:02 | 18 | 2.50% |
| C*05:01:01 | 12 | 1.66% |
| C*16:01:01 | 11 | 1.52% |
| C*03:02:01 | 10 | 1.38% |
| C*07:04:01 | 10 | 1.38% |
| C*17:01:01 | 7 | 0.97% |
| C*16:07:01 | 6 | 0.83% |
| C*17:03:01 | 5 | 0.69% |
| C*15:05:02 | 4 | 0.55% |
| C*15:13:01 | 4 | 0.55% |
| C*16:02:01 | 4 | 0.55% |
| C*15:04:01 | 3 | 0.41% |
| C*12:12 | 1 | 0.13% |
| C*07:18:01 | 1 | 0.13% |
| C*15:72 | 1 | 0.13% |
| C*08:03:01 | 1 | 0.13% |
| C*14:03:01 | 1 | 0.13% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caragea, A.M.; Ursu, R.-I.; Maruntelu, I.; Tizu, M.; Constantinescu, A.-E.; Tălăngescu, A.; Constantinescu, I. High Resolution HLA-A, HLA-B, and HLA-C Allele Frequencies in Romanian Hematopoietic Stem Cell Donors. Int. J. Mol. Sci. 2024, 25, 8837. https://doi.org/10.3390/ijms25168837
Caragea AM, Ursu R-I, Maruntelu I, Tizu M, Constantinescu A-E, Tălăngescu A, Constantinescu I. High Resolution HLA-A, HLA-B, and HLA-C Allele Frequencies in Romanian Hematopoietic Stem Cell Donors. International Journal of Molecular Sciences. 2024; 25(16):8837. https://doi.org/10.3390/ijms25168837
Chicago/Turabian StyleCaragea, Andreea Mirela, Radu-Ioan Ursu, Ion Maruntelu, Maria Tizu, Alexandra-Elena Constantinescu, Adriana Tălăngescu, and Ileana Constantinescu. 2024. "High Resolution HLA-A, HLA-B, and HLA-C Allele Frequencies in Romanian Hematopoietic Stem Cell Donors" International Journal of Molecular Sciences 25, no. 16: 8837. https://doi.org/10.3390/ijms25168837
APA StyleCaragea, A. M., Ursu, R.-I., Maruntelu, I., Tizu, M., Constantinescu, A.-E., Tălăngescu, A., & Constantinescu, I. (2024). High Resolution HLA-A, HLA-B, and HLA-C Allele Frequencies in Romanian Hematopoietic Stem Cell Donors. International Journal of Molecular Sciences, 25(16), 8837. https://doi.org/10.3390/ijms25168837






